Abstract
DEC1 is a transcription repressor that is induced by Hypoxia-Inducible Factor-α/β (HIF-α/β). In this study, we found that either hypoxic treatment or ectopic expression of DEC1 blocks induction of a master adipogenic transactivator, peroxisome proliferative activated receptor-γ2 (PPARγ2) in 3T3-L1 cells. DEC1 did not prevent C/EBPβ, which is an upstream transactivator for PPARγ2, from occupying the PPARγ2 promoter. DEC1 occupied the PPARγ2 promoter by interacting with DNA-bound C/EBPβ. DEC1 occupancy was accompanied by a reduction of acetylated histones and an increase in histone deacetylase 1 (HDAC1) occupancy on the PPARγ2 promoter. Based on the fact that DEC1 interacts with HDAC1, this study suggests that DEC1 blocks adipogenesis by reinforcing HDAC1 recruitment to the PPARγ2 promoter. This study implies that DEC1 is one of the mediators that reset the pattern of PPARγ2 expression in response to hypoxia.
Keywords: adipogenesis, C/EBPβ, DEC1, hypoxia, PPARγ
INTRODUCTION
Average partial pressure of oxygen (O2) in the human body has been estimated to vary from 9% to 1% depending on the balance between supply and consumption of O2 in the tissue. Recent studies have demonstrated that different lineages of adult stem cells prefer to reside in a certain microenvironment that contains a different oxygen concentration. These findings suggest that specific oxygen concentration is an important physiological condition that determines cell type specific differentiation (Mohyeldin et al., 2010). Previously, we showed that hypoxia (1–5% oxygen) inhibits adipogenesis by repressing the adipogenic master transcription factor, PPARγ2 (Park and Park, 2010). Transcriptional activation of the PPARγ2 gene is tightly controlled by sequential expression of upstream transcription factors, C/EBPs. Preadipocytes rapidly and transiently express C/EBPβ in response to adipogenesis-inducing hormones, 3-isobutyl-1-methylxanthine, dexamethasone, and insulin (MDI). C/EBPβ transcriptionally activates the expression of C/EBPα and PPARγ2 genes, which cooperate to induce the expression of many genes required for terminal differentiation (Fig. 1A) (Farmer, 2006).
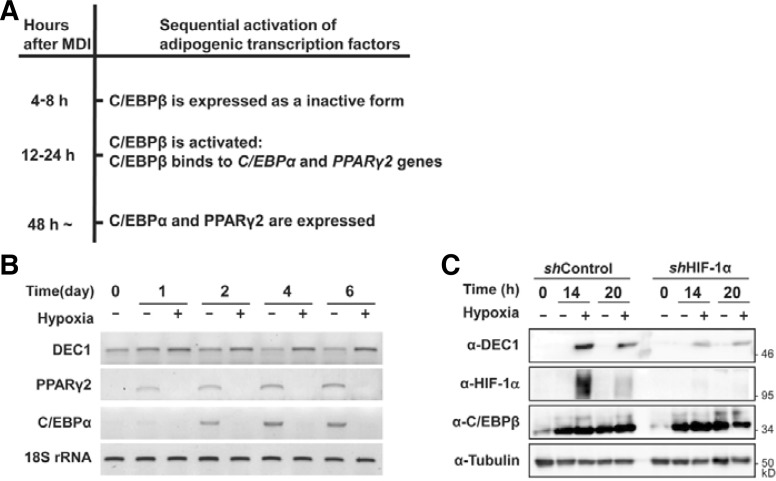
Fig. 1.
Hypoxia-induced DEC1 expression in 3T3-L1 cells. (A) The schematic diagram describing the sequential activation process of adipogenic transcription factors in response to MDI treatment. (B) The 3T3-L1 preadipocytes were differentiated to adipocytes in a normoxic (20–21% oxygen) or hypoxic conditions (0.1–0.5 % oxygen) for the indicated days. The levels of mRNAs were analyzed by RT-PCR using primer sets for DEC1, PPARγ2 and C/EBPα as described previously (Choi et al., 2008). The 18S rRNA was used as a loading control. (C) Adipogenesis was induced by MDI treatment in HIF-1α knockdown 3T3-L1 (shHIF-1α) cells and control 3T3-L1 (shControl) cells for the indicated times. DEC1, HIF-1α, and C/EBPβ protein levels were detected by western analyses. Tubulin was detected to check for equal loadings.
