Abstract
Sonic hedgehog (Shh) signaling regulates patterning, proliferation, and stem cell self-renewal in many organs. Smoothened (Smo) plays a key role in transducing Shh signaling into the nucleus by activating a glioma family of transcription factors; however, the cellular and molecular mechanisms underlying the role of sustained Smo activation in postnatal development are still unclear. In this study, we explored the effects of Shh signaling on bone development using a conditional knock-in mouse model that expresses a constitutively activated form of Smo (SmoM2) upon osteocalcin (OCN)-Cre-mediated recombination (SmoM2; OCN-Cre mice). We also evaluated the expression pattern of bone formation-related factors in primary calvarial cultures of mutant and control mice. The SmoM2;OCN-Cre mutant showed growth retardation and reduction of bone mineral density compared to control mice. Constitutively activated SmoM2 also repressed mRNA expression of Runx2, osterix, type I collagen, and osteocalcin. Further, sustained SmoM2 induction suppressed mineralization in calvarial primary osteoblasts cultures, whereas such induction did not affect cell proliferation in the mutant cultures as compared with SmoM2 only control cultures. These results suggest that sustained Smo activation inhibits postnatal development of bone by suppressing gene expression of bone formation regulatory factors in mice.
Keywords: constitutively activated form of Smoothened (SmoM2), gene expression, osteoblastic differentiation, postnatal bone development
INTRODUCTION
Hedgehog signaling plays a critical role in the regulation of growth, pattern formation, and stem cell maintenance and self-renewal in many organs during development (Baker, 2007; Mimeault and Batra, 2010). It has been implicated that alterations in hedgehog signaling cause abnormal regulation of cellular events, which is closely associated with tumorigenesis in several cancers (Gupta et al., 2010). It is also known that hedgehog-Patched1 (Ptch1) signaling plays a crucial role in postnatal bone homeostasis and is a potential target for the treatment of osteoporosis (Ohba et al., 2008). The finding that the inhibition of hedgehog signaling reduces bone loss in aged mice suggested its pivotal roles in regulating bone formation (Mak et al., 2008). Indian hedgehog (Ihh) signaling is believed to regulate chondrocyte proliferation and differentiation and osteoblast differentiation. Ihh-deficient mice showed a lack of endochondral bone formation due to the absence of osteoblasts, and runt-related transcription factor-2 (Runx2) expression was found to be absent in perichondrial cells before vascular invasion (Hilton et al., 2005). Ihh expression was also found to up-regulate the function of Runx2 through Gli2 (Shimoyama et al., 2007). In addition, ectopic Ihh expression in chondrocytes mediates Runx2 induction throughout the perichondrium, although Ihh alone is not sufficient to induce bone formation (Komori, 2012). Moreover, sonic hedgehog (Shh) is the most widely characterized of three analogous proteins in mammals and is essential for proper development in both embryonic and adult stages. Shh signaling involves both the cells expressing a member of the hedgehog family of secreted ligands and one or more patched family hedgehog receptors.
In the absence of a hedgehog ligand, the patched receptor inhibits the signaling activity of the downstream seven-pass transmembrane receptor Smoothened (Smo). Binding of Shh protein to its receptor causes Smo to activate signal transduction, which allows for the activation of a glioma (Gli) family of transcription factors and their translocation into the nucleus (Stanton and Peng, 2010). Eventually, the Smo-mediated Gli activation leads to the expression of specific genes that promote cell proliferation and differentiation. Smo is also frequently and ectopically expressed in human breast cancers (Visbal et al., 2011), while altered hedgehog signaling is implicated in approximately 20–25% of all cancers (Briscoe and Therond, 2005). Considerable evidence has shown that sustained Smo-mediated activation of hedgehog signaling leads to increased proliferation and abnormal ductal morphology of the mammary gland, although Smo activation in luminal mammary epithelial cells was found only in a small percentage (−5%) of glands (Moraes et al., 2007). It was also suggested that Smo stimulates proliferation via a non-cell autonomous paracrine or juxtacrine mechanism (Visbal et al., 2011). However, the precise mechanism by which altered Smo expression affects cell proliferation and postnatal development is not completely defined.
In this study, we examined the effects of Shh signaling on bone development using a conditional knock-in mouse model that expresses a constitutively activated form of Smo (SmoM2) upon osteocalcin (OCN)-Cre-mediated recombination (SmoM2; OCN-Cre mice). We investigated the expression pattern of several bone-related genes in primary calvarial osteoblasts derived from transgenic and control mice, and the potential of these osteoblasts to mineralize and proliferate was determined.
MATERIALS AND METHODS
Chemicals and laboratory wares
Unless specified otherwise, all chemicals and laboratory wares were obtained from Sigma Chemical Co. (USA) and Falcon Labware (Becton-Dickinson, USA), respectively.
