Abstract
Although linearly distant along mouse chromosome 7 and human chromosome 11, the mammalian β-globin gene is located in close proximity to the upstream locus control region enhancer when it is actively transcribed in the nuclear chromatin environment of erythroid cells. This organization is thought to generate a chromatin loop between the LCR, a powerful enhancer, and active globin genes by extruding intervening regions containing inactive genes. Loop formation in the β-globin locus requires erythroid specific transcriptional activators, co-factors and insulator-related factors. Chromatin structural features such as histone modifications and DNase I hypersensitive site formation as well as nuclear localization are all involved in loop formation in the locus through diverse mechanisms. Current models envision the formation of the loop as a necessary step in globin gene transcription activation, but this has not been definitively established and many questions remain about what is necessary to achieve globin gene transcription activation.
Keywords: β-globin locus, chromatin loops, LCR, transcription
INTRODUCTION
During the last ten years, evidence has been presented for chromatin loop formation between distant transcriptional regulatory elements, such as enhancers, insulators and locus control regions (LCRs) and the genes whose transcription they influence (Kadauke and Blobel, 2009). Experimental evidence supporting this idea is based on the chromosome conformation capture (3C) assay and its successors (Dekker et al., 2002; Ethier et al., 2012). The formation of looped structures is believed to be a critical step for the transcriptional activation of genes but why this is the case is still unclear. Mechanistic studies of chromatin loop formation have primarily addressed the proteins associated with the looping interactions and the chromatin structural features that appear to be involved.
Chromatin loop formation in mammalian nuclei was first reported for the β-globin locus, which has a complex structure and developmental gene activation pattern (Palstra et al., 2003; Tol-huis et al., 2002). This locus is the best studied from this perspective and remains one of the paradigms for understanding regulation of gene transcription by chromatin loop formation. In this review, we would like to present an overview of these studies and discuss the role of proteins involved in loop formation of the β-globin locus (Table 1) and how chromatin structure and nuclear positioning of the locus are involved.
Table 1.
Proteins involved in loop formation of the β-globin locus
| Erythroid specific transcriptional activators | EKLF, GATA-1, NF-E2 |
| Co-factors | NLI/Ldb1, Brg1 |
| Insulator-related proteins | CTCF, Rad21, SMC1, SMC3 |
Chromatin loop formation in the human and mouse β-globin loci
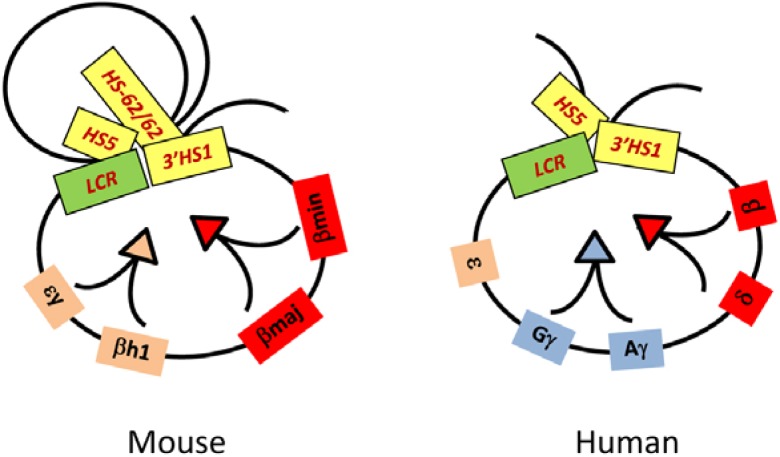
The mammalian β-globin loci each contain a family of genes that are activated at specific times in development and a far upstream powerful enhancer, the LCR, that is characterized by four DNase I hypersensitive sites (HSs) (Fig. 1). Transcription of the globin genes is regulated by the LCR in a tissue and developmental stage specific manner. In the early embryo, the mouse ɛy and βH1 genes and the human ɛ gene are transcribed while in the adult the mouse βmaj and βmin and the human δ and β genes are expressed. In humans, additional duplicated γ genes are expressed specifically at the fetal stage of development. In erythroid cells, the LCR achieves close proximity with the transcribed globin gene but not with the repressed genes, revealing a looped structure (Fig. 2). In the mouse β-globin locus, the transcriptionally active βmaj and βmin globin genes are closely positioned to the LCR HSs (Carter et al., 2002; Palstra et al., 2003; Tolhuis et al., 2002). HSs at both ends of the locus, 5′HS -62/-60, HS5 and 3′HS1, are also in close proximity to the active genes and LCR. However the inactive embryonic expressed globin genes and the intervening region between 5′HS -62/-60 and the LCR are looped out. The structure comprising the active globin gene and HSs has been named an active chromatin hub (Palstra et al., 2003).
