Abstract
Tiam-1 has been implicated in the development of the central nervous system. However, the in vivo function of Tiam-1 has not been fully determined in the developing mouse brain. In this study, we generated Tiam-1 knockout mice using a Tiam-1 gene-trapped embryonic stem cell line. Insertion of a gene trap vector into a genomic site downstream of exon 5 resulted in a mutant allele encoding a truncated protein fused with the β-geo LacZ gene. Primary mouse embryonic fibroblasts lacking Tiam-1 revealed a significant decrease in Rac activity and cell proliferation. In addition, whole-mount embryonic LacZ expression analysis demonstrated that Tiam-1 is specifically expressed in regions of the developing brain, such as the caudal telencephalon and rostral diencephalon. More importantly, mouse embryos deficient in Tiam-1 gene expression displayed a severe defect in embryonic brain development, including neural tube closure defects or a dramatic decrease in brain size. These findings suggest that embryonic Tiam-1 expression plays a critical role during early brain development in mice.
Keywords: early brain development, Rac, Tiam-1
INTRODUCTION
Tiam-1 was identified as an invasion-inducing gene that encodes a protein with homology to guanosine diphosphate (GDP)-guanosine triphosphate (GTP) exchange for Rho-like GTPases (Habets et al., 1994). Altered Tiam-1 expression (inactive mutation or overexpression) has been shown to be associated with a variety of human cancer types (Engers et al., 2001; Michiels et al., 1995; Minard et al., 2004). It was also shown that Tiam1-Rac signaling is critically involved in tumor initiation in mice. For example, Tiam-1 knockout mice had impaired carcinogen-induced skin carcinoma formation (Malliri et al., 2002). However, it was also suggested that Tiam-1 may act as a metastasis suppressor, since loss of Tiam-1 resulted in increased malignancy and invasion for the tumors that managed to form despite its loss of function.
Tiam-1 has also been implicated in the development of the nervous system (Iden and Collard, 2008; Mertens et al., 2006). For example, neurons overexpressing Tiam-1 extended axon-like neurites, whereas inhibition of Tiam-1 prevented axon formation, suggesting its role in neuronal polarization (Kunda et al., 2001). In addition, inhibition of Tiam-1 in the developing brain resulted in inhibition of radial migration of neurons, suggesting its additional role in cortical neuron migration (Kawauchi et al., 2003). Various functions of Tiam-1 seem to be dependent on its capacity to interact with various cell surface receptors. For example, Ephrin-B1, EphA2, and TrkB were shown to induce Tiam1-mediated neurite outgrowth (Miyamoto et al., 2006; Tanaka et al., 2004). It was also shown that N-Methyl-D-aspartate (NMDA) receptors or EphB receptors interacted with Tiam-1 to control dendritic spine development (Tolias et al., 2005; 2007). Similarly, partitioning-defective gene 3 (Par-3) balances with Tiam-1 to modulate the proper level of Rac-GTP and allows dendritic spine morphogenesis (Nishimura et al., 2005; Zhang and Macara, 2006). More recent studies revealed that Tiam-1 plays a critical role during the early mitotic phase. For example, Tiam1-Rac signaling retards centrosome separation to modulate proper separation, which is essential for the subsequent chromosome congression and segregation during mitosis (Woodcock et al., 2010). In summary, it is likely that Tiam-1 plays a central role in processes involving cell polarity or cell proliferation during development of the central nervous system.
In this study, we found that primary mouse embryonic fibroblasts (MEFs) lacking Tiam-1 gene expression exhibited a significant reduction in Rac activity and cell proliferation. In addition, lack of Tiam-1 expression caused abnormal brain development, such as neural tube closure defect, suggesting that embryonic Tiam-1 expression plays a critical role during early brain development.
