Abstract
Agrobacterium tumefaciens is widely utilized for delivering a foreign gene into a plant’s genome. We found the bacterial transposon Tn5393 in transgenic rice plants. Analysis of the flanking sequences of the transferred-DNA (T-DNA) identified that a portion of the Tn5393 sequence was present immediately next to the end of the T-DNA. Because this transposon was present in A. tumefaciens strain LBA4404, but not in EHA105 and GV3101, our findings indicated that Tn5393 was transferred from LBA4404 into the rice genome during the transformation process. We also noted that another bacterial transposon, Tn5563, is present in transgenic plants. Analyses of 331 transgenic lines revealed that 26.0% carried Tn5393 and 2.1% contained Tn5563. In most of the lines, an intact transposon was integrated into the T-DNA and transferred to the rice chromosome. More than one copy of T-DNA was introduced into the plants, often at a single locus. This resulted in T-DNA repeats of normal and transposon-carrying T-DNA that generated deletions of a portion of the T-DNA, joining the T-DNA end to the bacterial transposon. Based on these data, we suggest that one should carefully select the appropriate Agrobacterium strain to avoid undesirable transformation of such sequences.
Keywords: Agrobacterium, bacterial transposon, LBA4404, rice, T-DNA, Tn5393
INTRODUCTION
The soil-borne organism Agrobacterium tumefaciens induces crown gall tumorigenesis by transferring its own DNA, located on the transferred-DNA (T-DNA) region of a tumor-inducing (Ti) plasmid, into plant chromosomes. This capacity to move DNA into a plant genome has been exploited by researchers who have adopted this bacterium as a genetic engineering tool for plant transformation. In this way, any DNA molecule can be transferred when laid between two imperfect 25-bp repeats - the right border (RB) and the left border (LB). This is accomplished with the help of proteins encoded from virulence (vir) genes. In nature, both T-DNA and vir genes are located on the same Ti plasmid. Because such plasmids are difficult to manipulate directly in vitro due to their large size (about 200 kb), a binary vector system has been developed. One plasmid, disarmed, is the derivative of a Ti plasmid that contains vir genes but lacks the T-DNA region (Hoekema et al., 1983). The other, the binary vector, is a small-sized artificial plasmid that carries modified T-DNA containing cloning sites, a gene for selection of transformants, and the borders (An et al., 1985; Bevan, 1984). During the past 25 years numerous binary vectors have been developed (Kim et al., 2009; Komori et al., 2007; Lee and Gelvin, 2008), but only a few disarmed plasmids have been reported. The most widely used are Agrobacterium strains EHA105, GV3101, and LBA4404, which contain different disarmed plasmids (Hellens et al., 2000; Lee and Gelvin, 2008).
Although Agrobacterium transformation methods have been commonly applied in plant transformation, some concerns remain. For example, multiple T-DNA insertions frequently occur at one locus of a plant genome (De Buck et al., 1999; De Neve et al., 1997; Kim et al., 2003a). This T-DNA repeat structure may affect transgene expression through gene-silencing (Jorgensen et al., 1996). Another problem is that unexpected DNA can be integrated into a plant genome. A vector sequence outside of the T-DNA is often detected in transgenic plants (De Buck et al., 2000; Kim et al., 2003a; Kononov et al., 1997; Oltmanns et al., 2010). This is mainly due to the failure of T-DNA termination at the LB during T-DNA transformation, which then generates a read-through product. Agrobacterium chromosomal DNA has also been detected (Ülker et al., 2008).
In this study we discovered two bacterial transposons, Tn5393 and Tn5563, in transgenic rice. The former had first been reported from Erwinia amylovora, which causes fire blight in rosaceous plants. Tn5393 belongs to the Tn3 transposon family and is composed of 81 bp of inverted repeats (IRs), the transposase gene tnpA, a recombination site (res), the resolvase gene tnpR, the insertion sequence IS1133, and two streptomycin resistance genes - strA and strB (Chiou and Jones, 1993). The second transposon, Tn5563, was first identified from the pRA2 plasmid of Pseudomonas alcaligenes. It also belongs to the Tn3 family and has two 39-bp inverted repeats -- tnpA and tnpR – plus two putative mercuric ion transport genes, merP and merT (Yeo et al., 1998). Tn3 and its relatives are replicative DNA transposons.
