Abstract
Bax inhibitor-1 (BI-1) is an anti-apoptotic protein located in the endoplasmic reticulum (ER). The role of BI-1 has been studied in different physiopathological models including ischemia, diabetes, liver regeneration and cancer. However, fundamental knowledge about the effects of BI-1 deletion on the proteome is lacking. To further explore this protein, we compared the levels of different proteins in bi-1−/− and bi-1+/+ mouse tissues by two-dimensional electrophoresis (2-DE) and mass spectrometry (MS). In several bi-1−/− mice, glucose-regulated protein 75 (GRP75/mortalin/PBP74/mthsp70), peroxiredoxin6 (Prx6) and fumarylaceto-acetate hydrolase (FAH) showed a pI shift that could be attributed to post-translational modifications. Selenium-binding protein 2 (SBP2) and ferritin light chain 1 levels were significantly increased. Phosphatidylethanolamine-binding protein-1 (PEBP-1) was dramatically decreased in bi-1−/− mice, which was confirmed by Western blotting. The phosphorylation of GRP75, Prx6 and FAH were compared between bi-1+/+ and bi-1−/− mice using liver tissue lysates. Of these three proteins, only one exhibited modified phosphorylation; Tyr phosphorylation of Prx6 was increased in bi-1−/− mice. Our protein profiling results provide fundamental knowledge about the physiopathological function of BI-1.
Keywords: 2-DE, Bax inhibitor-1, ER stress, MS, proteomics
INTRODUCTION
Bax inhibitor-1 (BI-1) is an anti-apoptotic protein with six or seven transmembrane domains. BI-1 was discovered during a screen for suppressors of Bax-induced cell death. Overexpressed BI-1 provides protection against apoptosis, whereas BI-1 antisense RNA promotes apoptosis of some tumor cell lines (Xu and Reed, 1998). However, more recently the protective role of BI-1 was found to be focused on specific stress conditions such as endoplasmic reticulum (ER) stress, as opposed to all apoptotic stimuli (Chae et al., 2004).
Immunofluorescence microscopy and subcellular fractionation studies have demonstrated that BI-1 is located in the ER (Xu and Reed, 1998). ER is an important organelle where protein synthesis, folding and modification take place and the secretory pathway begins (Rutkowski and Kaufman, 2004). The ER can initiate apoptosis when the accumulation of unfolded proteins or the inhibition of ER-Golgi transport results in an ER stress response (Oyadomari et al., 2002). Reactive oxygen species (ROS) are generated in the ER through oxidative protein folding (Lee et al., 2007). ER responds to the accumulation of unfolded proteins in its lumen by activating intracellular signal transduction pathways cumulatively called the unfolded protein response (UPR) (Ron and Walter, 2007).
BI-1 can inhibit ROS accumulation in the ER by modifying heme oxygenase 1 (HO-1) expression (Lee et al., 2007) and reduce ROS products in the ER through regulation of cytochrome P450 2E1 (Kim et al., 2009). BI-1 also increases Ca2+ leakage from the ER (Kim et al., 2008), possibly in a pH-dependent manner. Although BI-1 regulates ER stress-related apoptosis, its role during ER stress and the resulting signal pathways are only now beginning to be revealed. Furthermore, recent efforts to understand the role of BI-1 have focused on ischemia/reperfusion using liver tissues from knock-out mouse models bi-1+/+ and bi-1−/−. The ischemic stress-associated damage was more severe in bi-1 knock-out mice compared to wild-type mice (Bailly-Maitre et al., 2006). When mice were exposed to hypoxic stress, the infarction size was significantly larger in bi-1−/− than it was in bi-1+/+ mice (Chae et al., 2004). Although one study found highly efficient regeneration of damaged liver in bi-1 knock-out mice (Bailly-Maitre et al., 2007), other studies have mainly focused on the protective functions of BI-1 in in vitro and in vivo systems (Bailly-Maitre et al., 2006; Chae et al., 2004; Dohm et al., 2006).
It is important to obtain basic information to elucidate the mechanisms of the effects of BI-1 on various disease models. Here we utilized a proteomics approach. This technology is ideal for detecting changes in protein expression as it allows for comparison of two or more samples on a relatively global level while requiring little or no knowledge about pathways influenced by the experimental conditions (Skynner et al., 2002).
This is particularly important for studies of genetically altered mice, since it is virtually impossible to predict all pathways that are likely to be affected. We used two-dimensional electrophoresis (2-DE) and mass spectrometry to identify changes in the proteomes of bi-1-genetically altered mouse models. In this study we identified differential protein expressions and modifications in tissues from bi-1 knockout compared to those of wild-type mice.
MATERIALS AND METHODS
Materials
Linear immobilized pH gradient (IPG) strips (24-cm, pH 3–10 NL) and IPG buffer (pH 3–10 NL) were purchased from GE Healthcare (Sweden). Trifluoroacetic acid (TFA), tributylphosphine (TBP), and acetonitrile (ACN) were purchased from Fluka (Switzerland). Antibodies against β-actin, GRP75, Prx6, SBP2 and phosphothreonine were acquired from Santa Cruz Biotechnology (USA). Anti-phosphotyrosine was obtained from BD Transduction Laboratories (USA). Anti-phosphoserine was purchased from Invitrogen Life Technologies (USA). Antibodies against FAH and ferritin light chain were purchased from Abcam (USA). Anti-PEBP-1 was purchased from Invitrogen Life Technologies (USA). All other chemicals used in 2-D electrophoresis were ultra pure grade and were purchased from Amresco (USA). All chemicals using in western blotting were of analytical grade and purchased from Sigma-Aldrich (USA).
Animals
The bi-1+/+ and bi-1−/− mice used in this study have been described previously (Chae et al., 2004). Mice were housed in cages in a temperature-controlled animal facility with a 12 h light/dark cycle.
Sample collection
Five independent tissues (brain, heart, lung, liver and kidney) from eight-week-old mice were snap frozen in liquid nitrogen and stored at −80°C for further analysis.
Preparation of protein extracts for proteomics studies
Frozen tissue samples were lysed in 0.5 ml 7 M-urea/2 M-thiourea buffer (7 M urea, 2 M thiourea, 4% CHAPS, 100 mM DTT, 40 mM Tris, and trace amounts of bromophenol blue) with 5 μl protease inhibitor mix (Amersham, USA). Briefly, tissues were sonicated ten times using an Ultrasonic processor VCX 130 (Sonic & Materials, USA) for approximately 10 s at 30 s intervals. Following sonication, 10 μl of 1,000 U/ml DNase was added to the lysate, and the mixture was vortexed for 30 min and then centrifuged at 12,000 rpm for 45 min to remove any undissolved material. The resulting supernatant was used as the protein lysate.
A 2-D clear-up kit (GE Healthcare, Sweden) was used to clarify the lysate according to the manufacturer’s protocol. The protein concentration was measured using the Bradford assay with BSA as the standard (Bradford, 1976). All of the samples were stored at −80°C.
Two-dimensional electrophoresis methodology
Two-dimensional electrophoresis (2-DE) was carried out for male and female (n = 5 each) bi-1+/+ mice, as well as for the corresponding bi-1−/− group (n = 5 for both males and females). To assure reproducibility and to prevent variations due to the technique, all 2-DE gels were carried out under exactly the same conditions with five independent tissue samples (brain, heart, lung, liver and kidney) from each mouse and two experimental replicates.