MATERIALS AND METHODS
Characterization of gene trap vector-insertion site, generation of Tiam-1 knockout mice, and polymerase chain reaction (PCR) genotyping
Embryonic stem (ES) cells harboring a gene trap vector (pGT0lxf) in the Tiam-1 locus (cell line ID CSJ260) were purchased from the Mutant Mouse Regional Resource Center. To determine the genomic insertion site of the gene trap vector into the Tiam-1 gene, forward primers were selected so that they were positioned every 2 kb apart between exon 5 and exon 6, and each forward primer was combined with a reverse primer from the gene trap vector to perform long-range PCR. The vector insertion site was successfully amplified from ES cell genomic DNA as an approximately 2.3-kb fragment, and the primers for this PCR product were as follows: forward, 5′-CAGAACCACCCTGTACTGTGAAG-3′; and reverse, 5′-GGTC ACAAGGTTCATATGGTGCC-3′. The PCR product was gel extracted using Wizard SV Gel and PCR Clean-Up System (Promega), and was used for mapping of restriction enzyme cleavage sites of the PCR fragment or DNA sequencing analysis in order to assess the insertion site.
The ES cells were then injected into C57BL/6 blastocyst to generate chimeric mice, and germline transmission was determined by mating male chimeric mice with wild-type 129/SvJ females. ES cell culture and blastocyst injections were performed essentially as described (Hogan, 1994). The presence of the transgene in the F1 agouti pups were analyzed by PCR analysis of tail genomic DNA. PCR analysis for genotyping was performed using the following primers: forward, 5′-ATTGT TGAAAGCTGCATTTCCTA-3′; wild-type reverse, 5′-GTAAGC TCTGCCACACACTCTTC-3′; and mutant reverse, 5′-GGTC ACAAGGTTCATATGGTGCC-3′. These primers amplified 160-bp and 250-bp bands for the mutant and wild-type allele, respectively.
MEFs, Rac activity, and cell proliferation assay
MEFs were isolated from E13.5 mouse embryos as previously described (Kim et al., 2010). In brief, embryos were separated, minced, and incubated in 0.25% trypsin EDTA at 37°C for 15 min. Cells were resuspended in Dulbecco’s modified Eagle’s medium (Hyclone), supplemented with 7% heat-inactivated fetal bovine serum (Hyclone) and 1% antibiotics (penicillin-streptomycin; Invitrogen). Harvested cells were plated onto 100-mm plates and maintained in a humidified incubator at 37°C in the presence of 5% CO2.
For the Rac activity assay, MEF cell lysates were prepared and incubated with Pak-1 binding domain (PBD)-glutathione S-transferase (GST) beads (Upstate Biotechnology), as described by the manufacture’s protocol, to pull down GTP-bound Rac. Activated or total Rac was detected by immunoblot using anti-Rac antibodies as previously described (Yoo et al., 2010).
Cell proliferation was assessed by MTT assay (Sigma) using third-passage MEF cells. The serum-starved 5 × 104 MEF cells were plated in 500-μl, 24-well flat bottom plates and then cultured for the indicated hours. At each time point, 100 μl of 5 mg/ml MTT solution in phosphate-buffered saline (PBS) were added to each well, and the plates were incubated at 37°C for an additional 4 h. Five hundred microliters of MTT solvent (4 mM HCl, 0.1% NP-40 in isopropanol) was then added to each well to dissolve the formazan for 15 min, and absorbance was read at 600 nm using a spectrophotometer (Eppendorf).
Whole-mount X-gal staining and sections
Embryos to be stained were dissected in PBS, fixed in 0.2% glutaraldehyde, and subjected to the washing and staining procedure described previously (Shim et al., 2007). After X-gal staining, embryos to be sectioned were fixed in 4% paraformaldehyde at 4°C, dehydrated in ethanol, and embedded in paraffin. Coronal sections were cut at 5–10 μm and counterstained with eosin.