MATERIALS AND METHODS
Plant materials and Agrobacterium strains
We used transgenic plants of rice (Oryza sativa var. Dongjin, Hwayoung, and Kitaake) produced by the binary vectors pGA2144, pGA2707, pGA2715, pGA2717, and pGA2772 (An et al., 2003; Jeon et al., 2000; Jeong et al., 2002; 2006; Lee et al., 2012; Ryu et al., 2004). Their backbone was derived from pGA472 (An et al., 1985; Kim et al., 2003b), having an RK2 replication origin. For DNA preparation, 10 seeds from individual primary transgenic plants were sterilized for 24 h in 0.025% (v/v) prochloraz diluted with water, then washed in tap water for 24 h. They were then sown in soil and grown in the greenhouse under natural light. Leaves of 20-day-old seedlings were sampled and used for DNA extraction by a modified cetyl trimethyl ammonium bromide (CTAB) method (Chen and Ronald, 1999). These DNAs were subjected to PCR analysis and Southern hybridization. Agrobacterium tumefaciens strains LBA4404, EHA105, and GV3101 were cultured in YEP liquid media without antibiotics, using a shaking incubator set at 28°C.
PCR analysis
All PCR reactions comprised 35 cycles of denaturation at 95°C, annealing at 57°C, and extension at 72°C, followed by a final elongation step at 72°C for 5 min. Amplifications were performed with the following primers: a (5′-GAGCTTCATGGTGTTCCAGAA-3′), b (5′-AGCCACGTCTCCGACCAAT-3′), c (5′-TGACCGCCTCATTTGGCTCAA-3′), d (5′-CATGATGCAGATCGCCATGTA-3′), e (5′-CTTGGAACGCGGATGGAGAA-3′), f (5′-CTGCGCTCCGATAAATTCGAT-3′), and g (5′-AGACTGCGAGCCATCGGCTTT-3′) on Tn5393; RB1 (5′-CCACAGTTTTCGCGATCCAGACT-3′), LB1 (5′-TCCGAAACTATCAGTGTCTAGCT-3′), 15F (5′-ATGATTAGAGTCCCGCAATT-3′), and 15R (5′-AGTGCTTGACATTGGGGAATTCAG-3′) on T-DNA; and 5563a (5′-AGGCTCTGTAAGCCGACCTT-3′) and 5563b (5′-GGTACGCTTAATCAAAGTTCAA-3′) on Tn5563.
Characterization of the Tn5393 flanking region in transgenic rice plants
Flanking regions of the bacterial transposons were amplified by iPCR as described previously (Kim et al., 2011). Briefly, genomic DNA was digested with SacI and self-ligated by T4 DNA ligase. Our iPCR was conducted with a set of primers located at the terminal regions of Tn5393: 5′-CATCTTCTGGAACACCATGAA-3′ and 5′-GCTTCAGCGCATGATGACGTT-3′ at the left end, and 5′-CTTGGAACGCGGATGGAGAA-3′ and 5′-CAAATCCGCTCCAGACAGAT-3′ at the right end, Nested PCR was performed with 5′-GAGGTCGTTGCAAACCAGAA-3′ and 5′-GAGTTCGGCATCCAGAATCCA-3′ for the left end, and 5′-GGAGCAACGCGATCTAGCTA-3′ and 5′-TTCCTCCTGCCAGTTGATCA-3′ for the right end.
Detection of transposition of Tn5393 into T-DNA in Agrobacterium
Agrobacterium cells harboring pGA2715 binary plasmid were plated on YEP-agar medium containing tetracycline (5 μg/ml). Single colonies were picked and grown in YEP liquid medium containing tetracycline for 36 h. After purification of DNA, PCR was performed with a pair of primers (one on T-DNA and the other on Tn5393). The PCR products were directly sequenced.
DNA-blot hybridization
Procedures were as described previously (Kang et al., 2010). Briefly, genomic DNA (2 μg) digested with SacI was separated on a 0.7% agarose gel, blotted onto a nylon membrane (GE Healthcare, USA), and hybridized with a 32P-labeled probe. For probe preparation, a portion of the streptomycin-resistant gene on Tn5393 was amplified by PCR with c and d primers. The PCR product was used for isotope-labeling with a Rediprime II random prime DNA labeling system (GE Healthcare).