Hypoxia stabilizes hypoxia-inducible factor-α (HIF-α) isoforms, 1α and 2α, which form heterodimers with HIF-1β. HIF-1α/β or HIF-2α/β heterodimers have been shown to play a central role in adaptive responses including erythropoiesis, angiogenesis, anaerobic metabolism, and cellular differentiation in response to hypoxia (Chandel and Simon, 2008; Jaakkola et al., 2001; Mohyeldin et al., 2010). Hypoxia induces a transcription repressor differentiated embryo chondrocyte 1 (DEC1/Stra13/BHLHB2/SHARP2) mainly through HIF-1α/β heterodimer (Miyazaki et al., 2002). DEC1 can repress gene expression by either directly binding to the E-box sequences (CACGTG) of its target genes or interacting with transactivators, such as CLOCK/BMAL1, sterol regulatory element binding protein-1c (SREBP-1c) and MyoD (Choi et al., 2008; Honma et al., 2002; Hsiao et al., 2009). We previously demonstrated that DEC1 represses the lipogenic transcription factor, SREBP-1c, and its target genes, including fatty acid synthase and SREBP-1. DEC1 bind to E-box, which is next to SREBP-1c biding sites, and interact with the SREBP-1c protein. By doing so, DEC1 prevent SREBP-1c from binding to the promoters of FAS and SREBP-1c (Choi et al., 2008).
Previously, we found that hypoxia inhibits adipogenesis through a HIF-1α-independent mechanism, by preventing C/EBPβ from binding to DNA. For C/EBPβ to bind to DNA, it must be dually phosphorylated by both ERK and GSK3β kinase (Kim et al., 2007; Tang et al., 2003). We found that hypoxia blocks the nuclear localization of GSK3β via a HIF-independent mechanism (Park and Park, 2010). Yun et al. suggested that hypoxia-induced DEC1/Stra13 is involved in hypoxic repression of PPARγ2 (Yun et al., 2002). However, the detailed mechanisms of how DEC1 inhibits adipogenesis are poorly understood. In addition to the HIF-independent mechanism, here, we show that hypoxia also represses PPARγ2 gene expression through a HIF-dependent mechanism by inducing DEC1, which are recruited to the promoter of PPARγ2 by interacting with C/EBPβ.
MATERIALS AND METHODS
Materials and plasmids
Antibodies against HIF-1α, flag-tag and myc-tag were purchased from Novus Biochemicals, Sigma-Aldrich (USA) and IG Therapy (Korea), respectively. Anti-C/EBPβ, anti-PPARγ, anti-HDAC1, and anti-tubulin antibodies were purchased from Santa Cruz Biotechnology. The anti-DEC1 antibody was kindly provided by Prof. Fuyuki Sato (Hirosaki University, Japan). cDNAs of DEC1 (AF010305) was subcloned into the pCMV-myc vector (Clontech, USA). cDNAs of C/EBPβ (NM_009883) was subcloned into the pCMV-3xflag vector (Sigma, USA) or the retroviral vector pBabe-puro encoding puromycin resistance, which was a generous gift from Garry Nolan (Stanford University School of Medicine, USA). The PPARγ2 promoter-driven luciferase reporter plasmid was constructed by subcloning the upstream regulatory region (−603 bp to +64 bp) of the mouse PPARγ2 promoter into the pGL3-Basic plasmid (Promega, USA).
Gene silencing using small interfering RNA (siRNA)
siRNAs specific for DEC1 and green fluorescent protein (GFP, for Control), were synthesized by Samchully Pharm. Co. (Korea) as described previously (Choi et al., 2008). Sequence of each siRNA was 5′-GAACGUGUCAGCACAAUUA-3′ for DEC1 and 5′-GUUCAGCGUGUCCGGCGAG-3′ for GFP. Transfection was carried out using PolyMAG according to the instructions of the manufacturer (Chemicell GmBH, Germany).
Cell culture and adipogenic differentiation
3T3-L1 (American Type Culture Collection, catalog no. CL-173) preadipocytes and NIH-3T3 cells were maintained and induced to differentiate into mature adipocytes by treatment of adipogenic hormones, MDI as described previously (Park and Park, 2010). For hypoxic treatment, cells were incubated in an anaerobic incubator (Model 1029, Forma Scientific, Inc.) in an atmosphere of 5% CO2, 10% H2, and 85% N2 at 37°C. This incubator maintains 0.1–0.5% oxygen concentration.
Retroviral infection
The stable HIF-1α-knock-down 3T3-L1 preadipocytes were generated by using a retroviral pSIREN-RetroQ vector system (BD Biosciences) as described previously (Park and Park, 2010). We generated NIH-3T3/Cβ cells that constitutively expressed flag-tagged C/EBPβ, by infecting NIH-3T3 mouse fibroblast cells with pBabe-puro retrovirus encoding flag-tagged C/EBPβ and puromycin resistance gene by using an Ampho-PackTM-293 cell line according to the manufacturer’s instructions (BD Biosciences). The infected NIH-3T3 cells were selected in the presence of 5 μg/ml puromycin over 6 days.