RESULTS
Generation of Tiam-1 null mutant mice using ES cell line CSJ260
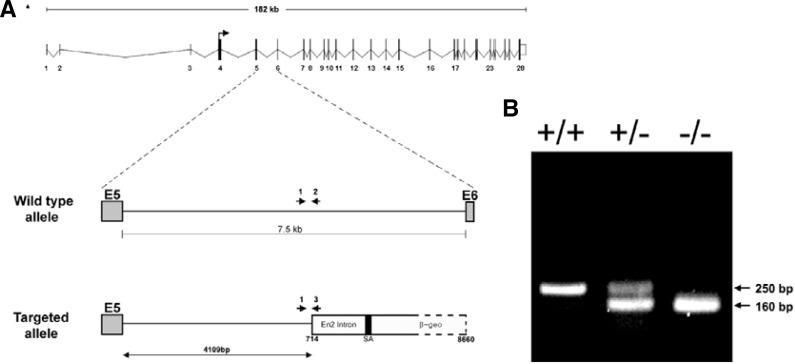
The gene trap database was searched for ES cell lines containing a targeted insertion within the Tiam-1 gene, and it was found that the CSJ260 cell line contained a gene trap vector inserted proximately to exon 5 of the 28-exon Tiam-1 gene (Fig. 1A). The CSJ260 ES cell line was further analyzed to identify the location of the gene trap vector insertion in a region spanning the full 7.5-kb intron between exon 5 and exon 6, revealing that the gene trap vector was specifically inserted into a site 4109 bp downstream of exon 5 (Fig. 1A). Chimeric mice were generated by injection of CSJ260 ES cells into C57BL/6 blastocysts, and their germline transmission was demonstrated by PCR amplification of the gene trap vector sequence in tail DNA (Fig. 1B).
Fig. 1.
Characterization of a gene-trapped Tiam-1 allele. (A) Schematic diagram representing Tiam-1 genomic locus (top) is shown together with the vector-integration site within the 5th intron of the mouse Tiam-1 gene in the ES clone CSJ260. PCR followed by sequencing analysis indicated that a gene trap vector was inserted into a genomic site 4100 bp downstream of exon 5. (B) A typical genotyping analysis showing the 250-bp PCR product for the wild-type allele and the 160-bp product for the mutant allele.
Rac activity and cell proliferation were reduced in Tiam-1-deficient MEF cells
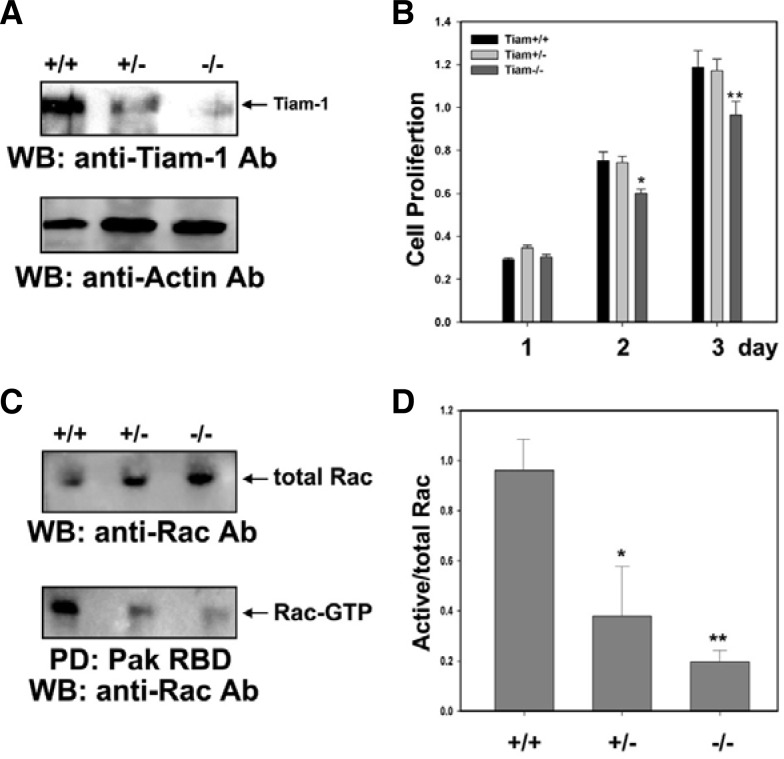
Heterozygous matings were set up to generate E13.5 littermate embryos, which were genotyped (Fig. 1B) and used for production of MEFs. Western blot analysis was performed to analyze the expression level of Tiam-1 protein in each MEF cell line, revealing that the level of Tiam-1 was significantly reduced in the heterozygous MEF cells and barely detectable in the Tiam-1 null mutant MEF cells (Fig. 2A). We further performed the colorimetric MTT assay to measure proliferation of each MEF cell line and consistently found that cell proliferation was significantly reduced in Tiam-1 null mutant MEF cell lines as compared with either wild-type or heterozygous MEF cell lines (Fig. 2B). Since Tiam-1 has guanine-nucleotide exchange activity toward Rac, we further examined whether the down-regulation of Tiam-1 affects the level of GTP-bound Rac. As expected, the PBD of PAK pull-down assay revealed that the level of GTP-bound Rac was reduced approximately 2.5-fold and 5-fold in heterozygous and homozygous MEF cells, respectively (Figs. 2C and 2D). Taken together, these results strongly suggest that downregulation of Tiam-1 decreases Rac signaling, thereby causing a partial impairment of cell proliferation.