Mouse strains and genotyping
The Animal Welfare Committee of Chonbuk National University approved all experimental procedures. The SmoM2 mice (Jeong et al., 2004) and R26R reporter mice (Soriano, 1999) were purchased from the Jackson Laboratory (USA). To generate SmoM2;OCN-Cre mice, SmoM2 mice were crossed with OCNCre mice (Tan et al., 2007). The offspring were genotyped by PCR analysis using primers described previously (Jeong et al., 2004; Tan et al., 2007). For analysis of Cre activity in OCN-Cre mice, OCN-Cre mice were crossed with R26R mice, and the mandibles of double transgenic mice were processed for X-gal staining as described previously (Hogan et al., 1994). SmoM2 only mice were used as a control of the SmoM2;OCN-Cre mutant mice in all experiments.
Micro-computed tomography
Tibias from 4-week-old control or mutant mice were scanned using a desktop scanner (1076 Skyscan Micro-CT, Skyscan, Belgium) and subsequently reconstructed and analyzed with CTscan software (Skyscan).
Preparation of primary calvarial osteoblasts
Primary osteoblasts were prepared from the calvariae of the control or mutant mice at 2, 3, 4, and 5 weeks of age. Calvariae were digested several times with 0.1% collagenase at 37°C for 30 min, and the last fraction was used as the source of primary osteoblasts. The primary osteoblasts were cultured in α-minimum essential medium (α-MEM) supplemented with 10% fetal bovine serum (FBS; HyClone, USA) and antibiotics. When the cells reached 90% confluence in a 100-mm culture dish, they were dissociated using Trypsin/EDTA and spread onto various culture plates according to the experimental design.
Real time RT-PCR
Total cellular RNA was prepared from primary osteoblasts using Trizol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Total RNA (1 μg) was subjected to cDNA synthesis using SuperScript Reverse Transcripatase II and oligo12–18 primers (Invitrogen). Power SYBR Green PCR Master Mix (Applied Biosystems, USA) was used to detect the accumulation of PCR product during cycling using the ABI 7500 Sequence Detection System (Applied Biosystems). The amplification conditions with a duration of 40 cycles were as follows: predenaturation at 95°C for 10 min, amplification using three-step cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. Oligonucleotide primers for real-time RT-PCR were designed with product sizes less than 200 bp using Primer Express 3.0 (Applied Biosystems). The primers specific to Runx2, osterix, osteocalcin (OCN), osteopontin (OPN), type I collagen (COL I), and alkaline phosphatase (ALP) were used in this study (Table 1).
Table 1.
Primers sequences and product size of bone-related genes
| Gene name | GenBank | Sequence | Product size (bp) | |
|---|---|---|---|---|
| GAPDH | NM_008084.2 | F | GACGGCCGCATCTTCTTGT | 65 |
| R | CACACCGACCTTCACCATTTT | |||
| RUNX2 | NM_009820.4 | F | GAGGGACTATGGCGTCAAACA | 70 |
| R | GGATCCCAAAAGAAGCTTTGC | |||
| Osterix | NM_130458.2 | F | TCAGCCGCCCCGATCTTCCA | 156 |
| R | AATGGGTCCACCGCGCCAAG | |||
| OCN | NM_001037939.1 | F | ACTCCGGCGCTACCTTGGGT | 109 |
| R | CCTGCACGTCTAGCCCTCTGC | |||
| OPN | NM_009263.1 | F | TGGTGGTGATCTAGTGGTGCCAA | 148 |
| R | CACCGGGAGGGAGGAGGCAA | |||
| COL I | NM_007742.3 | F | AAGACGGGAGGGCGAGTGCT | 121 |
| R | AACGGGTCCCCTTGGGCCTT | |||
| ALP | NM_007431.2 | F | GGGCCCTGCTGCTTCCACTG | 70 |
| R | GGCTTGTGGGACCTGCACCC |
Osteoblastic differentiation
Primary calvarial osteoblasts were spread onto 6-well culture plates and then incubated in osteoblast-inducing medium (OIM) supplemented with DAG (10 nM dexamethasone, 50 μM ascorbic acid, and 20 mM β-glycerophosphate). After 10 days of incubation, the cells were processed for the determination of osteoblastic differentiation and mineralization.
Alizarin red S staining
Calcium deposition was determined using alizarin red S staining. In brief, the culture medium was discarded, and cells were fixed for 30 min in 4% paraformaldehyde fluid, washed twice with distilled water, and then stained with 40 mM alizarin red S at pH 4.2 for 20 min with gentle agitation. The cells were briefly washed with distilled water and processed for light microscopic observation. To quantify the red dye, the stains were solubilized with 10% acetylpyridinium chloride by shaking for 30 min at room temperature, and the absorbance was measured at 540 nm using a microplate reader spectrophotometer (Bio-Tek instrument, USA).
Cell proliferation assay
Cell proliferation was assessed using a Cell Counting Kit (CCK)-8 (Dojindo Laboratories, Japan). In brief, single calvarial osteoblasts were resuspended in α-MEM and plated at a cell density of 1000 cells/well in a 96 multi-well plate. At 24 and 72 h after incubation, 10 μl of CCK-8 solution was directly added to each well followed by an additional 1 h-incubation. Finally, the absorbance was measured at 450 nm using a microplate reader.