Transient transfection and Luciferase assay
The 2 × 104 of 3T3-L1 cells were plated in a 24-well plate and transfected with the desired plasmids (250 ng) using the Lipofectamine reagent (Invitrogen). Either pCHO110 (50 ng) encoding β-galatosidase or pRL-TK (50 ng) encoding Ranilla luciferase was cotransfected to normalize the transfection efficiency. After 48 h transfection, cell extracts were prepared using the luciferase assay system (Promega). Luciferase activities were analyzed with a luminometer (Turner TD-20/20, Promega).
Western analyses and Co-immunoprecipitation
For Western blot analysis, 30 μg of protein from whole cell lysates were analyzed using the indicated antibodies. For immunoprecipitation, NIH-3T3/Cβ cells were transfected with the desired plasmids. The 300 μg of protein from the whole cell lysates were analyzed as described previously (Choi et al., 2008). The cleared extracts were incubated with 2 μg of the indicated antibodies at 4°C for overnight. The precipitated proteins were analyzed by Western blot analyses.
Chromatin immunoprecipitation (ChIP) assay
NIH-3T3/Cβ cells were transiently transfected with indicated plasmids and treated with MDI for 36 h. Thereafter the chromatin immunoprecipitation (ChIP) assays were performed as described previously (Park and Park, 2010). Briefly, the transfected cells were cross-linked in 1% formaldehyde at RT for 10 min and resuspended in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1]). Lysates were sonicated and then diluted 10-fold with ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl). The diluted lysates were immunoprecipitated with 2 μg of anti-C/EBPβ, anti-Flag or anti-Myc, anti-H3K9K14Ac, anti-HDAC1, or anti-immunoglobulin G (IgG). The precipitated DNA fragments were analyzed by PCR using the primers for the PPARγ2 promoter as described previously (Park and Park, 2010).
Immunofluorescence microscopy
NIH3T3/Cβ cells (2 × 104) were plated on glass coverslips in a 24-well culture plate and then transfected with 1 μg pDMV-myc-DEC1. Atter 36 h, the cells were treated with MDI for additional 36h as described above. The 4% paraformaldehyde fixed cells were incubated overnight with anti-PPARγ (1:200) or anti-Myc (1:500) antibodies at 4°C and then incubated with secondary antibodies conjugated to either AlexaFluor® 546 or AlexaFluor® 488 (Invitrogen, USA) for 1 h at room temperature. Cells were incubated with DAPI (Invitrogen) for 5 min and then the coverslips were mounted on slides, stained cells were observed under a Zeiss LSM510 inverted confocal microscope according to the manufacturer’s instructions.
RESULTS AND DISCUSSION
Hypoxia-induced DEC1 inhibits the activity of C/EBPβ
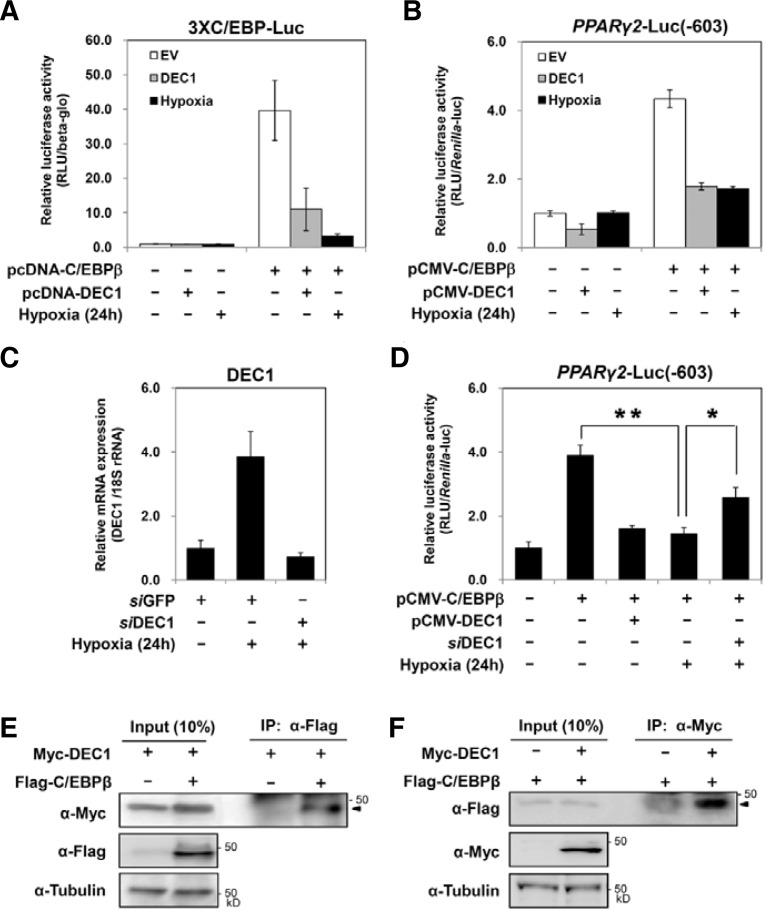
Mouse preadipocytes, 3T3-L1 cells, were treated with MDI to induce adipogenic differentiation. The expression of PPARγ2 and C/EBPα mRNA increased 1 day after MDI treatment. C/EBPβ, a common transactivator for both PPARγ2 and C/EBPα is induced 4 h after MDI treatment (Fig. 1A). Under hypoxic conditions, hormone-induced expression of either PPARγ2 or C/EBPα mRNA was not observed, whereas C/EBPβ is induced by MDI, even under hypoxia. Hypoxia induces DEC1 mRNA at an early time and continuously maintains its expression (Fig. 1B). Since DEC1 was known as a target gene of HIF-1α, we further investigated the effects of HIF-1α on the expression of DEC1 using HIF-1α knockdown 3T3-L1 