Fig. 2.
Analysis of Tiam-1 expression in primary MEF cells. (A) MEF cells were prepared from 13.5-day embryos with the indicated genotypes, and the expression level of Tiam-1 was analyzed by Western blot (WB) using an anti-Tiam1 antibody. (B) MEFs derived from each embryo were cultured for the indicated time and mixed with MTT reagent to measure viable cells. Data represent the means ± SE from three independent experiments. *, P < 0.01; **, P < 0.005. (C) Activity of Rac was measured by pull-down assay in each MEF cell. The GTP-bound form of Rac was precipitated by GST-Pak1-RBD and probed with an anti-Rac1 antibody. Total levels of Rac were visualized by WB using an anti-Rac1 antibody. (D) The ratio of GTP-Rac and total level of Rac was quantified and described as a bar graph with SE (n = 3). *, P < 0.01; **, P < 0.001.
Mice homozygous for the CSJ260 insertion display an exencephalic or anencephalic phenotype
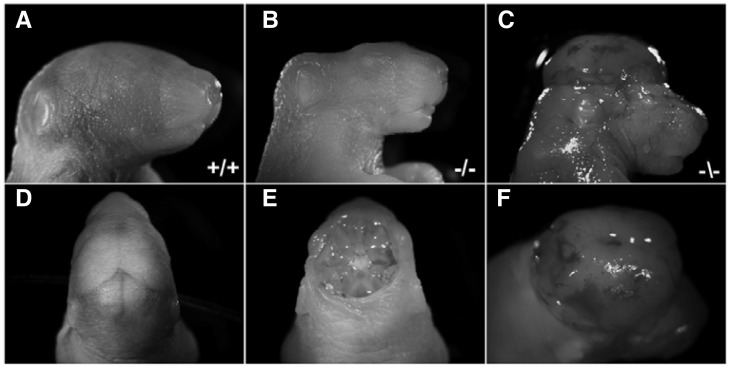
Heterozygous matings were performed to generate homozygous progeny, and we managed to recover a few from over 50 null mutant pups. Importantly, most of the homozygous null mutant pups displayed severe anencephaly wherein the exposed brain tissue was completely degenerated (Figs. 3B and 3E). In addition, some of null mutant pups showed exencephaly wherein the brain was located outside of the skull (Figs. 3C and 3F). These results suggest that Tiam-1 may be critically involved in the process of neural tube closure during early brain development.
Fig. 3.
Neonatal mice deficient for Tiam-1 expression display severe defects in brain development. (A, D) Lateral and dorsal view of wild-type littermate showing normal brain morphology, respectively. (B, E) Lateral and dorsal view of Tiam-1 null mutant littermate is shown, respectively. Note that the brain shows a typical anencephalic phenotype since the entire brain is missing and the base of skull is exposed. (C, F) Lateral and dorsal view of Tiam-1 null mutant littermate is shown, revealing an exencephalic phenotype. Note that the brain is located outside of the skull.