Statistical analysis
Unless specified otherwise, all data are expressed as the mean ± standard deviation (SD) from at least three independent experiments. A one-way analysis of variance (ANOVA) followed by the Scheffé’s test was used for multiple comparisons using the SPSS program (version 18.0). A value of p < 0.05 was considered statistically significant.
RESULTS
The SmoM2;OCN-Cre mutant shows growth retardation and reduction of bone mineral density compared to control mice
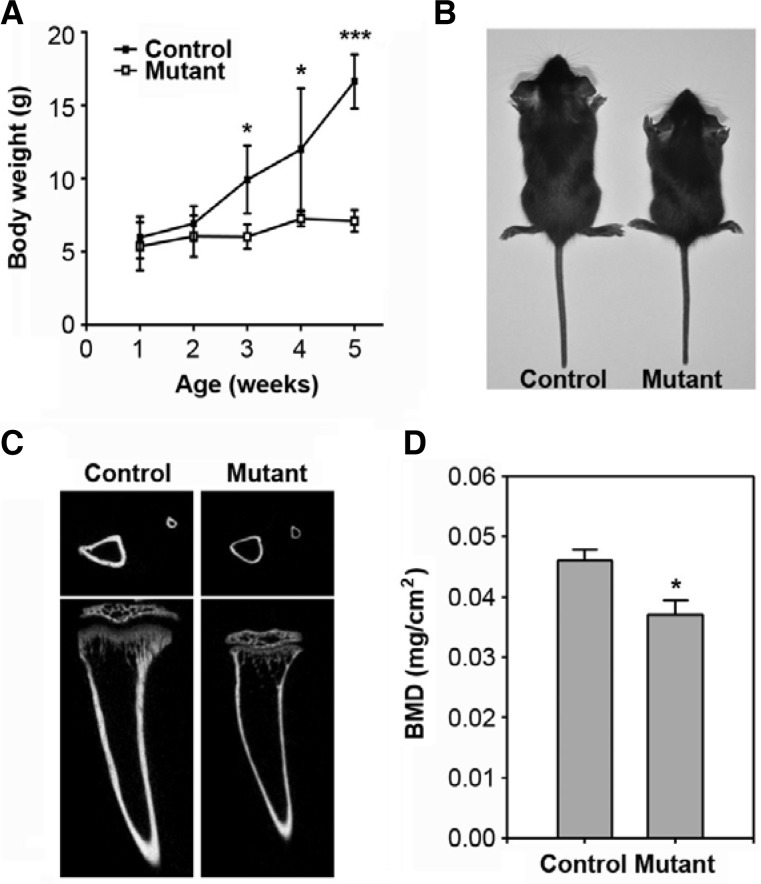
SmoM2;OCN-Cre mice were born normally and had similar body weight to controls until 2 weeks of age; however, the mutants showed a significant reduction in body weight by 3 weeks of age (Fig. 1A). This reduction was apparent according to age, such that the mutant mice were 39.3%, 39.6%, and 57.2% smaller than control mice at 3, 4, and 5 weeks of age, respectively. Figure 1B shows a smaller body size in the mutant mice at 4 weeks compared to the control mice. MicroCT analysis of the 4-week-old mutants revealed shorter and thinner tibias than that of the control (Fig. 1C). Similarly, there was a marked decrease in bone mineral density at 4 weeks of age in the SmoM2;OCN-Cre mutants compared to the controls (Fig. 1D). Constitutive SmoM2 expression also caused an impaired formation of the front teeth (data not shown). Most of the mutant mice died at 39 days of age (data not shown). Further, there was no gender difference in the mutant mice for the postnatal development (data not shown).
Fig. 1.
Postnatal retardation of growth and bone formation in SmoM2;OCN-Cre mutant mice. (A) Body weight of SmoM2 mutant and control mice was measured at the indicated weeks of age. The data indicate the mean ± SD (n = 5 in each group). (B) A representative image showing gross appearance of the mutant and control mice at 4 weeks of age. (C) MicroCT images of tibias from 4-week-old mice. (D) Bone mineral density was determined from the tibias of the two mice groups and is represented as the mean ± SD (n = 5 in each group). *P < 0.05 and ***P. < 0.001 indicate significant differences between the control and mutant mice.
Sustained SmoM2 induction changes dynamically the expression of bone-specific genes depending on the age
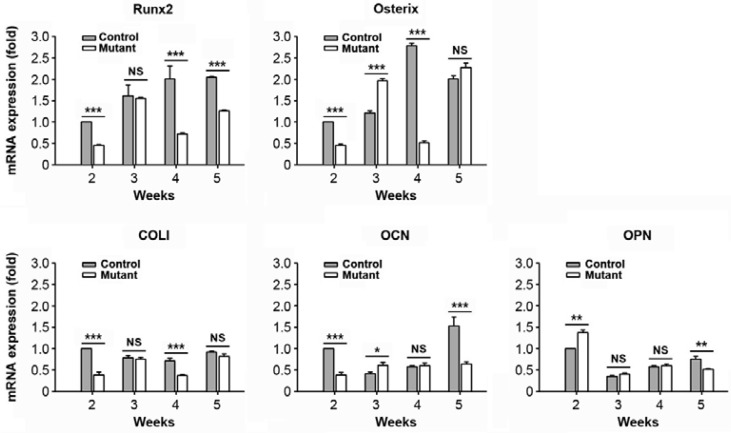
Real time RT-PCR was conducted to evaluate mRNA expression of bone-related factors in primary calvarial cells isolated from mutant and control mice at various ages (2, 3, 4, and 5 weeks). Calvarial cells from the control mice showed a gradual increase in the mRNA expression of Runx2, which was higher than the expression levels seen in the SmoM2;OCN-Cre mutant mice during the same experimental period (Fig. 2). While the genes specific for osterix, COL I, and OCN proteins showed dynamic expression patterns according to the age and type of mice, their expression levels at 2 weeks of age were greater in the control than in mutant mice. In contrast, mRNA expression levels of OPN at 2 weeks of age were higher in the mutant than in control mice.