cells (Park and Park, 2010). In the HIF-1α knockdown 3T3-L1 cells, hypoxia induced much less DEC1 expression compared to the control 3T3-L1 cells (Fig. 1C). The fact that DEC1 was also induced by HIF-2α suggests that the residual expression of DEC1 in HIF-1α knockdown 3T3-L1 cells can be induced by HIF-2α (Choi et al., 2008). In order to evaluate the contribution of DEC1 on the hypoxic repression of PPARγ2, we investigated whether DEC1 affected the activity of C/EBPβ. We used a reporter luciferase gene driven by three copies of C/EBP response elements (3xC/EBP-Luc) and another reporter gene driven by the PPARγ2 promoter (−603 to +64 bp) which contains the C/EBP response element (Figs. 2A and 2B). We confirmed that cotransfection of C/EBPβ increases the expression of these reporter genes. Hypoxia significantly inhibited C/EBPβ-mediated reporter activities. Notably, co-transfection with DEC1 was sufficient to decrease the transactivity of C/EBPβ even under normoxic conditions. These findings suggest that hypoxia can inhibit the transactivity of C/EBPβ by inducing DEC1. To examine the contribution of DEC1 on hypoxic inhibition of C/EBPβ, we knock-down the expression of DEC1 by using its specific siRNAs (Fig. 2C). Co-transfection of siRNA against DEC1 reverses the hypoxic inhibition of C/EBPβ in part, confirming that DEC1 mediates hypoxic inhibition of C/EBPβ (Fig. 2D). We next examined if DEC1 interacts with C/EBPβ proteins. Co-immunoprecipitation analyses showed that DEC1 interacts with C/EBPβ (Figs. 2E and 2F). These findings imply that DEC1 inhibits the transactivity of C/EBPβ by interacting with it.
Fig. 2.
Effects of DEC1 on the activity of C/EBPβ. (A) 3xC/EBP-Luc reporter plasmid and the indicated plasmids were transfected into 3T3-L1 cells with pCHO110. After 24 h, the transfected cells were further incubated under either normoxic or hypoxic conditions (0.1–0.5% oxygen) for 24 h before harvest. The luciferase activity of the cell lysate was measured and normalized based on β-galactosidase activity as described previously. (B) The PPARγ2-Luc reporter plasmids and the indicated plasmids were transfected into 3T3-L1 cells with the pRL-TK plasmids as described above. The luciferase activity was measured and normalized based on the Renilla luciferase activity. The values indicated averages and standard deviations of three independent experiments. (C) 3T3-L1 cells were transfected with siRNA against DEC1 as described. Before harvest, the transfected cells were exposed to hypoxia (0.1–0.5% oxygen) for 24 h. The levels of DEC1 mRNA were quantified by qRT-PCR. (D) The PPARγ2-Luc reporter plasmids were transfected with the indicated plasmid or siRNA into 3T3-L1 cells. The transfected cells were incubated in hypoxia (0.1–0.5% oxygen) for 24 h before harvesting, and luciferase assays were performed. Values represent the means and standard deviations of three experiments. P values for the indicated sets are calculated by Student’s t-test; *p < 0.05 and **p < 0.01, respectively. (E, F) NIH3T3 cells were transfected with the indicated plasmids. At 48 h after transfection, cell lysate was harvested and immunoprecipitated (IP) with either anti-flag antibody (E) or anti-myc antibody (F), then the resulting immunoprecipitant was analyzed by Western blot (WB) using the indicated antibodies. Input indicates the Western blot of 10% amount of the lysates which were used for IP.