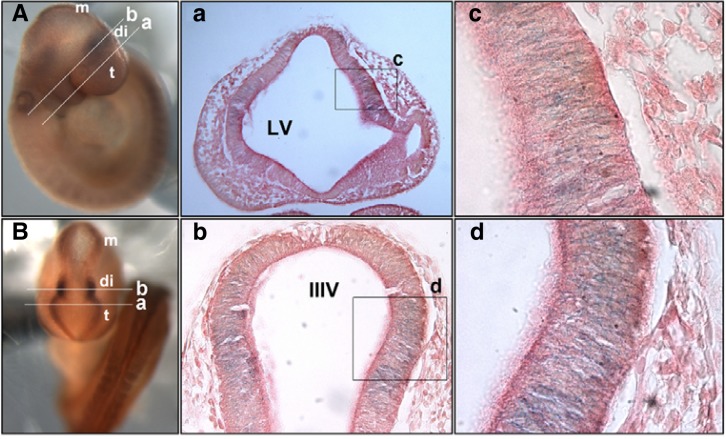
Both heterozygous and homozygous embryos express a fusion protein containing the translated regions upstream of the insertion site fused to the gene trap vector β-geo LacZ. Therefore, we performed whole-mount X-gal staining to examine expression of the fusion protein in homozygous embryos. LacZ expression was prominently detected in the forebrain of E9.5 embryos, in particular the caudal telencephalon and rostral diencephalon (Figs. 4A and 4B). High magnification of the coronal sections revealed that LacZ expression was restricted to the neuroepithelial cells present in the lateral region of the forebrain (Figs. 4a–4d). These results strongly suggest that Tiam-1 is expressed in a subset of neuroepithelial cells and its expression may be critical for the early development of the forebrain.
Fig. 4.
Analysis of LacZ expression in the Tiam-1 null mutant embryo. Tiam-1 mutant embryos at E9.5 were analyzed for LacZ activity by whole-mount X-gal staining. (A, B) Lateral and dorsal view of the embryo, respectively. Note that LacZ expression is restricted to the region corresponding to the caudal telencephalon and rostral diencephalon. m, mesencephalon; di, diencephalon; t, telencephalon. (a, b) Coronal sections were prepared from the caudal telencephalon and rostral diencephalon as marked in (A, B), respectively. Each section was subjected to eosin counterstaining to reveal X-gal-stained neuroepithelial cells. (b) Coronal section of the rostral diencephalon is shown [line b marked in (A, B)]. (c, d) Higher magnification of the boxes indicated in panels a and b, respectively. LV, lateral ventricle; IIIV, third ventricle.
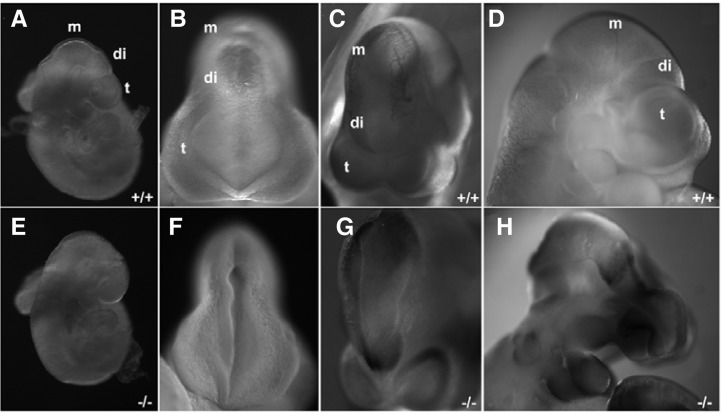
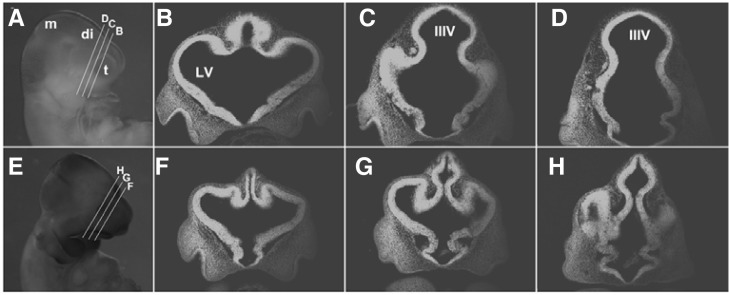
Consistent with the anencephalic or exencephalic phenotype observed in Tiam-1 null mutant neonatal pups, approximately 20% of the homozygous mutant embryos displayed a neural tube closure defect (Figs. 5E–5G), and an additional 10% of the null mutant embryos showed a collapsed forebrain phenotype with neural tube closure defect (Fig. 5H). We further selected littermate embryos with normal brain morphology and analyzed their anatomical structures in the coronal sections of the forebrain. It was consistently observed that the rostral region of the diencephalon but also its ventricular region were much smaller in the homozygous null mutant embryos (Figs. 6E–6H). Therefore, it is likely that a homozygous gene trap vector insertion in the Tiam-1 gene resulted in severe disruption of Tiam-1 gene expression, a possible cause for the defective growth of the neuroepithelial cells present in the lateral region of the forebrain.