Isoelectric focusing (IEF) was carried out with 24-cm, pH 3–10, non linear IPG strips at 20°C using the Ettan IPGphor II isoelectric focusing system (GE Healthcare, Sweden). Proteins (1 mg) were mixed with rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 100 mM DTT, 2% IPG buffer and trace amounts of bromophenol blue), and loaded onto first-dimension IPG gel strips. The IPG strips were rehydrated with the samples overnight (18–20 h). The IEF protocol was as follows: (i) 100 V, 3 h, step and hold mode; (ii) 200 V, 2 h, step and hold mode; (iii) 500 V, 1 h, step and hold mode; (iv) 1,000 V, 1 h, step and hold mode; (v) 2000 V, 1 h, step and hold mode; (vi) 4,500 V, 1 h, gradient mode; (vii) 8,000 V, 1 h, gradient mode; (viii) 8,000 V, 11 h, step and hold mode until an approximately 100,000 V total was reached. After IEF was completed, the strips were stored at −80°C.
Prior to second-dimension SDS/PAGE, the frozen strips were subjected to one-step equilibration for 25 min in TBP solution (6 M urea, 37.4 mM Tris-HCl, 20% glycerol, 2.5% acrylamide, 2% SDS, 5 mM TBP) in a shaker. The IPG strips were then positioned onto a 9–17% gradient 1.5 mm SDS/PAGE gel and fixed in place with a 0.5% agarose overlay. SDS/PAGE molecular weight standard (3 μl) on filter paper was embedded directly on the top of the gel. Gels were run in an Ettan DALTsix electrophoresis system (GE Healthcare, Sweden) at 15 mA/gel for 1 h and then at 60 mA/gel until the bromophenol blue dye reached the bottom of the gel.
In-gel protein visualization using Coomassie blue staining
After SDS/PAGE, the gels were washed in ultrapure water and fixed in 40% EtOH and 10% acetic acid solution for 1 h. The gels were then stained overnight with Coomassie blue G-250 staining solution (17% (w/v) ammonium sulfate, 3% (v/v) phosphoric acid, 34% (v/v) methanol, and 0.1% (w/v) Coomassie blue G-250 in ultrapure water). After staining, the gels were washed several times in ultrapure water over a period of more than 4 h.
Image analysis
Gels were scanned with a UMAX Powerlook 1120 scanner (Maxium Technologies, Taiwan). The images were saved in tagged image file format (TIFF) and imported to Melanie 7.0 Software (GeneBio, Switzerland). Image analysis software was used for spot detection, quantification, and analysis according to the manufacturer’s instructions. Briefly, the basic analysis scheme consisted of five steps: detection of spots, identification of landmark proteins, aligning and matching of gel spots, quantification of matched spots, and manual inspection of the spots to verify accuracy of the matching.
Spot detection parameters were carefully adjusted. First, the smooth parameter was set to a value of 2, allowing for the detection of all real spots and the differentiation of overlapping spots. The minimum area was then established to eliminate spots that had an area smaller than 30 pixels. Finally, the saliency parameter was experimentally adjusted to 2 in order to filter out artifacts. The detection parameters were recorded and applied to other gels. The spot volume was used as the analysis parameter for quantifying protein expression.
Spot excision and in-gel digestion
After identification of the spots of interest, protein spots were excised from Coomassie blue-stained second-dimension gels using a P200 yellow pipette tip with the end cut off. The excised gel plugs were approximately 2 mm in diameter and 1.5 mm in thickness. Digestion consisted of a series of washing and dehydrating steps using 50 mM ammonium bicarbonate in 50% ACN. Gel plugs were dried in a speed vacuum and were digested with 12.5 ng of trypsin at 37°C for 12 h.
Matrix assisted laser desorption ionisation time-of-flight mass spectrometry (MALDI-TOF MS) analysis and identification of tryptic peptides
The resulting sample solutions were desalted using GELoader tips (Eppendorf) and combined with matrix before spotting onto a clean MALDI target plate (Opti-TOF™ 384-well Insert; Applied Biosystems, USA). The MALDI matrix solution was prepared by mixing saturated α-cyano-4-hydroxycinnamic acid (CHCA) in ACN/water/TFA (50:50:0.1) solution.
Protein analysis was performed using Ettan MALDI-TOF (Amersham Biosciences, UK). Peptides were evaporated with a N2 laser at 337 nm, using a delayed extraction approach. They were accelerated with a 20 Kv injection pulse for time of flight analysis. Each spectrum was the cumulative average of 300 laser shots. The search program, ProFound, Rockefeller University (http://prowl.rockefeller.edu/prowl-cgi/profound.exe) was used for protein identification using peptide mass fingerprinting. Spectra were calibrated with trypsin auto-digestion ion peak m/z (842.510, 2211.1046) as the internal standard. Carbamidomethyl was selected as the fixed modification, while the variable modifications were oxidized methionine and deamidation.
Western blotting
For western blotting analysis, liver tissues were lysed in 1% TritonX-100, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 0.01 M sodium phosphate, 2 mM EDTA. Total protein was loaded onto 8% SDS-PAGE gels (for 40–75 kDa proteins) and 10% SDS-PAGE gels (for 20–25 kDa proteins). Gels were then transferred to PVDF (BIO-RAD, USA), blocked with skim milk (5%), and incubated at 4°C overnight with the indicated primary antibodies recognizing β-actin, GRP75, Prx6, FAH, PEBP-1 and ferritin light chain. Antibody detection was accomplished via horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse IgG at room temperature (RT) for one hour. The ECL reagents (GE Healthcare, UK) were used to detect signals and blots were exposed to X-ray film (Kodak).
Immunoprecipitation
The immunoprecipitation protocol has been described previously (Wu et al., 2009). Liver tissue lysates (∼1 mg of total protein) were incubated at 4°C for 2 h with anti-phosphothreonine (1:100), anti-phosphoserine (1:100) or anti-phosphotyrosine (1:100) followed by incubation with Protein G-conjugated agarose for an additional hour. As an internal control, 50 μg of protein was loaded onto the gel and labeled as the input. The beads were washed four times with lysis buffer. The pellet was mixed with 2× SDS sample buffer and then heated to 95°C for 5 min followed by 12% SDS/PAGE. Gels were transferred to PVDF, blocked with 5% BSA, and probed with primary antibodies at 4°C overnight followed by incubated with secondary antibodies. The membrane was exposed to X-ray film to detect signals.
Statistics
Statistical analysis of the 2-DE expression ratios of the proteins were evaluated using Melanie 7.0 Software. Statistical significance (P < 0.05) was determined based on qualitative (presence/absence) and quantitative ≥1.5-fold increase/decrease criteria. The spots had to be reproducible and to occur at a level of more than 50% in the five pairs of samples.