Fig. 2.
Lower mRNA expression levels of several bone-specific marker proteins in the SmoM2 mutant relative to the control. Primary calvarial cells were isolated from SmoM2;OCN-Cre mutant or controls at the indicated postnatal ages (2–5 weeks), and the expression patterns of bone-related genes were determined by real time RT-PCR. The results show the mean ± SD from three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 vs. the experiments. NS, not significant.
ALP expression pattern differs according to postnatal age
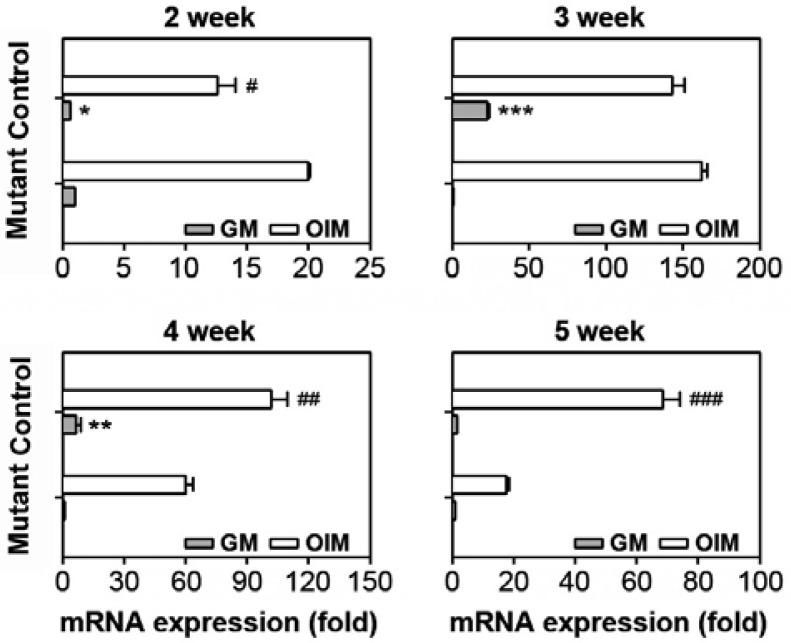
In order to investigate the potential of calvarial cells to differentiate into osteoblasts, the cells were incubated with and without DAG. After 10 days of differentiation, ALP mRNA levels were analyzed by real time RT-PCR. Control cultures from 2-weekold mice showed higher ALP mRNA expression levels than the mutant cultures in both the presence and absence of DAG (Fig. 3A). However, ALP mRNA levels in cultures from 3- or 4-weekold mice were higher in the mutant relative to the control cultures. Further, the mutant cultures showed higher ALP expression in response to DAG than in the control when the cells from 4- or 5-week-old mice were analyzed. This was in part consistent with the results from the ALP activity assay, where the activity was higher in the control relative to the mutant mice when the cells were isolated from 2- and 3-week-old mice (data not shown).
Fig. 3.
Different ALP mRNA expression levels according to week of age. Primary calvarial cells were isolated from the SmoM2;OCNCre mutant or the control at various postnatal stages (2–5 weeks). The cells were incubated in the presence or absence of DAG followed by an ALP mRNA expression assay. *P < 0.05, **P < 0.01, and ***P < 0.001 vs. the control cultures in GM. #P < 0.05, ##P < 0.01, and ###P < 0.001 vs. the control cultures in OIM.