Fig. 5.
Mouse embryos deficient for Tiam-1 expression display severe defects in early brain development. (A, E) Littermate embryos at E9.5 are shown with their genotypes. (B, F) Dorsal views of the embryos shown in (A, E), respectively. Note that the forebrain of the knockout embryo is still open as compared with the wild-type littermate. (C, G) Littermate embryos at E10.5 are shown with their genotypes. The knockout embryo reveals that its dien-mesence-phalon is not closed as compared with the wild-type littermate. (D, H) Littermate embryos at E10.5 are shown with their genotypes. The knockout embryo shows that the diencephalic vesicle collapsed during the developmental process. m, mesencephalon; di, diencephalon; t, telencephalon.
Fig. 6.
The size of the rostral diencephalon is significantly reduced in mouse embryos deficient for Tiam-1 expression. (A) Wild-type littermate embryos at E10.5 are shown with their coronal sections at different levels of the rostral diencephalon (B–D). (E) Knockout littermate embryos at E10.5 are shown with their coronal sections at different levels of rostral diencephalon (F-H). Note that the diencephalon of the knockout embryo is significantly reduced. m, mesencephalon; di, diencephalon; t, telencephalon.
DISCUSSION
We generated Tiam-1 null mutant mice using a Tiam-1 gene-trapped ES cell line. Mice carrying homozygous insertion downstream of exon 5 of the Tiam-1 gene are mostly inviable due to severe brain malformations, including anencephaly and exencephaly. In addition, an abnormally small diencephalon, neural tube closure defect, or collapsed neural tube was observed among homozygous mutant embryos. This suggests that Tiam-1 expression plays a crucial role during early brain development.
Previous studies showed that mice deficient in the Tiam-1 gene were resistant to Ras-induced skin tumors, but did not report any obvious phenotype for brain development (Malliri et al., 2002). However, a more recent study using a Tiam-1 knockdown approach indicated that Tiam1-Rac signaling is essential for proper centrosome separation followed by subsequent chromosome congression during the mitotic phase (Woodcock et al., 2010). Importantly, Tiam-1-depleted cells were shown to transit slowly through prometaphase and display chromosome congression errors. A similar process and mechanism may be adopted by the neuroepithelial cells expressing Tiam-1 in the embryonic forebrain. Disruption of Tiam-1 gene expression in those neuroepithelial cells may severely affect their cell proliferation, resulting in a significant decrease of the neural stem cell population in the caudal telencephalon and rostral diencephalon, thereby critically affecting subsequent brain development.
Like most gene-trapped ES cell lines, the CSJ260 ES cell has an insertion site in the intronic region. Therefore, we cannot rule out the possibility that alternative splicing takes place in CSJ260 ES-derived homozygous mutant animals, leading to lower levels of wild-type Tiam-1 transcripts and resulting in hypomorphic alleles. Consistent with this possibility, we observed variable expression levels of LacZ among homozygous mutant embryos, and some homozygous mutant embryos barely showed LacZ expression in the forebrain (unpublished observation). Furthermore, the degree of LacZ expression in the forebrain of the homozygous mutant embryos had a good correlation with that of the defective brain development. In this aspect, conditional knockout mice would be more desirable to investigate how Tiam-1 functions in the proliferating neuroepithelial cells during early brain development.
Acknowledgments
This study was supported by a grant from the Korean Health Technology R&D Project, Ministry for Health, Welfare and Family Affairs, Republic of Korea (A100078).