RESULTS
Bacterial Tn5393 is present in transgenic rice plants
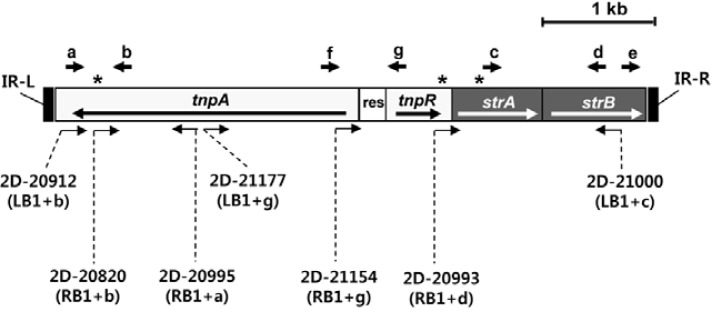
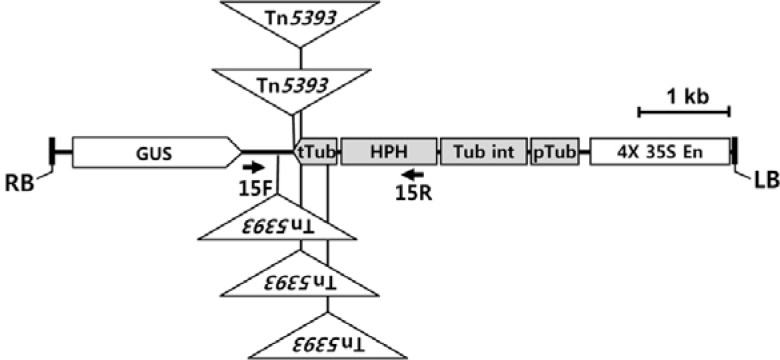
We generated numerous transgenic rice plants and determined the flanking sequences of T-DNA insertion sites (An et al., 2003; 2005; Jeon et al., 2000; Jeong et al., 2002; 2006; Ryu et al., 2004). During the analysis of those flanking sequences, we found DNA sequences for bacterial transposon Tn5393 (GenBank Accession No. M95402). To confirm its presence in our transgenic plants, we amplified DNA fragments containing this transposon, using a pair of specific primers in which one was located near the T-DNA border and the other was located on Tn5393. Sequencing the amplified fragments showed that the transposon was linked to the T-DNA ends, albeit with varying amounts of deletion (Fig. 1). For example, the transposon was found in the LB flanking region, and approximately 300 bp of the IR-L region of Tn5393 had been deleted in Line 2D-20912. Much larger deletions were noted in Lines 2D-21000 and 2D-21177. This transposon was also found at the RB, with varying amounts of deletions.
Fig. 1.
Schematic diagram of Tn5393 and deletion end points. Structure is based on report by Chiou and Jones (1993). T-DNA flanking sequences matching Tn5393 are indicated with arrows below map. Starting points of arrows are junctions between Tn5393 and T-DNA. Junction regions were PCR-amplified using T-DNA border primer and Tn5393 primer as indicated in parentheses. Asterisks (*) indicate restriction enzyme SacI sites. IR-L and IR-R, terminal inverted repeats; res, recombination site; strA and strB, streptomycin resistance genes; tnpA, transposase gene; tnpR, resolvase gene.
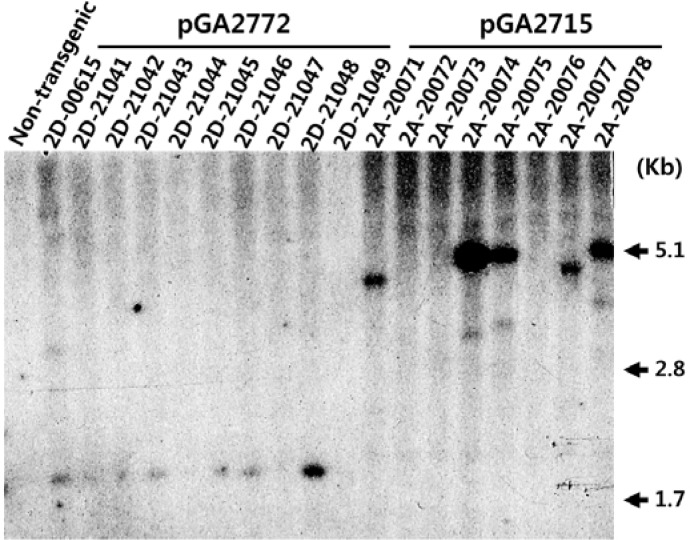
To confirm the presence of Tn5393 in transgenic plants, we performed DNA-blot analyses of randomly selected lines transformed with either pGA2772 or pGA2715. There, seven out of 10 lines from pGA2772 and five out of eight from pGA2715 had hybridized with the Tn5393 probe (Fig. 2).
Fig. 2.
DNA-blot hybridization experiment of transgenic rice plants with Tn5393 probe. Ten randomly selected pGA2772-transformed lines and eight pGA2715-transformed lines were examined. Genomic DNAs were digested with SacI, then blotted and hybridized with Tn5393 probe. Non-transgenic is wild type rice (O. sativa var. Dongjin) as a negative control.