Control mice show greater mineralization activity than mutant mice at 2 weeks of age
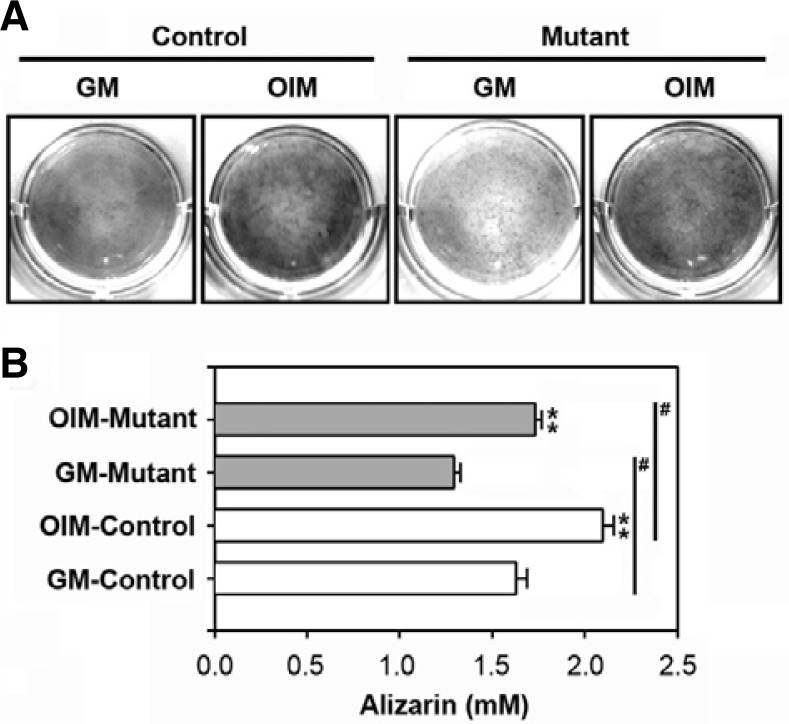
Calvarial cells isolated from 2-week-old mice were incubated for 10 days with and without DAG and subjected to the alizarin red S staining assay to examine the potential of these cells to accumulate intracellular calcium. While incubating the cultures with OIM increased mineralization in both the control and mutant cultures, the control cultures showed greater mineralization both in the presence and absence of DAG (Figs. 4A and 4B). However, there was no difference between the mice groups or between groups treated in the presence or absence of DAG in cultures from 3-, 4-, or 5-week-old mice (data not shown).
Fig. 4.
Different intracellular calcium accumulation levels depending on mouse type. Primary calvarial cells were isolated from the SmoM2;OCN-Cre mutant or the control at 2 weeks of age. (A) The cells were incubated in the presence or absence of DAG and then processed for alizarin red S staining. A representative result from triplicate experiments is shown. (B) Alizarin red S concentrations (mM) were determined using a standard curve of the agent. **P < 0.05 vs. the cultures in GM. #P < 0.05 vs. the experiments.
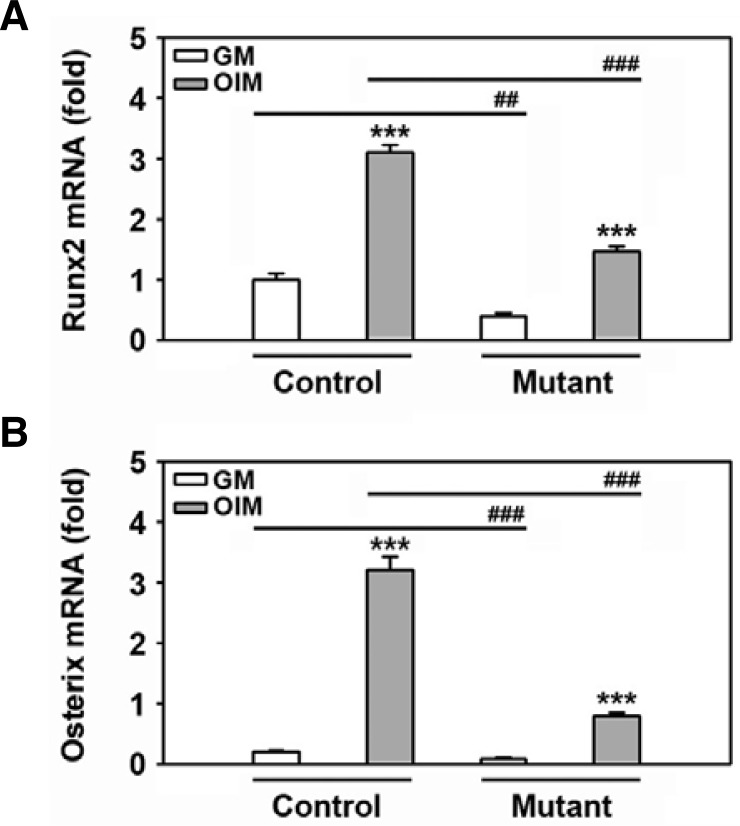
The expression of bone-specific transcription factors in response to DAG is higher in the control than the mutant mice
In order to explore whether there is a correlation between the expression pattern of bone-related factors and mineralization, the cells of 2-week-old mice were incubated with DAG for 24 h prior to RNA extraction. Real time RT-PCR revealed that Runx2 expression levels were higher in the control relative to the mutant mice (Fig. 5A). The DAG-mediated increase in Runx2 mRNA expression was also significantly greater in the control than in the mutant cells. These data coincide with the osterix expression pattern analyzed by real time RT-PCR, suggesting that expression of the transcription factors correlates with the capacity of calvarial cells to differentiate into osteoblasts (Fig. 5B). However, this tendency was not found when cells isolated from 3- or 5-week-old mice were examined (data not shown). Further, there was no difference found in the potential of these cells to proliferate when 2-week-old control or mutant cultures were explored (Fig. 6). This suggests that the retardation of postnatal development in the SmoM2;OCN-Cre mutant correlates more with impaired bone differentiation and formation rather than cell proliferation.