Fig. 1.
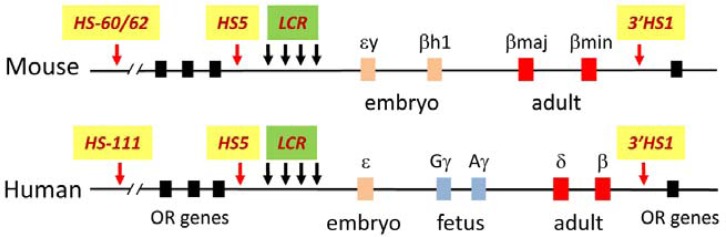
The β-globin loci in mouse and human. The β-globin loci consist of the LCR, the globin genes and insulator elements HS5 and 3′HS1. In the mouse locus, the ɛy and βH1 are transcribed the in embryo, while the βmaj and βmin are transcribed in the adult. In the human locus, the globin genes are transcribed in three developmental stages; ɛ in the embryo, Gγ and Aγ in the fetus, and δ and β in adult.
Fig. 2.
Chromatin loop formation it the β-globin loci. The globin genes are closely positioned with HS-60/62, HS5, LCR and 3′HS1 in the mouse β-globin locus when they are actively transcribed. In the human locus, the fetal genes and adult genes are in close proximity with HS5, LCR, and 3′HS1 when they are transcribed. The close positioning of active genes with HSs makes chromatin loops by extruding regions between them.
Similar chromatin loop structures are formed in the human β-globin locus. In this case it was possible to show that the active fetal γ-globin genes loop to the LCR during the period that are expressed in transgenic mouse erythroid cells and the close interactions switch to the β-globin gene and LCR later in development (Palstra et al., 2003). The active globin genes are also in proximity with 3′HS1 in transgenic mice (Fang et al., 2007; Palstra et al., 2003). However it is not clear whether the region that is equivalent to the mouse 5′HS -62/-60 is closely positioned with the LCR in an erythroid specific and/or globin gene transcription-related manner because this region (5′HS-111) showed modestly elevated interaction frequency with the LCR in both human erythroid K562 cells and non-erythroid GM06990 cells (Dostie et al., 2006). HS3 appears to be critical for LCR-β-globin loop formation as shown in transgenic mice where the HS3 was deleted (Fang et al., 2007; Patrinos et al., 2004). In these studies, deletion of the β-globin promoter did not interrupt loop formation, indicating that regions other than the promoter participate in the long range interactions. The relatively short linear distance between the LCR and the embryonic genes has, so far, precluded demonstration that they loop to the LCR when active, leaving open the possibility of other mechanisms such as enhancer tracking (Dean, 2006; Zhao and Dean, 2004).
Role of erythroid specific transcriptional activators in chromatin loop formation
Erythroid specific transcriptional activators including EKLF, GATA-1 and NF-E2 contribute to a high level of transcription of globin genes (Cantor and Orkin, 2002). Chromatin structural changes such as histone modifications and nucleosome remodeling are dependent on the binding of these transcriptional activators to the LCR HSs and globin gene promoters. These activators also play roles in forming chromatin loops in the β-globin locus.