DEC1 is recruited on the promoter of PPARγ2
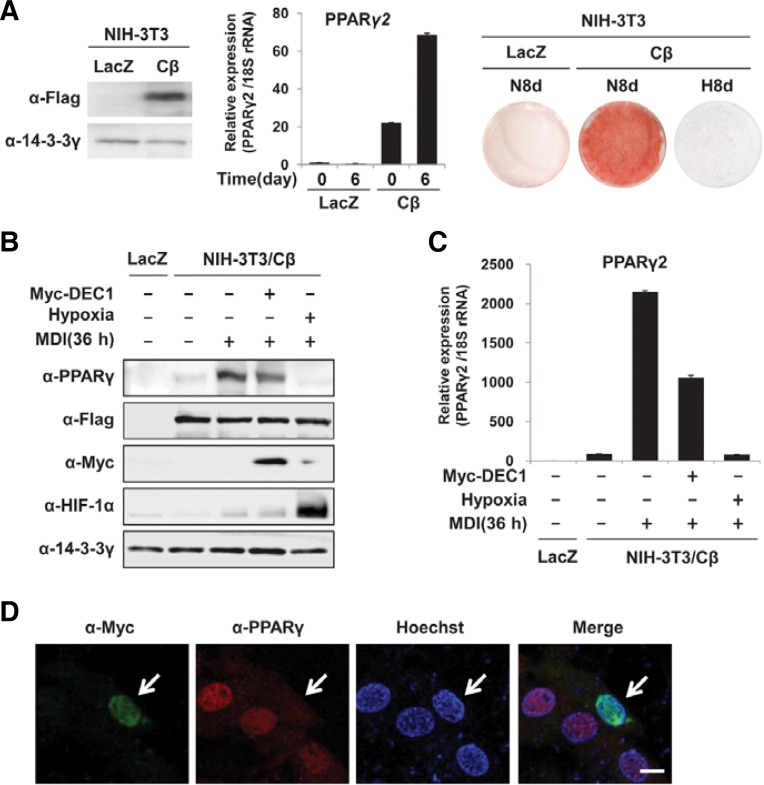
To investigate the effects of DEC1 on the promoter of the PPARγ2 gene, we generated flag-tagged C/EBPβ-expressing NIH-3T3 cells, named NIH-3T3/Cβ which can be differentiated into mature adipocytes by MDI treatment and which can be easily transfected with desired plasmids. Western blot analyses confirmed that the NIH-3T3/Cβ cells constitutively expressed flag-tagged C/EBPβ even in the absence of MDI. Quantitative RT-PCR (qRT-PCR) and Oil Red-O staining showed that MDI induced PPARγ2 mRNA and lipid accumulation in NIH-3T3/Cβ fibroblast but not in NIH-3T3 cells that were infected with the retrovirus encoding LacZ instead of C/EBPβ (Fig. 3A). To determine whether ectopic expression of DEC1 inhibits the induction of PPARγ, NIH-3T3/Cβ cells were transfected with myctagged DEC1 and then treated with MDI for 36 h. Western blot analyses (Fig. 3B) and qRT-PCR assays (Fig. 3C) showed that transfection with myc-DEC1 reduces induction of PPARγ2 in response to MDI. Additionally, we examined PPARγ expression by immunohistochemistry in NIH3T3/Cβ cells transfected with myc-tagged DEC1. The immunofluorescence staining showed that after 36 hr MDI treatment, PPARγ proteins are detected in untransfected cells but not in DEC1 transfected cells (Fig. 3D). These data suggest that DEC1 represses PPARγ expression even in NIH3T3 cells which constitutively express C/EBPβ.
Fig. 3.
Effects of DEC1 on the expression of PPARγ2 in NIH-3T3/Cβ cells. (A) The stably expressed flag-tagged C/EBPβ in NIH-3T3/Cβ cells was detected using an anti-flag antibody (left panel). NIH-3T3/LacZ or Cβ cells were differentiated to adipocytes for 6 or 8 days under either normoxic (20–21% oxygen) or hypoxic (0.1–0.5% oxygen) conditions. The expression of PPARγ2 mRNA was determined by qRT-PCR (middle panel). The lipid droplets were stained with Oil Red-O (right panel). (B, C) NIH-3T3/Cβ cells were transiently transfected with myc-tagged DEC1 then treated with MDI for 36 h. Western analyses were performed using the indicated antibodies. The 14-3-3γ was detected to check for equal loadings (B). mRNA levels of PPARγ2 of each sample was measured by quantitative RT-PCR analyses. Expression level of 18S rRNA was used for normalization (C). (D) Confocal microscopic images of PPARγ (red) or myc-tagged DEC1 (green) in NIH3T3/Cβ cells which were treated with MDI for 36 h. An arrow indicates a NIH3T3/Cβ cell which is transfected with myc-tagged DEC1.