RESULTS
Two-dimensional electrophoretic analysis of bi-1+/+ and bi-1−/− mice
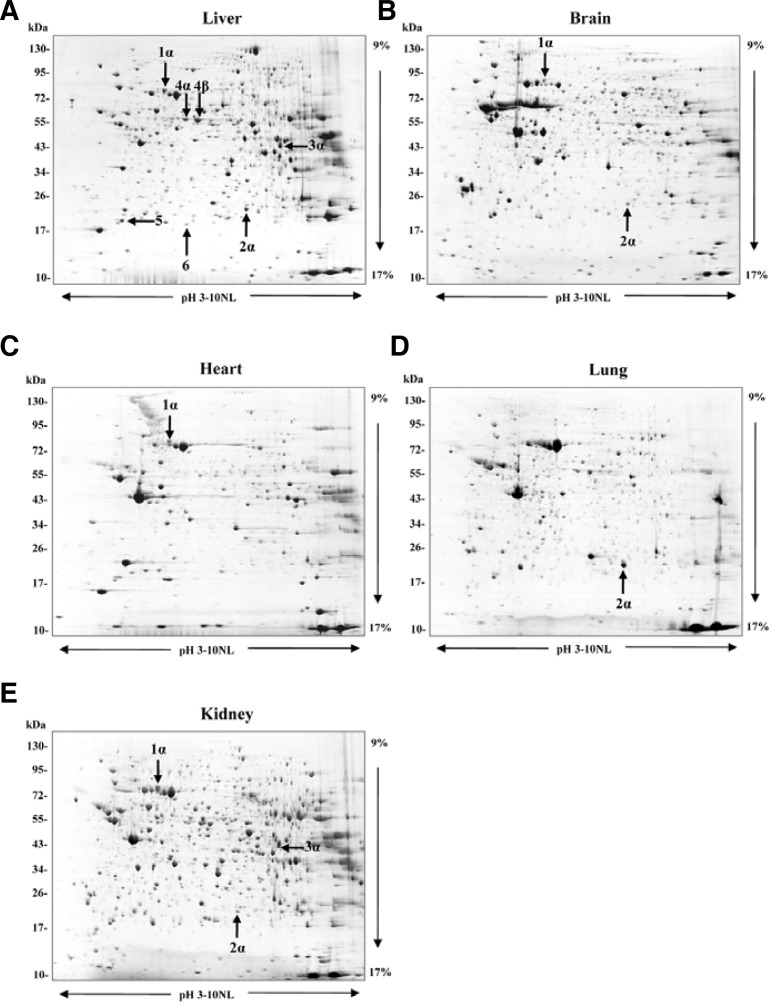
Two-dimensional electrophoretic analysis of the proteomes of bi-1+/+ and bi-1−/− mice was carried out (Fig. 1). Six spots showed consistently differential expression in bi-1−/− mice. MALDI-TOF MS identified these proteins as glucose-regulated protein 75 (GRP75/mortalin/PBP74/mthsp70), peroxiredoxin6 (Prx6), fumarylacetoacetate hydrolase (FAH), selenium-binding protein 2 (SBP2), phosphatidylethanolamine-binding protein-1 (PEBP-1) and ferritin light chain 1. Complete protein names, UniPROTKB accession numbers, subcellular location and biological process are listed in Table 1 and Table 2.
Fig. 1.
Two-dimensional electrophoresis gel maps of bi-1+/+ mouse tissues. Separation was performed with 1 mg of protein on pH 3–10 nonlinear immobilized pH-gradient strips, followed by a 9–17% gradient SDS-PAGE gel. The gel was stained with Coomassie blue G-250. Protein spots were identified using MALDI-TOF. Complete names and UniPROTKB accession numbers are provided in Table 1. (A) Liver, (B) Brain, (C) Heart, (D) Lung, (E) Kidney.
Table 1.
Differential protein spots identified by MALDI-TOF MS
| Spot no. | Protein name | Accession no.a | Gene name | Theoretical molecular weightb | Theoretical pIb |
|---|---|---|---|---|---|
| 1α, 1β | Glucose-regulated protein 75 (GRP75/mortalin/PBP74/mthsp70) | P38647 | Hspa9 | 73.80 | 5.9 |
| 2α, 2β | Peroxiredoxin-6 (Prx6) | O08709 | Prdx6 | 24.97 | 6.0 |
| 3α, 3β | Fumarylacetoacetate hydrolase (FAH) | P35505 | Fah | 46.42 | 6.9 |
| 4α, 4β | Selenium-binding protein 2 (SBP2) | Q63836 | Selenbp2 | 52.59 | 5.8 |
| 5 | Phosphatidylethanolamine-binding protein 1 (PEBP-1) | P70296 | Pebp1 | 20.98 | 5.2 |
| 6 | Ferritin light chain 1 | P29391 | Ftl1 | 20.84 | 5.6 |
Information from UniProtKB
Information from http://prowl.rockefeller.edu/prowl-cgi/profound.exe
Table 2.
Differential protein expression altered in bi-1−/− mice
| Spot no. | Protein name | Average ratio (bi-1−/−/bi-1+/+) | Tissue specificity (found in this study) | Subcellular locationa | Biological processa |
|---|---|---|---|---|---|
| 1α, 1β | Glucose-regulated protein 75 (GRP75/mortalin/PBP74/mthsp70) | pI shift | Liver, brain, kidney, heart | Mitochondria, cytoplasm | Protein export from nucleus, protein folding, response to stress |
| 2α, 2β | Peroxiredoxin-6 (Prx6) | pI shift | Liver, brain, kidney, heart, lung | Cytoplasm, lysosome, lung secretory organelles | Lipid catabolism, bleb formation, cell redox homeostasis, oxidation reduction, response to ROS |
| 3α, 3β | Fumarylacetoacetate hydrolase (FAH) | pI shift | Liver, kidney | Unknown | Phenylalanine, tyrosine, arginine catabolism |
| 4α, 4β | Selenium-binding protein 2 (SBP2) | 1.608 | Liver | Nucleus, cytoplasm, cytosol, membrane, peripheral membrane protein | Protein transport, selenium binding |
| 5 | Phosphatidylethanolamine-binding protein 1 (PEBP-1) | −2.860 | Liver | Cytoplasm | ATP-binding, lipid binding, protease and serine protease inhibitor |
| 6 | Ferritin light chain 1 | 1.610 | Liver | Unknown | Cellular iron ion homeostasis and transport |
Information from UniProtKB and NCBI
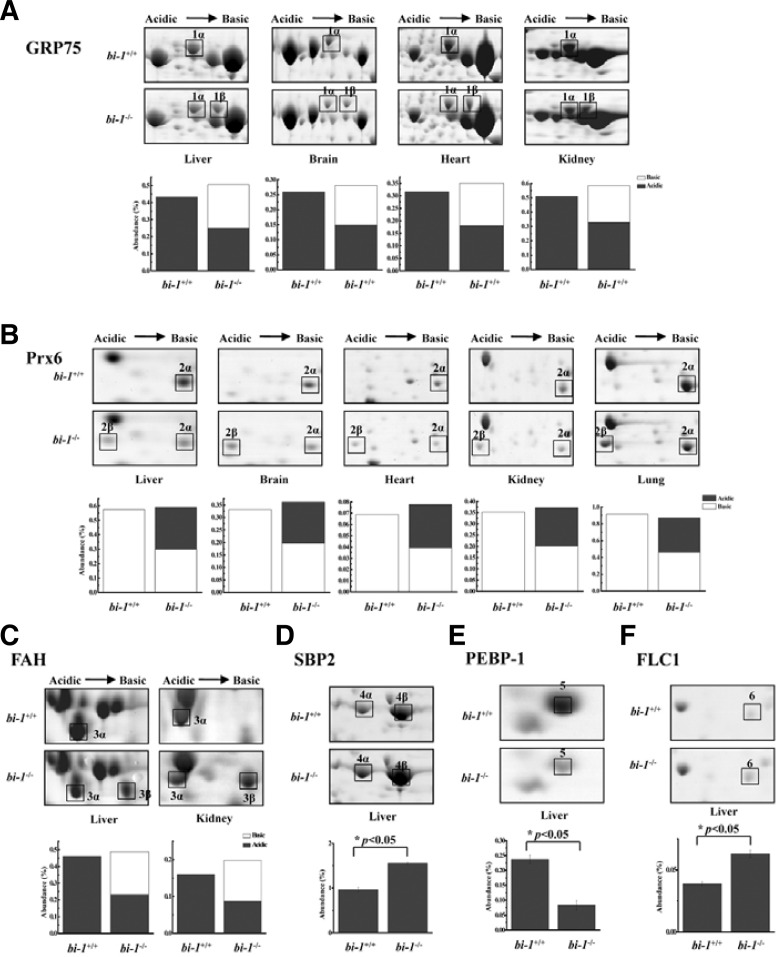
Liver, kidney, brain and heart tissues of bi-1−/− mice showed GRP75 as a 75 kDa doublet of spots with an approximate pI of 5.8–6.0 (Fig. 2A). GRP75 expression was also compared between bi-1+/+ and bi-1−/− lungs, but the GRP75 expression level was very low in lung tissue (data not shown). It appeared that a part of the acidic spot (1α) migrated to the basic area (1β) in bi-1−/− mice. The amounts of total protein were similar for bi-1+/+ and bi-1−/− mice, demonstrating that this basic spot might correspond to a genetic mutation or a post-translational modification such as phosphorylation, oxidation, acetylation, or glycosylation.