EKLF occupies the LCR HSs and the promoter of the βmaj globin gene and is required for loop formation between the gene and the LCR in mouse erythroid cells when this gene is actively transcribed (Drissen et al., 2004). However, EKLF is not necessary for formation of a chromatin hub consisting of the 5′HS -62/-60, LCR HS5 and 3′HS. GATA-1 is required for close positioning of LCR HS2 and the βmaj gene as shown by studies using GATA-1 null G1E cells (Vakoc et al., 2005). A similar requirement for GATA-1 for chromatin loop formation was seen in the human β-globin locus transcribing the γ-globin genes in K562 cells (Kim et al., 2011). GATA-1 is required for LCR/γ-globin gene proximity as shown by knock down experiments.
Studies in which NF-E2 was reduced by knock down of the p45 subunit also resulted in loss of LCR/γ-globin gene proximity; however, GATA-1 occupancy at these sites was not affected, indicating that GATA-1 is not sufficient for the long range interactions. Other investigators reduced NF-E2 by knocking down the MafK subunit in MEL cells and observed a similar loss of LCR/βmaj proximity despite normal GATA-1 binding at LCR HSs (Du et al., 2008). These observations from human and mouse erythroid cell lines reveal that NF-E2 has a role in looping between the LCR and active globin gene. However, a study in which p45/NF-E2 knockout mice were examined concluded that NF-E2 was dispensable for chromatin loop formation in the mouse β-globin locus (Kooren et al., 2007). These disparate results may be due to redundancy mechanisms present in vivo in mouse tissue that might be absent in the cell lines.
Role of co-factors in chromatin loop formation
Co-factors that do not directly bind to DNA are involved in chromatin loop formation in the β-globin locus. The widely expressed nuclear protein NLI/Ldb1 forms a complex with erythroid specific activators GATA-1 and SCL/TAL1 (Wadman et al., 1997) that binds to the LCR HSs of the human and mouse β-globin loci (Brand et al., 2004; Kiefer et al., 2011; Song et al., 2007). Reduction of NLI/Ldb1 by RNAi in MEL cells inhibits transcriptional activation of the βmaj globin gene with loss of spatial proximity between LCR HS2 and βmaj (Song et al., 2007). In addition, occupancy of GATA-1 and SCL/TAL1 at the LCR HSs and the β-globin gene promoter was decreased by reduction of NLI/Ldb1 (Song et al., 2010). Interestingly, in the human β-globin locus, the NLI/Ldb1 complex occupies a position 3′ of the γ-globin genes that is the site of an erythroid specific non-coding transcript (Kiefer et al., 2011). This region also comes into proximity with the LCR when the γ-globin genes are active, suggesting that not all LCR loops are directly to globin genes. NLI/Ldb1 appears to be the subunit of its complex that is responsible for loop formation and/or stabilization as its self-interaction domain is necessary and sufficient for loop formation (Krivega and Dean, in preparation).
Brg1, the ATPase component of nucleosome remodeling complex SWI/SNF, is required for the formation of a chromatin loop between the LCR and βmaj globin gene. This long range interaction was abrogated by mutant Brg1 in mouse fetal liver (Kim et al., 2009). It appears to be an indirect effect because LCR HSs lose hypersensitivity to DNase I in these mutant mice (Bultman et al., 2005). However, the binding of GATA-1 and NF-E2 at the LCR and gene promoter are not affected (Kim et al., 2007). This result is consistent with the function of NF-E2 as a ‘pioneer’ transcription factor that can bind to chromatin absent remodeling (Gui and Dean, 2003).