To investigate whether DEC1 prevents C/EBPβ from binding to the promoter of PPARγ2, we performed chromatin immunoprecipitation (ChIP) assays using nuclear extracts from NIH-3T3/Cβ cells that were transiently transfected with myc-tagged DEC1 or empty vector. ChIP analyses using either anti-C/EBPβ antibody or anti-flag antibody showed that ectopically expressed flag-tagged C/EBPβ binds to the PPARγ2 promoter in response to MDI. We found that overexpression of DEC1 did not prevented C/EBPβ from binding to the PPARγ2 promoter (Fig. 4A). ChIP analyses using anti-myc antibody revealed that ectopically expressed myc-tagged DEC1 also can be recruited to the PPARγ2 promoter (Fig. 4B). We analyzed 10 kb around the PPARγ2 transcription start site to identify the type B (CACGTG) E-box site which is a cognitive DNA sequences for DEC1 homodimers (Nakamura et al., 2008; St-Pierre et al., 2002). In silico analyses reveal that PPARγ2 gene did not contain type B E-box sites. In order to test whether DEC1 can directly bind to C/EBP binding sites in the PPARγ2 promoter, we performed an electrophoretic mobility shift assay (EMSA) using 32P-labeled probes containing the C/EBP binding site used for generating 3xC/EBP-Luc reporter (Fig. 2A). C/EBPβ and DEC1 were synthesized using an in vitro transcription/translation system, and incubated with the 32P-labeled probes. EMSA showed that C/EBPβ but not DEC1 directly binds to this probe, and that DEC1 does not alter the mobility of DNA and C/EBPβ binding complex (Fig. 4C). We presumed that DEC1 were indirectly recruited on the PPARγ2 promoter by interacting with C/EBPβ and/or other putative proteins on the PPARγ2 promoter. The previous finding that DEC1 can interact with histone deacetylase 1 (HDAC1), suggests that DEC1 on the PPARγ2 promoter can recruit HDAC1, which is involved in deacetylation of histones (Sun and Taneja, 2000). As shown in Fig. 4D, we transfected NIH3T3 cells with myc-tagged DEC1 and flag-tagged HDAC1. We immunoprecipitated the cell lysates with anti-myc antibody then detected the presence of HDAC1 with anti-flag antibody. Although a nonspecific band is detected at 55 kDa which corresponds to the molecular weight of both IgG and flag-tagged HDAC1, more protein is detected in the immune-precipitant from the cells transfected with myc-tagged DEC1. This result confirmed the previous finding that DEC1 interacts with HDAC1 (Sun and Taneja, 2000). Then, we tested whether the interaction between DEC1 and HDAC1 is maintained in MDI treated NIH3T3/Cβ cells. By using anti-myc antibody, we immunoprecipitated the cell lysates of NIH3T3/Cβ cells which were transfected with myc-tagged DEC1. Then we detected the presence of either HDAC1 or C/EBPβ in the precipitants. The results in Fig. 4D showed that MDI treatment increases the interaction between DEC1 and HDAC1, while interaction between DEC1 and C/EBPβ is not changed by MDI treatment (Fig. 4D). ChIP analyses using anti-H3K9K14Ac antibodies showed that in NIH-3T3/Cβ cells, the histone acetylation on the PPARγ2 promoter increases upon MDI treatment. Ectopic expression of DEC1 reverses the MDI-induced histone acetylation on the PPARγ2 promoter (Fig. 4E). Based on the findings that (i) DEC1 can interact with C/EBPβ (Figs. 2E and 2F), (ii) DEC1 also can interact with HDAC1 (Fig. 4D) and (iii) hypoxia induces DEC1 through a HIF-dependent mechanism (Fig. 1B), we can infer that the DEC1 induced by HIF, occupies the PPARγ2 promoter by interacting with the promoter-bound C/EBPβ, then DEC1 recruits HDAC1, which decreases the level of acetylated histones at the PPARγ2 promoter even in the presence of adipogenic hormones. Our findings suggest that DEC1 functions as a corepressor reinforcing the interaction between C/EBPβ and HDAC1 on the PPARγ2 promoter. However, hypoxia-induced DEC1, if any, may not be recruited on PPARγ2 promoter through interacting with promoter-bound C/EBPβ, since hypoxia prevents C/EBPβ from binding to PPARγ2 promoter by HIF-1α independent mechanism (Park and Park, 2010). The previous findings that DEC1 is induced by many other signals involved in mammalian cell differentiation, cell cycle, circadian rhythm and carcinogenesis suggest that DEC1 is one of the mediators which reset the pattern of PPARγ2 expression in response to not only hypoxia but also the other signals (Bhawal et al., 2011; Kovac et al., 2009; Noshiro et al., 2005; Sun and Taneja, 2000).