Tn5393 originates from Agrobacterium tumefaciens LBA4404
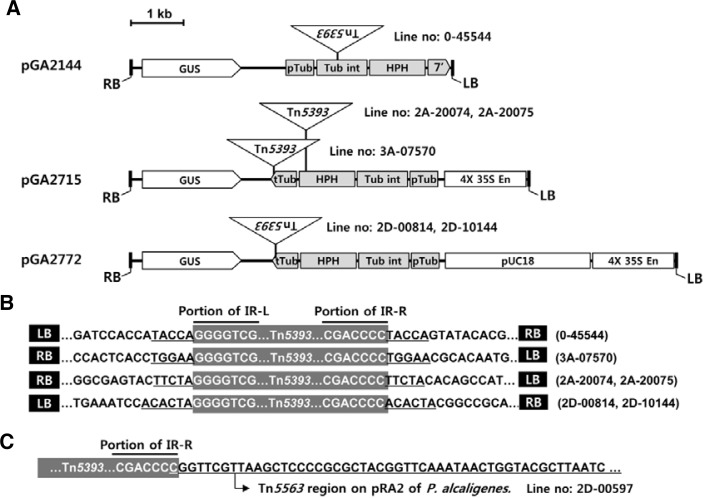
DNA hybridization and PCR analyses showed that Tn5393 was not present in wild-type rice, thus indicating that the sequence was instead introduced during generation of the transgenic plants (Fig. 3). We isolated its flanking regions from Line 0-45544, which had been transformed with pGA2144. Sequencing the flanking regions of this transposon showed that T-DNA sequences of the binary vector were located next to Tn5393. That transposon had been inserted at the OsTubA1 intron sequence, which is located between the OsTubA1 promoter and the hygromycin resistance gene HPH (Fig. 4A). At the junction points, 5-bp duplications of the target DNA were found (Fig. 4B).
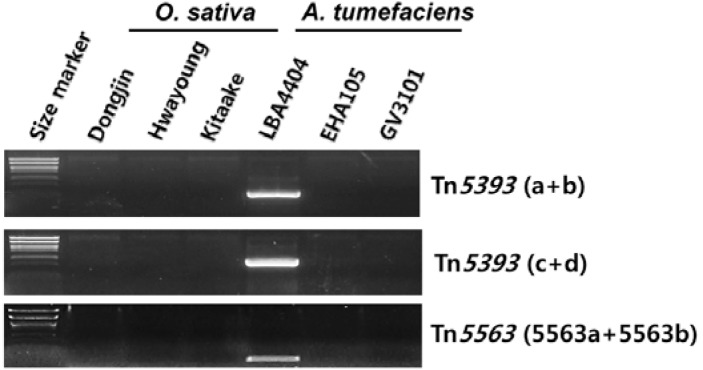
Fig. 3.
Detection of Tn5393 and Tn5563 from O. sativa and A. tumefaciens. Tests included 3 rice varieties (Dongjin, Hwayoung, and Kitaake) and 3 Agrobacterium strains (LBA4404, EHA105, and GV3101). Primers for amplification of Tn5393 are shown with Fig. 1. In amplifying Tn5563, only a portion detected from the transgenic plant was tested.
Fig. 4.
Locations of bacterial transposons within T-DNA in transgenic rice plants. (A) Structure of T-DNA and positioning of Tn5393. (B) Sequences at junctions between T-DNA and Tn5393. Underlined sequences (5–6 bp) are target duplications by Tn5393 insertion. (C) IR-R of Tn5393 was linked to another bacterial transposon, Tn5563, in Line 2D-00597. GUS, β-glucuronidase; HPH, hygromycin phosphotransferase; pUC18, plasmid pUC18; pTub, promoter of OsTubA1; tTub, terminator of OsTubA1; Tub int, first intron of OsTubA1; 4X 35S En, 4-fold transcription enhancer of CaMV 35S.
Similar results were obtained from transgenic plants generated with other vectors. Sequencing pGA2715-transformed lines identified two different insertions of Tn5393 in the T-DNA region of the vectors (Fig. 4A). In the first group, the transposon was inserted at the OsTubA1 terminator that was located after the hygromycin resistance coding sequence. In the second group, it was inserted to HPH and also caused target-site duplication (Fig. 4B). Analyses of the pGA2772-transformed lines showed that Tn5393 was inserted at the OsTubA1 terminator region (a different site from that associated with the first group of pGA2715 lines); there, 6 bp of duplications were observed (Figs. 4A and 4B).
Because the transposon-positioning was identical in more than one line transformed with the same vector, it is likely that Tn5393 was introduced into the binary vector before the plant cells could be transformed. If so, then A. tumefaciens strain LBA4404, used as a host for the binary vector, may have had the transposon. Therefore, we investigated for the presence of Tn5393 in strain LBA4404 that did not carry a binary vector, and discovered that it did indeed have the transposon (Fig. 3). Meanwhile, strains EHA105 and GV3101, carrying different disarmed plasmids, did not contain Tn5393. We examined whether Tn5393 was transposed to T-DNA before co-cultivation with rice. Analysis of 24 colonies resulted in identification of five independent transpositions to T-DNA (Fig. 5). This result indicates that Tn5393 was inserted into T-DNA in Agrobacterium.