Role of insulator-related proteins in chromatin loop formation
Insulators are nucleoprotein complexes that can block enhancer activity when placed between an enhancer and gene and can serve as boundary elements in chromatin to prevent the inappropriate spread of histone modifications (Bushey et al., 2008). In mammals, CTCF is the only known insulator binding protein. This protein is detected at (LCR) HS5 and 3′HS1 flanking the LCR and globin genes in the human and mouse β-globin loci and CTCF is required for interaction between them. The reduction of CTCF by conditional deletion of the CTCF gene decreased the interaction frequency between the two HSs in mouse erythroid progenitor cells (Splinter et al., 2006). A similar effect was observed in the mouse β-globin locus when CTCF binding sequences at 3′HS1 were mutated. Reduction of CTCF by shRNA or siRNA in K562 cells decreased the interaction frequency between HS5 and 3′HS1 of the human β-globin locus (Chien et al., 2011; Hou et al., 2010).
Recent data indicate that the cohesion complex interacts with CTCF and co-occupies CTCF sites in chromatin including at the β-globin locus (Hou et al., 2010; Wendt and Peters, 2009). This complex participates in chromatin loop formation in the β-globin locus. Subunits of the cohesin complex, Rad21, SMC1 and SMC3, are detected at HS5 and 3′HS1 in the human β-globin locus (Chien et al., 2011; Hou et al., 2010). Reduction of the subunits using shRNA or siRNA diminished the interaction between the two HSs in K562 cells. Interestingly, Rad21was also detected at the LCR HS2 and the promoter of the active β-globin gene in both mouse and human β-globin loci in MEL cells, mouse fetal liver, K562 cells and human CD34+ cells (Chien et al., 2011). Reduction of Rad21 binding by the removal of the cohesin loading factor Nipbl decreased proximity between LCR HS2 and β-globin gene, and reduced the proximity of HS5 with the gene and 3′HS1. Knockdown of SMC1 using siRNA resulted in similar effects on loop formation between the LCR and γ-globin gene in K562 cells. In these experiments, LCR-γ-globin loop formation was maintained when CTCF binding was reduced at HS5 and 3′HS1 suggesting the possibility that Rad21 binding at HS2 and the β-globin gene might be sufficient to maintain the long range interaction in the CTCF knockdown condition.
Role of active chromatin structure in chromatin loop formation
Chromatin loops are formed between DNase I HSs in the β-globin locus. When the HSs are not formed, the loop structure is not established. When HS3 and the β-globin promoter were deleted, DNase I hypersensitivity was lost at LCR HSs, and the LCR and βmaj globin gene were not in close proximity (Patrinos et al., 2004). Similarly, when LCR HSs were not hypersensitive to DNase I due to reduction of EKLF or mutation of Brg1, the chromatin loop between the LCR and βmaj globin gene did not form (Drissen et al., 2004; Kim et al., 2009). These observations imply that the establishment of HSs in the LCR is required for loop formation. Mechanistically, this might reflect the role of HSs in providing accessible binding sites for proteins mediating chromatin loop structure.
Histone acetylation often correlates with chromatin loop formation in the β-globin locus. Histone acetylation was less enriched in the LCR and β-globin gene after the deletion of HS3 and β-globin promoter and proximity between the LCR and the gene was decreased (Patrinos et al., 2004). Chromatin looping between the LCR and β-globin gene was disrupted and histone acetylation was decreased by mutation of Brg1 or knockdown of GATA-1 (Kim et al., 2009; 2011). However histone acetylation is not sufficient for the loop formation because the proximity between the LCR and Gγ-globin gene was reduced in NF-E2 knockdown K562 cells while there was no effect on histone acetylation (Kim et al., 2011). Inhibition of histone acetylation by knockdown of histone acetyltransferase activity might help explain the relationship between hyperacetylation in the LCR and active globin gene and chromatin loop formation between them.
The loop between HS5 and 3′HS1 appears to play a role in formation of an active chromatin domain. When the loop between insulators is disrupted by CTCF knockdown in K562 cells, H3K9me2 is increased through the locus (Hou et al., 2010). Other studies suggest the retention of the looped conformation between HS5 and 3′HS1 in NF-E2 knockdown K562 cells might contribute to the maintenance of high levels of histone H3 acetylation and H3K4 methylation (Kim et al., 2011). Thus these observations support a primary role of looping between insulators in forming a domain having active histone modifications, rather than directly activating gene transcription.