Fig. 5.
Different Runx2 and osterix mRNA expression levels in response to DAG between control and mutant mice. Primary calvarial cells were isolated from the SmoM2;OCN-Cre mutant or the control at 2 weeks of age. The cells were incubated in GM or OIM for 24 h and then processed for real time RT-PCR. The results show the mean ± SD from three independent experiments. ##P < 0.01 and ###P < 0.001 vs. the experiments.
Fig. 6.
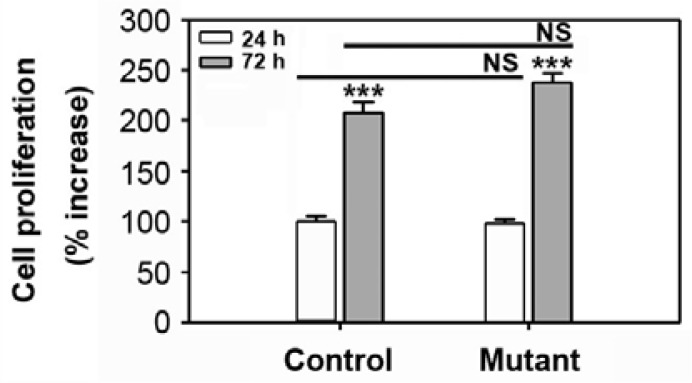
Proliferation rate of cultures from control and mutant mice. Primary calvarial cells were isolated from the SmoM2;OCN-Cre mutant or the control at 2 weeks of age. The cells were incubated in GM. Proliferation levels were measured at 24 h and 72 h of incubation. ***P < 0.01 vs. the cultures of 24 h incubation. NS, not significant.
DISCUSSION
Hedgehog signaling is known to regulate a variety of events such as patterning, proliferation, cell fate, and stem cell self-renewal in early and adult stages. Smo, like G-protein coupled receptors, plays a pivotal role in mediating hedgehog signaling into the nucleus by activating Gli family transcription factors (Peukert and Miller-Moslin, 2010). Gli1 itself may induce early osteoblast differentiation in a Runx2-independent manner and play key roles with Gli2 and Gli3 in osteogenesis (Hojo et al., 2012). Shh signaling is also involved in tumorigenesis in several cancers, particularly in breast cancers (Moraes et al., 2007). Because of this, considerable interest in elucidating the cellular and molecular mechanisms involved in Smo-mediated signaling has recently emerged. The early lethality of Shh or Smo mutants made it difficult to examine the roles of Shh or Smo signaling in postnatal tooth development. For example, null embryos specific for Shh (Chiang et al., 1996) or Smo (Zhang et al., 2001) died at birth or around E9, respectively. Both Shh (Dassule et al., 2000) and Smo conditional knockout mice (Gritli-Linde et al., 2002) died within 1 day after birth.
The construction of a SmoM2 mutant led to great advantages for the functional analysis of Smo signaling in postnatal development and to understand the role of Smo-mediated signaling. Jeong et al. (2004) demonstrated that hedgehog signaling in cranial neural crest cells is necessary for the formation of most of the head skeleton using SmoM2 mutant mice. Activation of the hedgehog pathway in Wnt1-Cre;R26SmoM2 mice also caused a mild hyperplasia of the facial processes at E10.5, which was contradictory to data resulting from the loss of hedgehog signaling. This indicated that Smo activation leads to altered proliferation and differentiation. It was also reported that the frequency of regenerative stem cells in MMTV-SmoM2 mutant epithelia is decreased compared to the wild-type (Visbal et al., 2011). Here we demonstrated that a SmoM2;OCN-Cre mutant with constitutive Smo expression represses skeletal bone development and is involved in the retarded growth phenotype. Our present findings support a negative role of sustained Smo signaling on postnatal bone development, which is similar to the finding showing that hedgehog signaling is downregulated during osteoblast differentiation, while signal activation suppresses osteoblastic differentiation in both human adipose-derived cells and bone marrow stromal cells (Plaisant et al., 2009).
The current findings reveal that SmoM2 mutants exhibit lower mRNA expression of genes encoding Runx2, osterix, COL I, and OCN proteins relative to the control at 2 weeks of age. Although there were dynamic expression patterns of osterix and OPN mRNAs at early ages, it was likely that constitutively expressed SmoM2 repressed the expression of bone-related specific factors during the experimental periods. The mutant also showed less mineralization at early ages compared to that of the control in in vitro studies. These results suggested that the differed postnatal development between the mutant and control mice was associated with the difference in the expression of bone-formation regulatory proteins.