REFERENCES
- Engers R., Springer E., Michiels F., Collard J.G., Gabbert H. E. Rac affects invasion of human renal cell carcinomas by up-regulating tissue inhibitor of metalloproteinases (TIMP)-1 and TIMP-2 expression. J. Biol. Chem. 2001;276:41889–41897. doi: 10.1074/jbc.M105049200. [DOI] [PubMed] [Google Scholar]
- Habets G.G., Scholtes E.H., Zuydgeest D., van der Kammen R.A., Stam J.C., Berns A., Collard J.G. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77:537–549. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- Hogan B. Manipulating the Mouse Embryo : A Laboratory Manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Iden S., Collard J.G. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat. Rev. Mol. Cell Biol. 2008;9:846–859. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]
- Kawauchi T., Chihama K., Nabeshima Y., Hoshino M. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J. 2003;22:4190–4201. doi: 10.1093/emboj/cdg413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee H., Kim Y., Yoo S., Park E., Park S. The SAM domains of Anks family proteins are critically involved in modulating the degradation of EphA receptors. Mol. Cell. Biol. 2010;30:1582–1592. doi: 10.1128/MCB.01605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P., Paglini G., Quiroga S., Kosik K., Caceres A. Evidence for the involvement of Tiam1 in axon formation. J. Neurosci. 2001;21:2361–2372. doi: 10.1523/JNEUROSCI.21-07-02361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malliri A., van der Kammen R.A., Clark K., van der Valk M., Michiels F., Collard J.G. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 2002;417:867–871. doi: 10.1038/nature00848. [DOI] [PubMed] [Google Scholar]
- Mertens A.E., Pegtel D.M., Collard J.G. Tiam1 takes PARt in cell polarity. Trends Cell Biol. 2006;16:308–316. doi: 10.1016/j.tcb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Michiels F., Habets G.G., Stam J.C., van der Kammen R.A., Collard J.G. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995;375:338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- Minard M.E., Kim L.S., Price J.E., Gallick G.E. The role of the guanine nucleotide exchange factor Tiam1 in cellular migration, invasion, adhesion and tumor progression. Breast Cancer Res. Treat. 2004;84:21–32. doi: 10.1023/B:BREA.0000018421.31632.e6. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y., Yamauchi J., Tanoue A., Wu C., Mobley W.C. TrkB binds and tyrosine-phosphorylates Tiam1, leading to activation of Rac1 and induction of changes in cellular morphology. Proc. Natl. Acad. Sci. USA. 2006;103:10444–10449. doi: 10.1073/pnas.0603914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Yamaguchi T., Kato K., Yoshizawa M., Nabeshima Y., Ohno S., Hoshino M., Kaibuchi K. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat. Cell Biol. 2005;7:270–277. doi: 10.1038/ncb1227. [DOI] [PubMed] [Google Scholar]
- Shim S., Kim Y., Shin J., Kim J., Park S. Regulation of EphA8 gene expression by TALE homeobox transcription factors during development of the mesencephalon. Mol. Cell. Biol. 2007;27:1614–1630. doi: 10.1128/MCB.01429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Ohashi R., Nakamura R., Shinmura K., Kamo T., Sakai R., Sugimura H. Tiam1 mediates neurite outgrowth induced by ephrin-B1 and EphA2. EMBO J. 2004;23:1075–1088. doi: 10.1038/sj.emboj.7600128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias K.F., Bikoff J.B., Burette A., Paradis S., Harrar D., Tavazoie S., Weinberg R.J., Greenberg M.E. The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron. 2005;45:525–538. doi: 10.1016/j.neuron.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Tolias K.F., Bikoff J.B., Kane C.G., Tolias C.S., Hu L., Greenberg M.E. The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proc. Natl. Acad. Sci. USA. 2007;104:7265–7270. doi: 10.1073/pnas.0702044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock S.A., Rushton H.J., Castaneda-Saucedo E., Myant K., White G.R., Blyth K., Sansom O.J., Malliri A. Tiam1-Rac signaling counteracts Eg5 during bipolar spindle assembly to facilitate chromosome congression. Curr. Biol. 2010;20:669–675. doi: 10.1016/j.cub.2010.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S., Shin J., Park S. EphA8-ephrinA5 signaling and clathrin-mediated endocytosis is regulated by Tiam-1, a Rac-specific guanine nucleotide exchange factor. Mol. Cells. 2010;29:603–609. doi: 10.1007/s10059-010-0075-2. [DOI] [PubMed] [Google Scholar]
- Zhang H., Macara I.G. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat. Cell Biol. 2006;8:227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]