Fig. 2.
Two-dimensional electrophoresis gel image enlargements of GRP75, Prx6, FAH, SBP2, PEBP-1 and ferritin light chain 1 in bi-1+/+ and bi-1−/− mouse liver tissues. (A) Representative tissue pairs are shown. Arrows indicate the protein spots corresponding to GRP75 (A), Prx6 (B), FAH (C), SBP2 (D), PEBP-1 (E) or ferritin light chain 1 (FLC1) (F). Bar chart represents protein abundance (%). The white bar represents the basic form, and the black bar represents the acidic form. The total height of each bar represents the sum of the two forms (A) to (C). Complete protein names and UniPROTKB accession numbers are provided in Table 1. *P < 0.05; significantly different from bi-1+/+ tissues.
Two 26-kDa Prx6 spots (2α, 2β) with approximate pIs of 5.0 and 5.7 were identified in bi-1−/− mice liver, brain, heart, lung and kidney tissues, while the tissues from bi-1+/+ mice only exhibited one spot (2α) (Fig. 2β). A part of the basic spot (2α) migrated to the acidic area (2β) in bi-1−/− mice. The observed pI shift of Prx6 suggests possible modifications including phosphorylation.
Liver and kidney tissues of bi-1−/− mice also showed two 46 kDa FAH spots (3α, 3β). Half of the acidic spot (3α) shifted to the basic area (3β) in bi-1−/− mice, possibly due to protein modification.
Protein spot 4 was identified as SBP2, and was significantly increased (∼40%) in the livers of bi-1−/− mice (Fig. 2D). In comparison with the proteins in bi-1+/+ mice, PEBP-1 (spot 5) decreased (∼65%) (Fig. 2E) and ferritin light chain 1 (spot 6) increased (∼40%) in the livers of bi-1−/− mice (Fig. 2F).
Protein analysis of bi-1+/+ and bi-1−/− mice through Western blotting
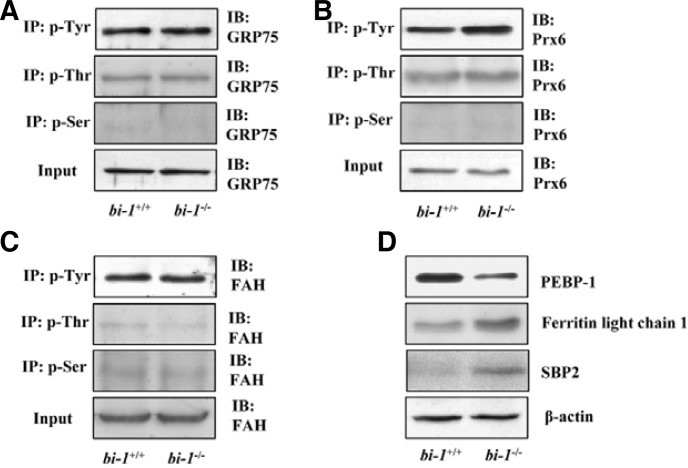
The presence of six proteins with different profiles in bi-1+/+ and bi-1−/− mice as illustrated through proteomics technology was confirmed using western blotting with liver tissues. To understand the pI shift of GRP75 in bi-1−/− mice, GRP75 phosphorylation at tyrosine (Tyr), serine (Ser) or threonine (Thr) residues was examined. However, we found no differences in amino acid phosphorylation of GRP75 between bi-1+/+ and bi-1−/− mice (Fig. 3A), indicating that phosphorylation was not causing the pI shift pattern of GRP75 expression in bi-1−/− mice.
Fig. 3.
The expression of GRP75, Prx6, FAH, SBP2, PEBP-1 and ferritin light chain 1 in bi-1+/+ and bi-1−/− mouse liver tissues. After preparing liver tissue lysates from bi-1+/+ and bi-1−/− mice, immunoprecipitation was performed with anti-p-Tyr, p-Thr or p-Ser antibodies and precipitates were immunoblotted with anti-GRP75 (A), Prx6 (B) or FAH antibodies (C). For liver tissue lysates from bi-1+/+ and bi-1−/− mice, immunoblotting was performed with anti-PEBP-1, ferritin light chain 1, SBP2 or β-actin antibodies (D).
The phosphorylation status of Prx6 was also examined. The phosphorylation of Tyr in Prx6 was greater in bi-1−/− than it was in bi-1+/+ mice (Fig. 3B); however, phosphorylation levels of Thr and Ser were not different between bi-1−/− and bi-1+/+ mouse livers. Finally, we observed no difference in phosphorylation of FAH between bi-1+/+ and bi-1−/− mice (Fig. 3C).
We also examined the expressions of other proteins, including PEBP-1, ferritin light chain 1 and SBP2. Ferritin light chain 1 and SBP2 were more highly expressed in bi-1−/− mice compared to bi-1+/+ mice. PEBP-1 was expressed at lower levels in bi-1−/− compared to bi-1+/+ mice (Fig. 3D).
DISCUSSION
In this study, we compared the levels of different proteins in bi-1−/− and bi-1+/+ mouse tissues through 2-DE and mass spectrometry. Six proteins were identified as having differences in expression or modifications including phosphorylation in bi-1−/− mice. GRP75, Prx6 and FAH showed a pI shift that could be attributed to protein post-translational modifications. The expression levels of SBP2, ferritin light chain 1 or PEBP-1 were dramatically affected in the knock-out mice. We focus our discussion on each protein.