Among various regulatory factors, Runx2 is an essential transcription factor for osteoblast differentiation and bone formation (Deng et al., 2008; Franceschi et al., 2007). Runx2 is considered to induce chondrocyte maturation and enhance chondrocyte proliferation through the direct induction of Ihh expression in the growth plate (Shimoyama et al., 2007). These findings, along with our present data, indicate a close association of reduced Runx2 expression with lowered osteogenic differentiation and mineralization in vitro as well as retarded postnatal development in vivo in the SmoM2;OCN-Cre mutant. Further, Ptch1+/− cells showed accelerated osteoblast differentiation, enhanced responsiveness to the Runx2, and reduced generation of the repressor Gli form, suggesting a relationship between hedgehog signaling and Runx2 in postnatal development (Ohba et al., 2008). However, a previous study showed an opposite finding demonstrating that Shh-expressing prostate cancer cells can directly and specifically induce differentiation in preosteoblasts via a Gli1-dependent mechanism that does not require transcriptional upregulation of Runx2 (Zunich et al., 2009). Additional experiments will be needed to clarify why the constitutively activated Smo signaling represses postnatal bone development. In addition, understanding the exact role of Runx2 in Smo-mediated signaling, especially in bone metabolism, will be the main goal of our future studies.
In addition to Runx2, osterix is another osteogenic transcription factor essential for bone formation and osteoblast differentiation (Nakashima et al., 2002; Qiao et al., 2011). Our previous study reported that bone-specific transcription factors such as Runx2 and osterix have important roles in LiCl-mediated osteoblastic differentiation (Heo et al., 2010). It is known that osterix acts downstream of Runx2 (Franceschi et al., 2007). Considerable evidence has revealed that Wnt up-regulates the expression of both Runx2 and osterix (Bennett, 2005; Rodda and McMahon, 2006). Consequently, it is likely that suppression of osterix is associated with a reduction in postnatal development in mutant mice. By contrast, it was recently suggested that osterix and Runx2 inhibit osteoblast differentiation at a late stage in an independent manner of Runx2 and osterix, respectively, where osterix positively regulates its own promoter (Yoshida et al., 2012). In this study, we showed that unlike Runx2, the mRNA expression of osterix in mutant mice was quite dynamic and thus its level was higher in the mutant than control mice at 2 weeks of age. Additional experiments to clarify the relationship between Runx2 and osterix will be needed.
In conclusion, the current findings suggest that Smo-mediated signaling is closely involved in postnatal bone development, where constitutively activated Smo expression in SmoM2; OCN-Cre mutant mice resulted in the reduction of body weight and skeletal bone formation. We also reveal that primary osteoblast cultures from mutant mice have less potential in both osteoblastic differentiation and mineralization than in the control, although the results were different according to mouse age. Collectively, it is likely that reduced activation of Runx2 and osterix is related to the repressed postnatal development of SmoM2 mice. Additional experiments will be needed in order to clarify a relationship between these transcriptional factors and other bone-specific regulatory factors in the mutant mice.
Acknowledgments
This work was supported by the Mid-career Researcher Program through National Research Foundation grant by the MEST, Republic of Korea (2009-0084511). We thank Jong-Suk Han for technical assistance.
REFERENCES
- Baker N.E. Patterning signals and proliferation in Drosophila imaginal discs. Curr. Opin. Genet. Dev. 2007;17:287–293. doi: 10.1016/j.gde.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Bennett C.N., Ouyang H., Ma Y.L., Zeng Q., Gerin I., Sousa K.M., Lane T.F., Krishnan V., Hankenson K.D., MacDougald O.A. Wnt10b increases postnatal bone formation by enhancing osteoblast differentiation. J. Bone Miner. Res. 2007;22:1924–1932. doi: 10.1359/jbmr.070810. [DOI] [PubMed] [Google Scholar]
- Briscoe J., Thérond P. Hedgehog signaling: from the Drosophila cuticle to anti-cancer drugs. Dev Cell. 2005;8:143–151. doi: 10.1016/j.devcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Chiang C., Litingtung Y., Lee E., Young K.E., Corden J.L., Westphal H., Beachy P.A. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Dassule H.R., Lewis P., Bei M., Maas R., McMahon A.P. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- Deng Z.L., Sharff K.A., Tang N., Song W.X., Luo J., Luo X., Chen J., Bennett E., Reid R., Manning D., et al. Regulation of osteogenic differentiation during skeletal development. Front. Biosci. 2008;13:2001–2021. doi: 10.2741/2819. [DOI] [PubMed] [Google Scholar]
- Franceschi R.T., Ge C., Xiao G., Roca H., Jiang D. Transcriptional regulation of osteoblasts. Ann. NY Acad. Sci. 2007;1116:196–207. doi: 10.1196/annals.1402.081. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A., Bei M., Maas R., Zhang X.M., Linde A., McMahon A.P. Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development. 2002;129:5323–5337. doi: 10.1242/dev.00100. [DOI] [PubMed] [Google Scholar]