Fig. 5.
Schematic diagram of Tn5393 insertions in T-DNA of pGA2715. Primers 15F and 15R were used together with b and e primers in Fig. 1 for amplification of the junction regions between Tn5393 and T-DNA.
In Line 2D-00597, the Tn5563 transposon occurred in the flanking sequence of the T-DNA insertion site. This bacterial transposon has also been found on plasmid pRA2 from Pseudomonas alcaligenes (GenBank Accession No. U88088.2; Yeo et al., 1998). Sequencing the surrounding region of Tn5563 showed that it was located within Tn5393 (Fig. 4C). Therefore, these results indicate that Tn5563 was inserted into Tn5393 before both were transferred into the plant chromosome. Furthermore, Tn5563 was not detected in wild-type rice, but was found in A. tumefaciens LBA4404 (Fig. 3).
Intact Tn5393 is inserted within the T-DNA of most transgenic rice lines examined here
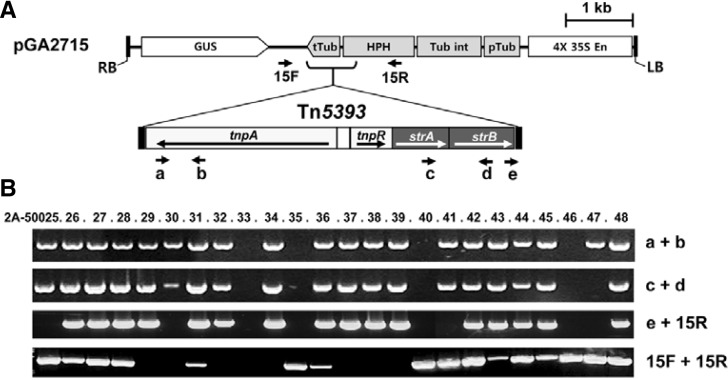
To elucidate whether Tn5393 was intact in our transgenic rice, we examined 24 lines that had been transformed with pGA2715. Amplification of the end regions of the transposon showed that 19 out of 24 lines contained the entire Tn5393 (Fig. 6). In Line 2A-50047, only one end of Tn5393 had been amplified, suggesting that the transposon was truncated. In 16 lines, the junction region was amplified between the IR-R of Tn5393 and the T-DNA, indicating that, for the majority of lines, this transposon was located near the terminator of OsTubA1 on the T-DNA. However, no PCR band was detectable in three lines (2A-50025, 2A-50030, and 2A-50041), implying that Tn5393 either was located elsewhere or had an inverse orientation. If Tn5393 was not in fact present at the locus, then the T-DNA sequence surrounding the insertion site should have been amplified. As expected, the T-DNA region was amplified in Lines 2A-50025 and 2A-50041, but not in Line 2A-50030. This suggested the possibility that the transposon was set at an inverse orientation. Interestingly, the T-DNA sequence surrounding the Tn5393 insertion site was amplified in 10 of 16 lines carrying the transposon at that locus. This demonstrated that more than one T-DNA insertion had occurred in the lines: one with Tn5393 and the other without a transposon.
Fig. 6.
Detection of Tn5393-inserted T-DNA. (A) Schematic diagrams of pGA2715 T-DNA and Tn5393. Arrows below maps indicate primers for PCR. (B) PCR-amplifications of Tn5393 (a + b, c + d), T-DNA (15F + 15R), and their junction regions (e + 15R) from 24 randomly selected plants transformed with pGA2715.
We used PCR to determine the frequency of bacterial transposon insertions in our transgenics. Approximately 26% (86 lines) out of a randomly selected 331 lines contained Tn5393 (Table 1). Although the Tn5563 sequence was found at a much lower frequency (2.1%), it did occur in the lines transformed with different vectors examined here (Table 1).
Table 1.
Detection frequencies of Tn5393 and Tn5563 from rice T-DNA-tagged lines
| Tagging vector | Number of lines tested | Number of lines bearing transposon
|
|
|---|---|---|---|
| Tn5393 (%) | Tn5563 (%) | ||
| pGA2707 | 37 | 5 (13.5) | 1 (2.7) |
| pGA2715 | 232 | 48 (20.7) | 5 (2.2) |
| pGA2717 | 33 | 12 (36.4) | 0 (0.0) |
| pGA2772 | 29 | 21 (72.4) | 1 (3.4) |
|
| |||
| Total | 331 | 86 (26.0) | 7 (2.1) |
PCR was performed with f and g primers for Tn5393 and 5563a and 5563b primers for Tn5563.