Fig. 4.
Proteins bound to the PPARγ2 promoter. NIH3T3/Cβ cells were transiently transfected with myc-tagged DEC1 and then treated with MDI for 36 h. By using nuclear extracts from the transfected cells, ChIP assays were performed with the indicated antibodies and a primer set for PPARγ2 promoter region (−474 bp to −185 bp). (A) ChIP assays using either anti-C/EBPβ (top panel) or anti-Flag antibodies (bottom panel). (B) ChIP assay using anti-Myc antibody. (C) EMSA was performed as described using in vitro transcribed and translated C/EBPβ and DEC1 (Park and Park, 2010). Oligonucleotides containing the C/EBP binding element (5′-TGC AGA TTG CGC AAT CTG CA-3′) were radiolabeled with [α-32P]-dATP. (D) NIH3T3 cells were transfected with myc-tagged DEC1 and flag-tagged HDAC1. The cell lysates were harvested and immunoprecipitated (IP) with anti-myc antibody then the resulting immunocomplex was analyzed by Western blot (WB) using indicated antibodies. Input indicates the western blot of 10% amount of the lysates which were used for IP (left panel). NIH3T3/Cβ cells were transfected with myc-tagged DEC1 and treated with MDI. At 36 h after treated MDI, cell lysate was harvested and immunoprecipitated (IP) with anti-myc antibody. The resulting immunocomplex was analyzed by Western blot (WB) using indicated antibodies. Input indicates the Western blot of 10% amount of the lysates which were used for IP (right panel). (E) ChIP assay using anti-HDAC1 antibody or anti-acetylated K9/K14 residues of histone3 (H3K9K14Ac). (F) Hypothetical mechanism by which hypoxia-induced DEC1 represses PPARγ2 gene.
DEC1 also enhances the recruitment of HDAC1 to the ATP-binding cassette transporter A1 (ABCA1) gene, a target gene of retinoid X receptor (RXR)/liver X receptor heterodimer by interacting with RXR (Cho et al., 2009). Although DEC1 prevents neither C/EBPβ nor RXR from binding to its target gene, DEC1 prevents bHLH domain containing transactivators such as BMAL1, MyoD and SREBP-1c, from binding to their target DNA by either protein-protein interaction or by directly binding to their responsive elements containing an E-box (Azmi et al., 2004; Choi et al., 2008; Li et al., 2004). Recently, Gulbagci et al. demonstrated that DEC2, isoform of DEC1, also interacts with C/EBPβ and increases the recruitment of histone methyltransferase G9a to the promoter of PPARγ2 (Gulbagci et al., 2009). Our results suggested the fact that not only DEC2 but also DEC1 is involved in repression of PPARγ2 gene. The fact that DECs are induced by HIF-α/β heterodimers in response to hypoxia suggests that hypoxia can block the expression of PPARγ2 gene through induction of DEC2 as well as DEC1.
Acknowledgments
We thank Dr. Shizuo Akira, Dr. Nicolai A. Timchenko, and Dr. Jae Bum Kim for providing cDNAs for C/EBPβ, C/EBPα, and PPARγ2, respectively. This study was supported by a grant of the Korea Healthcare technology R&D Project (A090616) to H. Park.