- Gupta S., Takebe N., Lorusso P. Targeting the Hedgehog pathway in cancer. Ther. Adv. Med. Oncol. 2010;2:237–250. doi: 10.1177/1758834010366430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton M.J., Tu X., Cook J., Hu H., Long F. Ihh controls cartilage development by antagonizing Gli3, but requires additional effectors to regulate osteoblast and vascular development. Development. 2005;132:4339–4351. doi: 10.1242/dev.02025. [DOI] [PubMed] [Google Scholar]
- Heo J.S., Lee S.Y., Lee J.C. Wnt/β-catenin signaling enhances osteoblastogenic differentiation from human periodontal ligament fibroblasts. Mol Cells. 2010;30:449–454. doi: 10.1007/s10059-010-0139-3. [DOI] [PubMed] [Google Scholar]
- Hogan B., Beddington R., Costantini F., Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2nd eds. Cold Spring Harbor New York: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Hojo H., Ohba S., Yano F., Saito T., Ikeda T., Nakajima K., Komiyama Y., Nakagata N., Suzuki K., Takato T., et al. Gli1 protein participates in Hedgehog-mediated specification of osteoblast lineage during endochondral ossification. J. Biol. Chem. 2012;287:17860–17869. doi: 10.1074/jbc.M112.347716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Mao J., Tenzen T., Kottmann A.H., McMahon A.P. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordial. Genes Dev. 2004;18:937–951. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T. Signaling networks in RUNX2-dependent bone development. J. Cell. Biochem. 2011;112:750–755. doi: 10.1002/jcb.22994. [DOI] [PubMed] [Google Scholar]
- Mak K.K., Bi Y., Wan C., Chuang P.T., Clemens T., Young M., Yang Y. Hedgehog signaling in mature osteoblasts regulates bone formation and resorption by controlling PTHrP and RANKL expression. Dev Cell. 2008;14:674–688. doi: 10.1016/j.devcel.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Mimeault M., Batra S.K. Frequent deregulations in the hedgehog signaling network and cross-talks with the epidermal growth factor receptor pathway involved in cancer progression and targeted therapies. Pharmacol. Rev. 2010;62:497–524. doi: 10.1124/pr.109.002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes R.C., Zhang X., Harrington N., Fung J.Y., Wu M.F., Hilsenbeck S.G., Allred D.C., Lewis M.T. Constitutive activation of smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development. 2007;134:1231–1242. doi: 10.1242/dev.02797. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J.M., Behringer R.R., de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Ohba S., Kawaguchi H., Kugimiya F., Ogasawara T., Kawamura N., Saito T., Ikeda T., Fujii K., Miyajima T., Kuramochi A., et al. Patched1 haploinsufficiency increases adult bone mass and modulates Gli3 repressor activity. Dev Cell. 2008;14:689–699. doi: 10.1016/j.devcel.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Peukert S., Miller-Moslin K. Small-molecule inhibitors of the hedgehog signaling pathway as cancer therapeutics. Chem. Med. Chem. 2010;5:500–512. doi: 10.1002/cmdc.201000011. [DOI] [PubMed] [Google Scholar]
- Plaisant M., Fontaine C., Cousin W., Rochet N., Dani C., Peraldi P. Activation of hedgehog signaling inhibits osteoblast differentiation of human mesenchymal stem cells. Stem Cells. 2009;27:703–713. doi: 10.1634/stemcells.2008-0888. [DOI] [PubMed] [Google Scholar]
- Qiao L.J., Kang K.L., Heo J.S. Simvastatin promotes osteogenic differentiation of mouse embryonic stem cells via canonical Wnt/β-catenin signaling. Mol Cells. 2011;32:437–444. doi: 10.1007/s10059-011-0107-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda S.J., McMahon A.P. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- Shimoyama A., Wada M., Ikeda F., Hata K., Matsubara T., Nifuji A., Noda M., Amano K., Yamaguchi A., Nishimura R., et al. Ihh/Gli2 signaling promotes osteoblast differentiation by regulating Runx2 expression and function. Mol. Biol Cell. 2007;18:2411–2418. doi: 10.1091/mbc.E06-08-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stanton B.Z., Peng L.F. Small-molecule modulators of the Sonic Hedgehog signaling pathway. Mol. Biosyst. 2010;6:44–54. doi: 10.1039/b910196a. [DOI] [PubMed] [Google Scholar]
- Tan X., Weng T., Zhang J., Wang J., Li W., Wan H., Lan Y., Cheng X., Hou N., Liu H., et al. Smad4 is required for maintaining normal murine postnatal bone homeostasis. J. Cell Sci. 2007;120:2162–2170. doi: 10.1242/jcs.03466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visbal A.P., LaMarca H.L., Villanueva H., Toneff M.J., Li Y., Rosen J.M., Lewis M.T. Altered differentiation and paracrine stimulation of mammary epithelial cell proliferation by conditionally activated Smoothened. Dev. Biol. 2011;352:116–127. doi: 10.1016/j.ydbio.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C.A., Komori H., Maruyama Z., Miyazaki T., Kawasaki K., Furuichi T., Fukuyama R., Mori M., Yamana K., Nakamura K., et al. SP7 inhibits osteoblast differentiation at a late stage in mice. PLoS One. 2012;7:e32364. doi: 10.1371/journal.pone.0032364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.M., Ramalho-Santos M., McMahon A.P. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell. 2001;106:781–792. [PubMed] [Google Scholar]
- Zunich S.M., Douglas T., Valdovinos M., Chang T., Bushman W., Walterhouse D., Iannaccone P., Lamm M.L. Paracrine sonic hedgehog signalling by prostate cancer cells induces osteoblast differentiation. Mol Cancer. 2009;8:12. doi: 10.1186/1476-4598-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]