Role of nuclear localization in chromatin loop formation
The β-globin locus migrates from the nuclear periphery to a more central position when the β-globin gene is transcriptionally activated as erythroid differentiation progresses (Ragoczy et al., 2006). Similarly, in uninduced MEL cells, the β-globin locus is positioned at the nuclear periphery and relocates to the nuclear interior after induction of β-globin transcription (Francastel et al., 2001). The relocation in mouse fetal liver cells requires the LCR (Ragoczy et al., 2006). GATA-1, NLI/Ldb1 and EKLF, factors that mediate chromatin loop formation between the LCR and β-globin gene, are also required for migration of the locus away from the nuclear periphery (Lee et al., 2009; 2011; Song et al., 2010). A simple interpretation of these results is that looping precedes migration.
However the temporal order of chromatin loop formation and nuclear relocation remains to be further clarified. A recent kinetic analysis carried out in G1E GATA-1 null cells showed that nuclear relocation precedes transcription of the βmaj globin gene (Lee et al., 2011). However, another study using mouse fetal liver cells showed that the transcription of the βmaj globin gene begins at the nuclear periphery prior to relocation (Ragoczy et al., 2006). Based on the common acceptance of the idea that looping is required for β-globin transcription activation (see below), these data could be consistent with looping before or after migration. It also remains unclear whether looping/transcription activation and nuclear relocalization are linked or entirely separate processes.
Role of chromatin loop formation in the globin gene transcription
Transcription of the β-like globin genes is accompanied by chromatin looping between the LCR and target genes in the β-globin locus. Proximity between HS2 and the βmaj globin gene correlates with increased pol II recruitment to the promoter and transcription of the gene (Kim et al., 2009; Vakoc et al., 2005). The close proximity between the LCR and gene is reduced when transcription is abolished by the remove of transcriptional activators or co-factors (Chien et al., 2011; Drissen et al., 2004; Kim et al., 2009; 2011; Kooren et al., 2007; Song et al., 2007; Vakoc et al., 2005). Furthermore, the looped structure and transcriptional activation of the gene is recovered by restoration of GATA-1 (Vakoc et al., 2005). These results support the idea that chromatin looping between the LCR and target genes is required for the β-globin gene transcription. There is no report that the globin genes are actively transcribed without LCR loop formation.
Interestingly, the chromatin loop can be maintained without gene transcription in the β-globin locus. In a transgenic human β-globin locus, if the β-globin gene promoter is deleted, transcription is abolished but the LCR looped structure is maintained essentially unchanged (Patrinos et al., 2004). In addition, inhibition of transcription initiation/elongation of pol II using α-amanitin or DRB does not change the chromatin loop structure of the mouse β-globin locus in primary erythroid cells (Mitchell and Fraser, 2008; Palstra et al., 2008). These observations indicate that ongoing transcription of the gene itself is not necessary for the maintenance of loop structure and that the loop structure is not the result of transcription.
In addition to the loop between the LCR and active gene, another loop is formed between LCR HS5 and 3′HS1 in a CTCF binding dependent manner. It is not clear whether loop formation between these insulators is necessary for the transcription of globin genes. Transcription of the βmaj globin gene is activated during differentiation of wild type ES cells and in ES cells with mutations in 3′HS1 that preclude CTCF binding and loop formation with HS5 (Splinter et al., 2006). However the disruption of loop formation between HS5 and 3′HS1 by CTCF knockdown in K562 cells using shRNA reduced transcription of the γ-globin genes and increased H3K9me2, a repressive histone modification (Hou et al., 2010). This result suggests the importance of CTCF mediated loop formation more broadly within chromosome 11 to the transcription of globin genes. Other investigators recently reported no transcriptional alteration of the γ-globin genes when the LCR loop was disrupted by reduction of CTCF in K562 cells (Chien et al., 2011). The reasons for these differences are unclear and require further study.