GRP75
GRP75 (also known as mortalin, PBP74 and mthsp70) is a member of the heat shock protein 70 (HSP70) family of stress proteins (Mizzen et al., 1989; Wadhwa et al., 1993). It has general cytoprotective effects against some stressful conditions including glucose deprivation (GD), oxidative injury, and ultraviolet (UV) irradiation (Liu et al., 2005; Yang et al., 2008).
GRP75 resides in multiple subcellular sites including mitochondria, ER, plasmamembrane, cytoplamic vesicles and cytosol. It is differentially distributed in normal and cancerous cells. In cancer cells, GRP75 is expressed in a different subcellular niche with a different binding partner (Kaul et al., 2002; Ran et al., 2000). Since BI-1 is mainly located in the ER, GRP75 may be binding to ER-localized BI-1. The flexible location of GRP75 may explain the expression of GRP75 in the bi-1−/− mice. Binding of BI-1 and GRP75 may also be possible in cells, where it could affect the phosphorylation of GRP75. GRP75 may undergo tyrosine phosphorylation when intact cells are subjected to oxidative stress, such as exposure to peroxovanidium compounds (Hadari et al., 1997). Because the observed pI modification may represent different phosphorylation states, the phosphorylation status of GRP75 was confirmed through western blotting. However, there were no differences in phosphorylation between samples from bi-1+/+ and bi-1−/− mice (Fig. 3A).
The proteomic change of GRP75 was described previously. Genetically altered mice showed a pI shift between the two isoforms of GRP75. This observed change indicated that stress-responsive protein may be altered in transgenic mice (Skynner et al., 2002). It has been demonstrated that this GRP75 pI shift was not due to different states of phosphorylation but to a single amino acid change in Egr1 genetically modified mice (Chardonnet et al., 2007). This allelic difference can introduce variations into the differential proteomic analysis of genetically modified mouse strains. The possibility needs to be further examined.
Peroxiredoxin6
Peroxiredoxins (Prxs) are a family of peroxidases with molecular sizes of 20–30 kDa that appear to be present in all organisms. Prxs have been divided into two classes based on whether the proteins contain one or two conserved catalytic cysteine (cys) residues. Prx1-4 are classified as 2-cys, Prx5 as atypical 2-cys, and Prx6 as 1-cys (Rhee et al., 2001). Prxs vary in subcellular localization, Prx6 is found in the cytoplasm (Cox et al., 2010).
Prxs exert their antioxidant functions through peroxidase activity (Wood et al., 2003). Prx6 is unique as it can reduce phospholipid hydroperoxides (Fisher et al., 1999). Prx6 has been shown to protect lungs from oxidative stress caused by paraquat administration and hypoxia (Wang et al., 2003; 2004) and to protect the myocardium from oxidative stress in a model of ischemia-reperfusion injury (Nagy et al., 2006). Prx6 is the most abundant member of its family in the lung. In addition to its peroxidatic activity, Prx6 has phospholipase A2 (PLA2) activity that is necessary for the recycling and synthesis of lung phospholipids (Fisher and Dodia, 1996; 1997; Kim et al., 1998). Both of these activities are critical to the maintenance of lung physiology. In this study, we observed the modification of Prx6 in lungs of bi-1−/− mice. The expression of BI-1 has been previously correlated with lung development (Jean et al., 1999). Throughout development from embryo to adult, the expression of BI-1 decreases, suggesting that the presence of BI-1 is important for normal lung development.
The bi-1−/− mice used in this study showed a strong shift between basic and acidic forms of Prx6. The basic form had a pI corresponding to the predicted value of 5.7, while the acidic spot had a pI of approximately 5.0. Compared with other Prx proteins, the phosphorylation of Prx6 has not been studied well, and we therefore made this one of the focuses of our study. Recently, mass spectroscopic analysis of in vitro phosphorylated Prx6 revealed a unique phosphorylation site at Thr-177, and mutation of this residue abolished protein phosphorylation and the increase in mitogen-activated protein kinase (MAPK)-mediated activity (Wu et al., 2009). We found that Tyr phosphorylation of Prx6 was greatly increased in bi-1−/− mice (Fig. 3B). However, the levels of phosphorylation of other amino acids were not different between bi-1+/+ and bi-1−/− mice. Prx6 is localized in the cytoplasm. The main location of BI-1 is in the ER, where it has an antioxidant and cell protective function, with its C-terminal extruding out of the ER membranes. The C-terminal has been reported to mediate the key function of BI-1 on its own (Ahn et al., 2009) and through its binding proteins (Kim et al., 2009; Lisbona et al., 2009). We cannot rule out interactions between BI-1 and Prx6 in our study, and at least, the C-terminal motif of BI-1 may affect the expression of the cytoplasmic Prx6. In addition, BI-1 shows unique characteristics including an acidic pH-sensitive Ca2+ channel-like function and Ca2+/H+ antiporter. How acidic pH relates to BI-1 function is not yet fully understood. Such a pH-sensitive Ca2+ channel-like function of BI-1 may be involved in the modification of Prx6. The physiological role of Prx6 Tyr phosphorylation in bi-1 knock-out mice should be further investigated.
Another possible cause of the pI shift of Prx6 could be oxidation of the active site of the cysteine residue by sulfenic acid (Choi et al., 1998; Kang et al., 1998). Most Prxs are known to be converted into variants with a lower pI upon exposure to hydroperoxides (Mitsumoto et al., 2001). The acidic shift reflects phosphorylation or sulfenic acid formation (Chang et al., 2002; Chevallet et al., 2003). These modifications of Prx6 indicate oxidative stress. The anti-oxidant role of BI-1 has also been studied. BI-1 is known to protect against ER stress-induced cell death through the expression of heme oxygenase-1 (HO-1) (Lee et al., 2007) or through the regulation of the interaction between NPR and P450 2E1 (Kim et al., 2009). In bi-1 knock-out mice, this oxidative stress-associated modification might be enhanced. The molecular mechanisms that give rise to Prx6 and its roles in stress responses are currently under investigation.
Fumarylacetoacetate hydrolase (FAH)
FAH is a protein involved in the pathogenesis of hereditary tyrosinemia 1 (HT1), a rare and potentially lethal autosomal recessive disorder that causes severe hepatorenal disease. FAH catalyzes the hydrolysis of 4-fumarylacetoacetate (FAA) into fumarate and acetoacetate, the final step in the tyrosine catabolic pathway (Kvittingen, 1986). A defect in FAH results in accumulation of FAA that can lead to oxidative stress and severe liver and kidney disease (Dieter et al., 2003). Although the post-translational modification of FAH was confirmed through the protein’s basic pI shift (Fig. 2C), the phosphorylation status of FAH did not differ between the two groups (Fig. 3C). Considering previous studies of the relationship of FAH deficiency to pathological conditions such as tyrosinemia (Bergeron et al., 2003; Jorquera and Tanguay, 2001; Orejuela et al., 2008), posttranslational modification of FAH protein may result in the accumulation of FAA and the resultant pathological conditions in bi-1−/− mice under ER stress. The pI shift of FAH protein has been reported, but the reason is still not clear yet (Hernandez-Fernaud and Salido, 2010).