Occurrence of transposon sequences in the T-DNA flanking region is due to T-DNA repeat structures
Because Tn5393 was located within the T-DNA, we did not expect to identify its sequence from the T-DNA flanking region. Kim et al. (2003a) have previously reported that more than one T-DNA can be inserted into a single locus as a tandem or inverse repeat. During the transformation process, a portion of the T-DNA might be deleted, connecting a T-DNA border to the middle of the next T-DNA. If such a deletion occurs from one T-DNA end to the sequence near or at the bacterial transposon, the transposon sequence can then be detected in that flanking region.
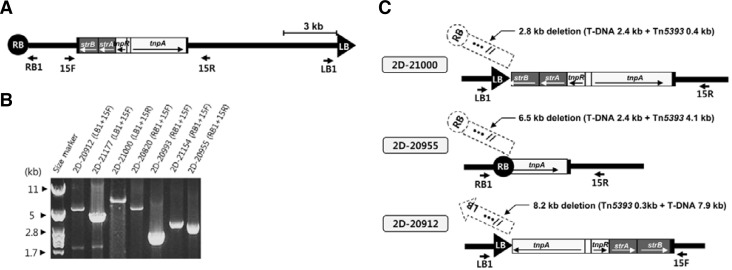
To examine this hypothesis, we selected seven lines described in Fig. 1. Genomic DNA isolated from them was amplified, using one primer located near the T-DNA borders (RB1 or LB1 in Fig. 7A) and another from within the T-DNA where Tn5393 was inserted (15F or 15R in Fig. 7A). As predicted, T-DNA repeat structures containing Tn5393 were amplified (Fig. 7B). Based on the size of the amplified DNA and the sequence at the junction between T-DNA and Tn5393, we estimated that 2.8–11.4 kb of T-DNA had been deleted. Those deduced structures are illustrated in Fig. 7C.
Fig. 7.
Formation of T-DNA end::Tn5393 through deletion of a portion of T-DNA. (A) Schematic diagram of T-DNA of pGA2772 with Tn5393 insertion. PCR primers are indicated below map. (B) PCR-amplification of T-DNA end::Tn5393 region. Primer pairs are shown in parentheses. (C) Schematic diagrams for formation of T-DNA end::Tn5393. Deleted regions are shown in dotted boxes.
DISCUSSION
We observed that the Tn5393 sequence was often present at the flanking regions of T-DNA tagging lines. Inserting multiple copies of T-DNA at a single locus and subsequence rearrangement had brought Tn5393 next to the T-DNA border. At the junction points between that border and Tn5393, micro homology (1–4 bp) was present, often along with small filler DNA. These contexts are similar to that of general T-DNA repeats (Kim et al., 2003a; Kumar and Fladung, 2000).
Tn5393 was previously discovered in Arabidopsis transformed with LBA4404 (Zhao et al., 2009). Because we also used LBA4404 as a host for our binary vectors, we supposed that the transposon had originated from those bacteria. The Agrobacterium strain carries disarmed Ti plasmid pAL4404, a derivative of the octopine Ti plasmid pTiAch5 (Hellens et al., 2000). To generate insertional mutations in the Ti plasmid, bacterial transposon Tn904 conferring streptomycin resistance has been introduced to that plasmid (Klapwijk et al., 1980). Screening of T-DNA deletion mutants has resulted in the identification of a Ti plasmid designated as pAL4404 (Ooms et al., 1982). The strain carrying the mutant plasmid is named LBA4404. Therefore, Tn5393 is a derivative of Tn904.
We gained evidence here that Tn5393 is inserted into the T-DNA of binary vectors in Agrobacterium before it is transferred to plant chromosomes. This transposon is likely derived from the pAL4404 plasmid based on our finding that other Agrobacterium strains carrying a different disarmed Ti plasmid did not contain the transposon.
We often detected normal T-DNA and Tn5393-carrying T-DNA within one transgenic plant, perhaps because of transfection of one plant cell by more than one Agrobacterium: one with normal T-DNA, the other with T-DNA carrying Tn5393. Alternatively, this might be explained by the fact that normal as well as Tn5393-inserted binary plasmids were present in the same Agrobacterium cells. The vectors used in this study had an RK2 replication origin, which generally entails seven to 10 copies per cell in A. tumefaciens (Oltmanns et al., 2010).
Transferring Agrobacterium DNA into a plant via the Ti plasmid appears to be a common phenomenon. Although it happened here at low frequency, we also found a second transposon, Tn5563. Both Tn5393 and Tn5563 have previously exhibited transposition activity into another replicon (Chiou and Jones, 1993; Yeo et al., 1998).