CONCLUSIONS
Clearly, progress has been made in understanding the proteins/protein complexes involved directly in mediating chromatin looping in the β-globin loci. The presence of active histone modification marks and relocalization to the nuclear interior have been correlated with LCR/β-globin looping. However, a clear cause and effect relationship between looping and transcription activation has not been established and remains a challenge going forward. Likewise, we do not yet understand how specific loops are initially formed in chromatin. It will also be important to address the mechanism by which the LCR activates the embryonic genes. These genes are too close to the LCR to be studied by 3C and it remains possible that tracking of RNA pol II from the LCR to these proximal genes is involved in their activation. Finally, the potential involvement of the BGL3 non-coding transcript in chromatin looping is intriguing in the light of recent studies reporting the function of non-coding enhancer transcripts in gene activation (Kim et al., 2010; Orom et al., 2010). Future efforts are required to elucidate these remaining mysteries about the mechanism by which gene are transcribed in a nuclear chromatin environment.
Acknowledgments
This work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH (A.D.) and the Korea Research Foundation Grant funded by the Korean Government [Ministry of Education and Human Resources Development (MOEHRD)] (KRF-2007-331-C00193) (A.K.).
REFERENCES
- Brand M., Ranish J.A., Kummer N.T., Hamilton J., Igarashi K., Francastel C., Chi T.H., Crabtree G.R., Aebersold R., Groudine M. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat. Struct. Mol. Biol. 2004;11:73–80. doi: 10.1038/nsmb713. [DOI] [PubMed] [Google Scholar]
- Bultman S.J., Gebuhr T.C., Magnuson T. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in β-globin expression and erythroid development. Genes Dev. 2005;19:2849–2861. doi: 10.1101/gad.1364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey A.M., Dorman E.R., Corces V.G. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol. Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor A.B., Orkin S.H. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- Carter D., Chakalova L., Osborne C.S., Dai Y., Fraser P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- Chien R., Zeng W., Kawauchi S., Bender M.A., Santos R., Gregson H.C., Schmiesing J.A., Newkirk D.A., Kong X., Ball A.R., Jr., et al. Cohesin mediates chromatin interactions that regulate mammalian b-globin expression. J. Biol. Chem. 2011;286:17870–17878. doi: 10.1074/jbc.M110.207365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean A. On a chromosome far, far away: LCRs and gene regulation. Trends Genet. 2006;22:38–45. doi: 10.1016/j.tig.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., et al. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R., Palstra R.J., Gillemans N., Splinter E., Grosveld F., Philipsen S., de Laat W. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M.J., Lv X., Hao D.L., Zhao G.W., Wu X.S., Wu F., Liu D.P., Liang C.C. MafK/NF-E2 p18 is required for β-globin genes activation by mediating the proximity of LCR and active b-globin genes in MEL cell line. Int. J. Biochem. Cell. Biol. 2008;40:1481–1493. doi: 10.1016/j.biocel.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Ethier S.D., Miura H., Dostie J. Discovering genome regulation with 3C and 3C-related technologies. Biochim. Biophys. Acta. 2012;1819:401–410. doi: 10.1016/j.bbagrm.2011.12.004. [DOI] [PubMed] [Google Scholar]
- Fang X., Xiang P., Yin W., Stamatoyannopoulos G., Li Q. Cooperativeness of the higher chromatin structure of the b-globin locus revealed by the deletion mutations of DNase I hypersensitive site 3 of the LCR. J. Mol. Biol. 2007;365:31–37. doi: 10.1016/j.jmb.2006.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francastel C., Magis W., Groudine M. Nuclear relocation of a transactivator subunit precedes target gene activation. Proc. Natl. Acad. Sci. USA. 2001;98:12120–12125. doi: 10.1073/pnas.211444898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui C.Y., Dean A. A major role for the TATA box in recruitment of chromatin modifying complexes to a globin gene promoter. Proc. Natl. Acad. Sci. USA. 2003;100:7009–7014. doi: 10.1073/pnas.1236499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C., Dale R., Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl. Acad. Sci. USA. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadauke S., Blobel G.A. Chromatin loops in gene regulation. Biochim. Biophys. Acta. 2009;1789:17–25. doi: 10.1016/j.bbagrm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer C.M., Lee J., Hou C., Dale R.K., Lee Y.T., Meier E.R., Miller J.L., Dean A. Distinct Ldb1/NLI complexes orchestrate γ-globin repression and reactivation through ETO2 in human adult erythroid cells. Blood. 2011;118:6200–6208. doi: 10.1182/blood-2011-06-363101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.I., Bultman S.J., Jing H., Blobel G.A., Bresnick E.H. Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation. Mol. Cell. Biol. 2007;27:4551–4565. doi: 10.1128/MCB.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.I., Bultman S.J., Kiefer C.M., Dean A., Bresnick E.H. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc. Natl. Acad. Sci. USA. 2009;106:2259–2264. doi: 10.1073/pnas.0806420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S., et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.W., Kim S., Kim C.G., Kim A. The distinctive roles of erythroid specific activator GATA-1 and NF-E2 in transcription of the human fetal γ-globin genes. Nucleic Acids Res. 2011;39:6944–6955. doi: 10.1093/nar/gkr253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooren J., Palstra R.J., Klous P., Splinter E., von Lindern M., Grosveld F., de Laat W. β-globin active chromatin hub formation in differentiating erythroid cells and in p45 NF-E2 knock-out mice. J. Biol. Chem. 2007;282:16544–16552. doi: 10.1074/jbc.M701159200. [DOI] [PubMed] [Google Scholar]
- Lee H.Y., Johnson K.D., Fujiwara T., Boyer M.E., Kim S.I., Bresnick E.H. Controlling hematopoiesis through sumoylation-dependent regulation of a GATA factor. Mol. Cell. 2009;36:984–995. doi: 10.1016/j.molcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.Y., Johnson K.D., Boyer M.E., Bresnick E.H. Relocalizing genetic loci into specific subnuclear neighborhoods. J. Biol. Chem. 2011;286:18834–18844. doi: 10.1074/jbc.M111.221481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.A., Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22:20–25. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q., et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra R.J., Tolhuis B., Splinter E., Nijmeijer R., Grosveld F., de Laat W. The β-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- Palstra R.J., Simonis M., Klous P., Brasset E., Eijkelkamp B., de Laat W. Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS One. 2008;3:e1661. doi: 10.1371/journal.pone.0001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrinos G.P., de Krom M., de Boer E., Langeveld A., Imam A.M., Strouboulis J., de Laat W., Grosveld F.G. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 2004;18:1495–1509. doi: 10.1101/gad.289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy T., Bender M.A., Telling A., Byron R., Groudine M. The locus control region is required for association of the murine b-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.-H., Hou C., Dean A. A positive role for NLI/Ldb1 in long-range b-globin locus control region function. Mol. Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.-H., Kim A., Ragoczy T., Bender M.A., Groudine M., Dean A. Multiple functions of Ldb1 required for β-globin activation during erythroid differentiation. Blood. 2010;116:2356–2364. doi: 10.1182/blood-2010-03-272252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E., Heath H., Kooren J., Palstra R.J., Klous P., Grosveld F., Galjart N., de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the b-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W. Looping and interaction between hypersensitive sites in the active b-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Vakoc C.R., Letting D.L., Gheldof N., Sawado T., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A. Proximity among distant regulatory elements at the b-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Wadman I.A., Osada H., Grutz G.G., Agulnick A.D., Westphal H., Forster A., Rabbitts T.H. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt K.S., Peters J.M. How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res. 2009;17:201–214. doi: 10.1007/s10577-008-9017-7. [DOI] [PubMed] [Google Scholar]
- Zhao H., Dean A. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nucleic Acids Res. 2004;32:4903–4919. doi: 10.1093/nar/gkh832. [DOI] [PMC free article] [PubMed] [Google Scholar]