REFERENCES
- Azmi S., Ozog A., Taneja R. Sharp-1/DEC2 inhibits skeletal muscle differentiation through repression of myogenic transcription factors. J. Biol. Chem. 2004;279:52643–52652. doi: 10.1074/jbc.M409188200. [DOI] [PubMed] [Google Scholar]
- Bhawal U.K., Sato F., Arakawa Y., Fujimoto K., Kawamoto T., Tanimoto K., Ito Y., Sasahira T., Sakurai T., Kobayashi M., et al. Basic helix-loop-helix transcription factor DEC1 negatively regulates cyclin D1. J. Pathol. 2011;224:420–429. doi: 10.1002/path.2878. [DOI] [PubMed] [Google Scholar]
- Chandel N.S., Simon M.C. Hypoxia-inducible factor: roles in development, physiology, and disease. Cell Death Differ. 2008;15:619–620. doi: 10.1038/cdd.2008.11. [DOI] [PubMed] [Google Scholar]
- Cho Y., Noshiro M., Choi M., Morita K., Kawamoto T., Fujimoto K., Kato Y., Makishima M. The basic helix-loophelix proteins differentiated embryo chondrocyte (DEC) 1 and DEC2 function as corepressors of retinoid X receptors. Mol. Pharmacol. 2009;76:1360–1369. doi: 10.1124/mol.109.057000. [DOI] [PubMed] [Google Scholar]
- Choi S.M., Cho H.J., Cho H., Kim K.H., Kim J.B., Park H. Stra13/DEC1 and DEC2 inhibit sterol regulatory element binding protein-1c in a hypoxia-inducible factor-dependent mechanism. Nucleic Acids Res. 2008;36:6372–6385. doi: 10.1093/nar/gkn620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbagci N.T., Li L., Ling B., Gopinadhan S., Walsh M., Rossner M., Nave K.A., Taneja R. SHARP1/DEC2 inhibits adipogenic differentiation by regulating the activity of C/EBP. EMBO Rep. 2009;10:79–86. doi: 10.1038/embor.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma S., Kawamoto T., Takagi Y., Fujimoto K., Sato F., Noshiro M., Kato Y., Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- Hsiao S.P., Huang K.M., Chang H.Y., Chen S.L. P/CAF rescues the Bhlhe40-mediated repression of MyoD transactivation. Biochem. J. 2009;422:343–352. doi: 10.1042/BJ20090072. [DOI] [PubMed] [Google Scholar]
- Jaakkola P., Mole D.R., Tian Y.M., Wilson M.I., Gielbert J., Gaskell S.J., Kriegsheim A., Hebestreit H.F., Mukherji M., Schofield C.J., et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Kim J.W., Tang Q.Q., Li X., Lazne M.D. Effect of phosphorylation and S-S bond-induced dimerization on DNA binding and transcriptional activation by C/EBPbeta. Proc. Natl. Acad. Sci. USA. 2007;104:1800–1804. doi: 10.1073/pnas.0611137104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovac J., Husse J., Oster H. A time to fast, a time to feast: the crosstalk between metabolism and the circadian clock. Mol. Cells. 2009;28:75–80. doi: 10.1007/s10059-009-0113-0. [DOI] [PubMed] [Google Scholar]
- Li Y., Song X., Ma Y., Liu J., Yang D., Yan B. DNA binding, but not interaction with Bmal1, is responsible for DEC1-mediated transcription regulation of the circadian gene mPer1. Biochem. J. 2004;382:895–904. doi: 10.1042/BJ20040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K., Kawamoto T., Tanimoto K., Nishiyama M., Honda H., Kato Y. Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J. Biol. Chem. 2002;277:47014–47021. doi: 10.1074/jbc.M204938200. [DOI] [PubMed] [Google Scholar]
- Mohyeldin A., Garzon-Muvdi T., Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7:150–161. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Nakamura H., Tanimoto K., Hiyama K., Yunokawa M., Kawamoto T., Kato Y., Yoshiga K., Poellinger L., Hiyama E., Nishiyama M. Human mismatch repair gene, MLH1, is transcriptionally repressed by the hypoxia-inducible transcription factors, DEC1 and DEC2. Oncogene. 2008;27:4200–4209. doi: 10.1038/onc.2008.58. [DOI] [PubMed] [Google Scholar]
- Noshiro M., Furukawa M., Honma S., Kawamoto T., Hamada T., Honma K., Kato Y. Tissue-specific disruption of rhythmic expression of Dec1 and Dec2 in clock mutant mice. J. Biol. Rhythms. 2005;20:404–418. doi: 10.1177/0748730405280195. [DOI] [PubMed] [Google Scholar]
- Park Y.K., Park H. Prevention of CCAAT/enhancerbinding protein beta DNA binding by hypoxia during adipogenesis. J. Biol. Chem. 2010;285:3289–3299. doi: 10.1074/jbc.M109.059212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre B., Flock G., Zacksenhaus E., Egan S.E. Stra13 homodimers repress transcription through class B E-box elements. J. Biol. Chem. 2002;277:46544–46551. doi: 10.1074/jbc.M111652200. [DOI] [PubMed] [Google Scholar]
- Sun H., Taneja R. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Proc. Natl. Acad. Sci. USA. 2000;97:4058–4063. doi: 10.1073/pnas.070526297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q.Q., Otto T.C., Lane M.D. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc. Natl. Acad. Sci. USA. 2003;100:850–855. doi: 10.1073/pnas.0337434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun Z., Maecker H.L., Johnson R.S., Giaccia A.J. Inhibition of PPAR gamma 2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev. Cell. 2002;2:331–341. doi: 10.1016/s1534-5807(02)00131-4. [DOI] [PubMed] [Google Scholar]