Agrobacterium chromosomal DNA sequences are often found in transgenic plants generated by that organism (Ülker et al., 2008). Transposable element IS426, located on linear chromosomes, is detected at relatively high frequency. It has been proposed that processed T-DNA is first integrated into a nonessential region, such as transposons on the Agrobacterium chromosome, before that region is transferred into the plant genome (Ülker et al., 2008). In such a model, a border-like sequence should be present near the T-DNA insertion site on the Agrobacterium chromosome. By contrast, we present an alternative model here. Briefly, chromosomal DNA is first inserted into a binary plasmid before it is transferred into the plant cell. The Agrobacterium genome carries 25 IS elements (Wood et al., 2001), which might often be transferred into a binary vector along with surrounding chromosomal DNA.
The presence of Agrobacterium DNA is not desirable in transgenic plants and can be disadvantageous to genetic engineering efforts, especially those applied toward the production of commercial transgenic plants. However, this problem could be reduced by using Agrobacterium strains freshly engineered to carry binary vectors. In that way, the chance of transposon movement to T-DNA can be diminished. Avoiding helper plasmids containing transposable elements would also minimize the risk. Ultimately, development of transposon-free Agrobacterium strains will resolve this challenge.
Acknowledgments
We thank our laboratory members involved in the generation of T-DNA tagging lines and isolation of T-DNA flanking sequences. We are also grateful to Jong-Seong Jeon for kindly providing the Agrobacterium strains. This work was supported by a post-doctoral fellowship grant from Kyung Hee University in 2011 (KHU-20110205).
REFERENCES
- An G., Watson B.D., Stachel S., Gordon M.P., Nester E.W. New cloning vehicles for transformation of higher plants. EMBO J. 1985;4:277–284. doi: 10.1002/j.1460-2075.1985.tb03626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S., Park S., Jeong D.H., Lee D.Y., Kang H.G., Yu J.H., Hur J., Kim S.R., Kim Y.H., Lee M., et al. Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol. 2003;133:2040–2047. doi: 10.1104/pp.103.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G., Lee S., Kim S.H., Kim S.R. Molecular genetics using T-DNA in rice. Plant Cell Physiol. 2005;46:14–22. doi: 10.1093/pcp/pci502. [DOI] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.H., Ronald P.C. A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Mol. Biol. Rep. 1999;17:53–57. [Google Scholar]
- Chiou C.S., Jones A.L. Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other gram-negative bacteria. J. Bacteriol. 1993;175:732–740. doi: 10.1128/jb.175.3.732-740.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Neve M., De Buck S., Jacobs A., van Montagu M., Depicker A. T-DNA integration patterns in cotransformed plant cells suggest that T-DNA repeats originate from co-integration of separate T-DNAs. Plant J. 1997;11:15–29. doi: 10.1046/j.1365-313x.1997.11010015.x. [DOI] [PubMed] [Google Scholar]
- De Buck S., Jacobs A., van Montagu M., Depicker A. The DNA sequences of T-DNA junctions suggest that complex T-DNA loci are formed by a recombination process resembling T-DNA integration. Plant J. 1999;20:295–304. doi: 10.1046/j.1365-313x.1999.t01-1-00602.x. [DOI] [PubMed] [Google Scholar]
- De Buck S., De Wilde C., van Montagu M., Depicker A. T-DNA vector backbone sequences are frequently integrated into the genome of transgenic plants obtained by Agrobacterium-mediated transformation. Mol. Breed. 2000;6:459–468. [Google Scholar]
- Hellens R., Mullineaux P., Klee H. A guide to Agrobacterium binary Ti vectors. Trends Plant Sci. 2000;5:446–451. doi: 10.1016/s1360-1385(00)01740-4. [DOI] [PubMed] [Google Scholar]
- Hoekema A., Hirsch P.R., Hooykaas P.J.J., Schilperoort R.A. A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature. 1983;303:179–180. [Google Scholar]
- Jeon J.S., Lee S., Jung K.H., Jun S.H., Jeong D.H., Lee J., Kim C., Jang S., Yang K., Nam J., et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- Jeong D.H., An S., Kang H.G., Moon S., Han J.J., Park S., Lee H.S., An K., An G. T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 2002;130:1636–1644. doi: 10.1104/pp.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.H., An S., Park S., Kang H.G., Park G.G., Kim S.R., Sim J., Kim Y.O., Kim M.K., Kim S.R., et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- Jorgensen R.A., Cluster P.D., English J., Que Q., Napoli C.A. Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol. Biol. 1996;31:957–973. doi: 10.1007/BF00040715. [DOI] [PubMed] [Google Scholar]