Treatment of Chinese hamster lung cells with FAA similarly induces ER stress, including the induction of the major ER chaperone GRP78 and phosphorylation of eIF2α (p-eIF2α), as well as the induction of pro-apoptotic CHOP and caspase-12 activation (Bergeron et al., 2006). The pI shift of this enzyme in bi-1−/− mice needs to be further investigated.
Selenium-binding protein 2 (SBP2)
SBP2, also known as 56 kDa acetaminophen-binding protein, may be involved in the detection of reactive xenobiotics in the cytoplasm and in intra-Golgi protein transport. SBP2 specifically binds selenium and is mainly detectable in the liver (Lanfear et al., 1993). SBP2 levels decreased dramatically in livers of C57BL/6J mice with peroxisome proliferator-activated receptor-binding properties for selenium, and SBP2 is mainly detectable in liver cell-specific pleiotropic responses, including the development of liver tumors (Chu et al., 2004). SBP2 is also relevant for hepatic fibrosis (Henkel et al., 2005) and has been identified as a major hepatic target for acetaminophen, while demonstrating gender differences in protein abundance (Mattow et al., 2006). In bi-1−/− mice, SBP2 expression was relatively higher compared to bi-1+/+ mice. The SBP2 levels might indicate a stress response and changes in xenobiotic metabolism in bi-1−/− mice livers.
Phosphatidylethanolamine-binding protein 1 (PEBP-1)
PEBP-1, also known as Raf kinase inhibitory protein (RKIP), was identified through a yeast two-hybrid screen for suppressors of raf-1 kinase activity and MAPK signaling in fibroblasts. This protein can sequester inactive Raf-1 and MEK1 (Yeung et al., 2000) and is expressed at greatly reduced levels in tumor-derived liver cell lines (Wirth et al., 1995). Rats subjected to chronic corticosterone treatment showed a decrease in hippo-campal PEBP-1 expression and impaired cognition (Feldmann et al., 2008). In our study, PEBP-1 was also reduced in the livers of bi-1−/− mice.
Ferritin light chain 1
Ferritin plays a central role in the delicate maintenance of intracellular iron balance. Ferritin is a ubiquitous and highly conserved cytosolic iron-binding protein composed of two subunits, the heavy (H) and light (L) chains (Harrison and Arosio, 1996). Depending on the tissue type and the physiological status of the cell, the ratio of H to L subunits in ferritin can vary greatly, from predominantly L in such tissues as liver and spleen to predominantly H in heart and kidney (Arosio et al., 1976). The H subunit plays a role in rapid iron detoxification, whereas the L subunit facilitates iron nucleation, mineralization and long-term storage (Harrison and Arosio, 1996). The H-to-L ratio is not fixed but is readily modified in many inflammatory and infectious conditions and in response to xenobiotic stress, cellular differentiation, and developmental transitions, as well as other stimuli (Torti and Torti, 2002). Increases in ferritin synthesis result in oxidative stress, conferring resistance to subsequent insults (Balla et al., 1992; Cairo et al., 1995). These functions of ferritin suggest that it might serve as a cytoprotective protein, minimizing oxygen free radical formation by sequestering intracellular iron. The increased abundance of ferritin light chain 1 in the livers of bi-1−/− mice might be a response to environmental stress.
Six proteins in bi-1−/− mice that showed different expression levels compared to wild type mice were identified using 2-DE and Western blotting. The changed migration patterns of GRP75, Prx6 and FAH and the modified expression of SBP2, PEBP-1 and ferritin light chain 1 may be associated with metabolic, oxidative and ER stress responses. These results provide fundamental information about the pathophysiological roles of BI-1. The results will be useful in studies of BI-1-associated cell/disease systems. Further studies of this system will provide insight into the BI-1-associated regulatory mechanisms in disease models.
Acknowledgments
This work was supported by Korea Science and Engineering Foundation grants (R01-2007-000-20275-0) and partially by the National Research Foundation (2010-0087202, 2010-0029497) and by the National Institutes of Health, USA (NIH grant AG15393).
REFERENCES
- Ahn T., Yun C.H., Chae H.Z., Kim H.R., Chae H.J. Ca2+/H+ antiporter-like activity of human recombinant Bax inhibitor-1 reconstituted into liposomes. FEBS J. 2009;276:2285–2291. doi: 10.1111/j.1742-4658.2009.06956.x. [DOI] [PubMed] [Google Scholar]
- Arosio P., Yokota M., Drysdale J.W. Structural and immunological relationships of isoferritins in normal and malignant cells. Cancer Res. 1976;36:1735–1739. [PubMed] [Google Scholar]
- Bailly-Maitre B., Fondevila C., Kaldas F., Droin N., Luciano F., Ricci J.E., Croxton R., Krajewska M., Zapata J.M., Kupiec-Weglinski J.W., et al. Cytoprotective gene bi-1 is required for intrinsic protection from endoplasmic reticulum stress and ischemia-reperfusion injury. Proc. Natl. Acad. Sci. USA. 2006;103:2809–2814. doi: 10.1073/pnas.0506854103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Maitre B., Bard-Chapeau E., Luciano F., Droin N., Bruey J.M., Faustin B., Kress C., Zapata J.M., Reed J.C. Mice lacking bi-1 gene show accelerated liver regeneration. Cancer Res. 2007;67:1442–1450. doi: 10.1158/0008-5472.CAN-06-0850. [DOI] [PubMed] [Google Scholar]
- Balla G., Jacob H.S., Balla J., Rosenberg M., Nath K., Apple F., Eaton J.W., Vercellotti G.M. Ferritin: a cytoprotective antioxidant strategem of endothelium. J. Biol. Chem. 1992;267:18148–18153. [PubMed] [Google Scholar]
- Bergeron A., Jorquera R., Tanguay R.M. Hereditary tyrosinemia: an endoplasmic reticulum stress disorder? Med. Sci. (Paris) 2003;19:976–980. doi: 10.1051/medsci/20031910976. [DOI] [PubMed] [Google Scholar]
- Bergeron A., Jorquera R., Orejuela D., Tanguay R.M. Involvement of endoplasmic reticulum stress in hereditary tyrosinemia type I. J. Biol. Chem. 2006;281:5329–5334. doi: 10.1074/jbc.M506804200. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cairo G., Tacchini L., Pogliaghi G., Anzon E., Tomasi A., Bernelli-Zazzera A. Induction of ferritin synthesis by oxidative stress. Transcriptional and post-transcriptional regulation by expansion of the “free” iron pool. J. Biol. Chem. 1995;270:700–703. doi: 10.1074/jbc.270.2.700. [DOI] [PubMed] [Google Scholar]
- Chae H.J., Kim H.R., Xu C., Bailly-Maitre B., Krajewska M., Krajewski S., Banares S., Cui J., Digicaylioglu M., Ke N., et al. BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol. Cell. 2004;15:355–366. doi: 10.1016/j.molcel.2004.06.038. [DOI] [PubMed] [Google Scholar]
- Chang T.S., Jeong W., Choi S.Y., Yu S., Kang S.W., Rhee S.G. Regulation of peroxiredoxin I activity by Cdc2-mediated phosphorylation. J. Biol. Chem. 2002;277:25370–25376. doi: 10.1074/jbc.M110432200. [DOI] [PubMed] [Google Scholar]
- Chardonnet S., Decottignies P., Amar L., Le Caer J.P., Davis S., Laroche S., Le Marechal P. New mortalin and histidyl tRNA synthetase isoforms point out a pitfall in proteomic analysis of Egr1 genetically modified mice. Proteomics. 2007;7:289–298. doi: 10.1002/pmic.200600513. [DOI] [PubMed] [Google Scholar]
- Chevallet M., Wagner E., Luche S., van Dorsselaer A., Leize-Wagner E., Rabilloud T. Regeneration of peroxiredoxins during recovery after oxidative stress: only some over-oxidized peroxiredoxins can be reduced during recovery after oxidative stress. J. Biol. Chem. 2003;278:37146–37153. doi: 10.1074/jbc.M305161200. [DOI] [PubMed] [Google Scholar]
- Choi H.J., Kang S.W., Yang C.H., Rhee S.G., Ryu S.E. Crystal structure of a novel human peroxidase enzyme at 2.0 A resolution. Nat. Struct. Biol. 1998;5:400–406. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- Chu R., Lim H., Brumfield L., Liu H., Herring C., Ulintz P., Reddy J.K., Davison M. Protein profiling of mouse livers with peroxisome proliferator-activated receptor alpha activation. Mol. Cell. Biol. 2004;24:6288–6297. doi: 10.1128/MCB.24.14.6288-6297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A.G., Winterbourn C.C., Hampton M.B. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem. J. 2010;425:313–325. doi: 10.1042/BJ20091541. [DOI] [PubMed] [Google Scholar]
- Dieter M.Z., Freshwater S.L., Miller M.L., Shertzer H.G., Dalton T.P., Nebert D.W. Pharmacological rescue of the 14CoS/14CoS mouse: hepatocyte apoptosis is likely caused by endogenous oxidative stress. Free Radic. Biol. Med. 2003;35:351–367. doi: 10.1016/s0891-5849(03)00273-9. [DOI] [PubMed] [Google Scholar]
- Dohm C.P., Siedenberg S., Liman J., Esposito A., Wouters F.S., Reed J.C., Bahr M., Kermer P. Bax inhibitor-1 protects neurons from oxygen-glucose deprivation. J. Mol. Neurosci. 2006;29:1–8. doi: 10.1385/JMN:29:1:1. [DOI] [PubMed] [Google Scholar]
- Feldmann R.E., Jr, Maurer M.H., Hunzinger C., Lewicka S., Buergers H.F., Kalenka A., Hinkelbein J., Broemme J.O., Seidler G.H., Martin E., et al. Reduction in rat phosphatidylethanolamine binding protein-1 (PEBP1) after chronic corticosterone treatment may be paralleled by cognitive impairment: a first study. Stress. 2008;11:134–147. doi: 10.1080/10253890701649904. [DOI] [PubMed] [Google Scholar]
- Fisher A.B., Dodia C. Role of phospholipase A2 enzymes in degradation of dipalmitoylphosphatidylcholine by granular pneumocytes. J. Lipid Res. 1996;37:1057–1064. [PubMed] [Google Scholar]
- Fisher A.B., Dodia C. Role of acidic Ca2+-independent phospholipase A2 in synthesis of lung dipalmitoyl phosphatidylcholine. Am. J. Physiol. 1997;272:L238–243. doi: 10.1152/ajplung.1997.272.2.L238. [DOI] [PubMed] [Google Scholar]
- Fisher A.B., Dodia C., Manevich Y., Chen J.W., Feinstein S.I. Phospholipid hydroperoxides are substrates for non-selenium glutathione peroxidase. J. Biol. Chem. 1999;274:21326–21334. doi: 10.1074/jbc.274.30.21326. [DOI] [PubMed] [Google Scholar]
- Hadari Y.R., Haring H.U., Zick Y. p75, a member of the heat shock protein family, undergoes tyrosine phosphorylation in response to oxidative stress. J. Biol. Chem. 1997;272:657–662. doi: 10.1074/jbc.272.1.657. [DOI] [PubMed] [Google Scholar]
- Harrison P.M., Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- Henkel C., Roderfeld M., Weiskirchen R., Scheibe B., Matern S., Roeb E. Identification of fibrosis-relevant proteins using DIGE (difference in gel electrophoresis) in different models of hepatic fibrosis. Z. Gastroenterol. 2005;43:23–29. doi: 10.1055/s-2004-813911. [DOI] [PubMed] [Google Scholar]
- Hernandez-Fernaud J.R., Salido E. Differential expression of liver and kidney proteins in a mouse model for primary hyperoxaluria type I. FEBS. J. 2010;277:4766–4774. doi: 10.1111/j.1742-4658.2010.07882.x. [DOI] [PubMed] [Google Scholar]
- Jean J.C., Oakes S.M., Joyce-Brady M. The Bax inhibitor-1 gene is differentially regulated in adult testis and developing lung by two alternative TATA-less promoters. Genomics. 1999;57:201–208. doi: 10.1006/geno.1999.5761. [DOI] [PubMed] [Google Scholar]
- Jorquera R., Tanguay R.M. Fumarylacetoacetate, the metabolite accumulating in hereditary tyrosinemia, activates the ERK pathway and induces mitotic abnormalities and genomic instability. Hum. Mol. Genet. 2001;10:1741–1752. doi: 10.1093/hmg/10.17.1741. [DOI] [PubMed] [Google Scholar]
- Kang S.W., Baines I.C., Rhee S.G. Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J. Biol. Chem. 1998;273:6303–6311. doi: 10.1074/jbc.273.11.6303. [DOI] [PubMed] [Google Scholar]
- Kaul S.C., Taira K., Pereira-Smith O.M., Wadhwa R. Mortalin: present and prospective. Exp. Gerontol. 2002;37:1157–1164. doi: 10.1016/s0531-5565(02)00135-3. [DOI] [PubMed] [Google Scholar]
- Kim T.S., Dodia C., Chen X., Hennigan B.B., Jain M., Feinstein S.I., Fisher A.B. Cloning and expression of rat lung acidic Ca(2+)-independent PLA2 and its organ distribution. Am. J. Physiol. 1998;274:L750–761. doi: 10.1152/ajplung.1998.274.5.L750. [DOI] [PubMed] [Google Scholar]
- Kim H.R., Lee G.H., Ha K.C., Ahn T., Moon J.Y., Lee B.J., Cho S.G., Kim S., Seo Y.R., Shin Y.J., et al. Bax Inhibitor-1 Is a pH-dependent regulator of Ca2+ channel activity in the endoplasmic reticulum. J. Biol. Chem. 2008;283:15946–15955. doi: 10.1074/jbc.M800075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.R., Lee G.H., Cho E.Y., Chae S.W., Ahn T., Chae H.J. Bax inhibitor 1 regulates ER-stress-induced ROS accumulation through the regulation of cytochrome P450 2E1. J. Cell Sci. 2009;122:1126–1133. doi: 10.1242/jcs.038430. [DOI] [PubMed] [Google Scholar]
- Kvittingen E.A. Hereditary tyrosinemia type I--an overview. Scand. J. Clin. Lab. Invest. Suppl. 1986;184:27–34. [PubMed] [Google Scholar]