- Kang K., Lee K., Park S., Lee S., Kim Y.S., Back K. Overexpression of rice ferrochelatase I and II leads to increased susceptibility to oxyfluorfen herbicide in transgenic rice. J. Plant Biol. 2010;53:291–296. [Google Scholar]
- Kim S.R., Lee J., Jun S.H., Park S., Kang H.G., Kwon S., An G. Transgene structures in T-DNA-inserted rice plants. Plant Mol. Biol. 2003a;52:761–773. doi: 10.1023/a:1025093101021. [DOI] [PubMed] [Google Scholar]
- Kim S.R., Lee S., Kang H.G., Jeon J.S., Kim K.M., An G. A complete sequence of the binary vector. J. Plant Biol. 2003b;46:211–214. [Google Scholar]
- Kim S.R., Lee D.Y., Yang J.I., Moon S., An G. Cloning vectors for rice. J. Plant Biol. 2009;52:73–78. [Google Scholar]
- Kim S.R., Jeon J.S., An G. Development of an efficient inverse PCR method for isolating gene tags from T-DNA insertional mutants in rice. Meth. Mol. Biol. 2011;678:139–146. doi: 10.1007/978-1-60761-682-5_11. [DOI] [PubMed] [Google Scholar]
- Klapwijk P.M., van Breukelen J., Korevaar K., Ooms G., Schilperoort R.A. Transposition of Tn904 encoding streptomycin resistance into the octopine Ti plasmid of Agrobacterium tumefaciens. J. Bacteriol. 1980;141:129–136. doi: 10.1128/jb.141.1.129-136.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T., Imayama T., Kato N., Ishida Y., Ueki J., Komari T. Current status of binary vectors and superbinary vectors. Plant Physiol. 2007;145:1155–1160. doi: 10.1104/pp.107.105734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononov M.E., Bassuner B., Gelvin S.B. Integration of T-DNA binary vector “backbone” sequences into the tobacco genome: evidence for multiple complex patterns of integration. Plant J. 1997;11:945–957. doi: 10.1046/j.1365-313x.1997.11050945.x. [DOI] [PubMed] [Google Scholar]
- Kumar S., Fladung M. Transgene repeats in aspen: molecular characterization suggests simultaneous integration of independent T-DNAs into receptive hotspots in the host genome. Mol. Gen. Genet. 2000;264:20–28. doi: 10.1007/s004380000296. [DOI] [PubMed] [Google Scholar]
- Lee L.Y., Gelvin S.B. T-DNA binary vectors and systems. Plant Physiol. 2008;146:325–332. doi: 10.1104/pp.107.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Ryoo N., Jeon J.S., Guerinot M.L., An G. Activation of rice Yellow Stripe1-Like 16 (OsYSL16) enhances iron efficiency. Mol. Cells. 2012;33:117–126. doi: 10.1007/s10059-012-2165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltmanns H., Frame B., Lee L.Y., Johnson S., Li B., Wang K., Gelvin S.B. Generation of backbone-free, low transgene copy plants by launching T-DNA from the Agrobacterium chromosome. Plant Physiol. 2010;152:1158–1166. doi: 10.1104/pp.109.148585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms G., Hooykaas P.J., Van Veen R.J., Van Beelen P., Regensburg-Tuïnk T.J., Schilperoort R.A. Octopine Ti-plasmid deletion mutants of Agrobacterium tumefaciens with emphasis on the right side of the T-region. Plasmid. 1982;7:15–29. doi: 10.1016/0147-619x(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Ryu C.H., You J.H., Kang H.G., Hur J., Kim Y.H., Han M.J., An K., Chung B.C., Lee C.H., An G. Generation of T-DNA tagging lines with a bidirectional gene trap vector and the establishment of an insertion-site database. Plant Mol. Biol. 2004;54:489–502. doi: 10.1023/B:PLAN.0000038257.93381.05. [DOI] [PubMed] [Google Scholar]
- Ülker B., Li Y., Rosso M.G., Logemann E., Somssich I.E., Weisshaar B. T-DNA-mediated transfer of Agrobacterium tumefaciens chromosomal DNA into plants. Nat. Biotechnol. 2008;26:1015–1017. doi: 10.1038/nbt.1491. [DOI] [PubMed] [Google Scholar]
- Wood D.W., Setubal J.C., Kaul R., Monks D.E., Kitajima J.P., Okura V.K., Zhou Y., Chen L., Wood G.E., Almeida N.F., Jr, et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;14:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- Yeo C.C., Tham J.M., Kwong S.M., Yiin S., Poh C.L. Tn5563, a transposon encoding putative mercuric ion transport proteins located on plasmid pRA2 of Pseudomonas alcaligenes. FEMS Microbiol. Lett. 1998;165:253–260. doi: 10.1111/j.1574-6968.1998.tb13154.x. [DOI] [PubMed] [Google Scholar]
- Zhao Z., Zhu Y., Erhardt M., Ruan Y., Shen W.H. A non-canonical transferred DNA insertion at the BRI1 locus in Arabidopsis thaliana. J. Integr. Plant Biol. 2009;51:367–373. doi: 10.1111/j.1744-7909.2009.00821.x. [DOI] [PubMed] [Google Scholar]