- Lanfear J., Fleming J., Walker M., Harrison P. Different patterns of regulation of the genes encoding the closely related 56 kDa selenium- and acetaminophen-binding proteins in normal tissues and during carcinogenesis. Carcinogenesis. 1993;14:335–340. doi: 10.1093/carcin/14.3.335. [DOI] [PubMed] [Google Scholar]
- Lee G.H., Kim H.K., Chae S.W., Kim D.S., Ha K.C., Cuddy M., Kress C., Reed J.C., Kim H.R., Chae H.J. Bax inhibitor-1 regulates endoplasmic reticulum stress-associated reactive oxygen species and heme oxygenase-1 expression. J. Biol. Chem. 2007;282:21618–21628. doi: 10.1074/jbc.M700053200. [DOI] [PubMed] [Google Scholar]
- Lisbona F., Rojas-Rivera D., Thielen P., Zamorano S., Todd D., Martinon F., Glavic A., Kress C., Lin J.H., Walter P., et al. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol. Cell. 2009;33:679–691. doi: 10.1016/j.molcel.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu W., Song X.D., Zuo J. Effect of GRP75/mthsp70/PBP74/mortalin overexpression on intracellular ATP level, mitochondrial membrane potential and ROS accumulation following glucose deprivation in PC12 cells. Mol. Cell. Biochem. 2005;268:45–51. doi: 10.1007/s11010-005-2996-1. [DOI] [PubMed] [Google Scholar]
- Mattow J., Demuth I., Haeselbarth G., Jungblut P.R., Klose J. Selenium-binding protein 2, the major hepatic target for acetaminophen, shows sex differences in protein abundance. Electrophoresis. 2006;27:1683–1691. doi: 10.1002/elps.200500703. [DOI] [PubMed] [Google Scholar]
- Mitsumoto A., Takanezawa Y., Okawa K., Iwamatsu A., Nakagawa Y. Variants of peroxiredoxins expression in response to hydroperoxide stress. Free Radic. Biol. Med. 2001;30:625–635. doi: 10.1016/s0891-5849(00)00503-7. [DOI] [PubMed] [Google Scholar]
- Mizzen L.A., Chang C., Garrels J.I., Welch W.J. Identification, characterization, and purification of two mammalian stress proteins present in mitochondria, grp 75, a member of the hsp 70 family and hsp 58, a homolog of the bacterial groEL protein. J. Biol. Chem. 1989;264:20664–20675. [PubMed] [Google Scholar]
- Nagy N., Malik G., Fisher A.B., Das D.K. Targeted disruption of peroxiredoxin 6 gene renders the heart vulnerable to ischemia-reperfusion injury. Am. J. Physiol. Heart Circ. Physiol. 2006;291:H2636–2640. doi: 10.1152/ajpheart.00399.2006. [DOI] [PubMed] [Google Scholar]
- Orejuela D., Jorquera R., Bergeron A., Finegold M.J., Tanguay R.M. Hepatic stress in hereditary tyrosinemia type 1 (HT1) activates the AKT survival pathway in the fah−/− knock-out mice model. J. Hepatol. 2008;48:308–317. doi: 10.1016/j.jhep.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Oyadomari S., Araki E., Mori M. Endoplasmic reticulum stress-mediated apoptosis in pancreatic beta-cells. Apoptosis. 2002;7:335–345. doi: 10.1023/a:1016175429877. [DOI] [PubMed] [Google Scholar]
- Ran Q., Wadhwa R., Kawai R., Kaul S.C., Sifers R.N., Bick R.J., Smith J.R., Pereira-Smith O.M. Extramitochondrial localization of mortalin/mthsp70/PBP74/GRP75. Biochem. Biophys. Res. Commun. 2000;275:174–179. doi: 10.1006/bbrc.2000.3237. [DOI] [PubMed] [Google Scholar]
- Rhee S.G., Kang S.W., Chang T.S., Jeong W., Kim K. Peroxiredoxin, a novel family of peroxidases. IUBMB Life. 2001;52:35–41. doi: 10.1080/15216540252774748. [DOI] [PubMed] [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rutkowski D.T., Kaufman R.J. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Skynner H.A., Rosahl T.W., Knowles M.R., Salim K., Reid L., Cothliff R., McAllister G., Guest P.C. Alterations of stress related proteins in genetically altered mice revealed by two-dimensional differential in-gel electrophoresis analysis. Proteomics. 2002;2:1018–1025. doi: 10.1002/1615-9861(200208)2:8<1018::AID-PROT1018>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Torti F.M., Torti S.V. Regulation of ferritin genes and protein. Blood. 2002;99:3505–3516. doi: 10.1182/blood.v99.10.3505. [DOI] [PubMed] [Google Scholar]
- Wadhwa R., Kaul S.C., Ikawa Y., Sugimoto Y. Identification of a novel member of mouse hsp70 family. Its association with cellular mortal phenotype. J. Biol. Chem. 1993;268:6615–6621. [PubMed] [Google Scholar]
- Wang X., Phelan S.A., Forsman-Semb K., Taylor E.F., Petros C., Brown A., Lerner C.P., Paigen B. Mice with targeted mutation of peroxiredoxin 6 develop normally but are susceptible to oxidative stress. J. Biol. Chem. 2003;278:25179–25190. doi: 10.1074/jbc.M302706200. [DOI] [PubMed] [Google Scholar]
- Wang Y., Feinstein S.I., Manevich Y., Ho Y.S., Fisher A.B. Lung injury and mortality with hyperoxia are increased in peroxiredoxin 6 gene-targeted mice. Free Radic. Biol. Med. 2004;37:1736–1743. doi: 10.1016/j.freeradbiomed.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Wirth P.J., Hoang T.N., Benjamin T. Micropreparative immobilized pH gradient two-dimensional electrophoresis in combination with protein microsequencing for the analysis of human liver proteins. Electrophoresis. 1995;16:1946–1960. doi: 10.1002/elps.11501601321. [DOI] [PubMed] [Google Scholar]
- Wood Z.A., Schroder E., Robin Harris J., Poole L.B. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- Wu Y., Feinstein S.I., Manevich Y., Chowdhury I., Pak J.H., Kazi A., Dodia C., Speicher D.W., Fisher A.B. Mitogen-activated protein kinase-mediated phosphorylation of peroxiredoxin 6 regulates its phospholipase A(2) activity. Biochem. J. 2009;419:669–679. doi: 10.1042/BJ20082061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Reed J.C. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol. Cell. 1998;1:337–346. doi: 10.1016/s1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- Yang L., Liu X., Hao J., Yang Y., Zhao M., Zuo J., Liu W. Glucose-regulated protein 75 suppresses apoptosis induced by glucose deprivation in PC12 cells through inhibition of Bax conformational change. Acta Biochim. Biophys. Sin. (Shanghai) 2008;40:339–348. doi: 10.1111/j.1745-7270.2008.00409.x. [DOI] [PubMed] [Google Scholar]
- Yeung K., Janosch P., McFerran B., Rose D.W., Mischak H., Sedivy J.M., Kolch W. Mechanism of suppression of the Raf/MEK/extracellular signal-regulated kinase pathway by the raf kinase inhibitor protein. Mol. Cell. Biol. 2000;20:3079–3085. doi: 10.1128/mcb.20.9.3079-3085.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]