Abstract
BRCA1 is a well-known tumor suppressor implicated in familial breast and ovarian cancer. Since its cloning in 1994, numerous studies have established BRCA1’s role in diverse cellular and biochemical processes, such as DNA damage repair, cell cycle control, and transcriptional regulation as well as ubiquitination. In addition, a number of recent studies have functionally linked this tumor suppressor to another important cellular regulator, microRNAs, which are short (19–22 nt) RNAs that were discovered in the nematode in 1993. Soon their presence and function were validated in mammals, and since then, the role of microRNAs has been actively investigated in almost all biological processes, including cancer. In this review, we will describe recent progress in the understanding of the BRCA1 function through microRNAs and the role of microRNAs in regulating BRCA1, with emphasis on the implication of these processes on the development and progression of cancer. We will also discuss the therapeutic potential of microRNA mimics or inhibitors of microRNAs to affect BRCA1 function.
Keywords: antagomir, breast cancer, BRCA1, microRNA mimic, microRNAs
INTRODUCTION
In United States, one in eight women will develop breast cancer in her lifetime. Worldwide, breast cancer accounts for almost 23% of all cancers in women, excluding non-melanoma skin cancers (Jones and Buzdar, 2004). Breast cancer not only affects the lives of the women who have it, but it also changes forever the lives of their family members. Moreover, because of its high incidence and the associated, rapidly increasing cost of treatment, breast cancer has a broad-reaching impact on society as a whole. In this regard, preventing or eliminating this disease represents a significant and urgent challenge.
The breast cancer-associated 1 (BRCA1) gene is one of the well-characterized cancer susceptibility genes associated with hereditary breast and ovarian cancer (Fackenthal and Olopade, 2007; Ramus and Gayther, 2009). A mutation in BRCA1 increases the lifetime risk of developing the disease by up to 54% over the risk found in the general population (Easton et al., 1995). Many aspects of this multifunctional protein have been revealed since its cloning in 1994 (Miki et al., 1994), including its essential role in DNA damage repair, E3 ubiquitin ligase activity, and transcriptional activation/repression of downstream genes (Huen et al., 2010). We have recently discovered a novel function of BRCA1 in the epigenetic control of an oncogenic microRNA (miRNA), miR-155 (Chang et al., 2011). MiRNAs are small RNAs (19–22 nt) that are generated by a series of enzymatic processes in the nucleus and as well as in the cytoplasm (Murchison and Hannon, 2004). Approximately 1,400 miRNAs are thought to exist in humans; about 1,100 miRNAs have already been identified (http://www.microrna.org). Recent studies have revealed an intricate relationship between BRCA1 and miRNAs: BRCA1 regulates the expression of miRNAs, which may in turn regulate the expression of BRCA1. In this review, we describe the complex regulatory network of BRCA1 and its associated miRNAs. We also discuss the implication of this network in BRCA1-mediated tumorigenesis as well as in the development of novel therapeutic tools.
MIRNAS TARGETING THE BRCA1 UTR
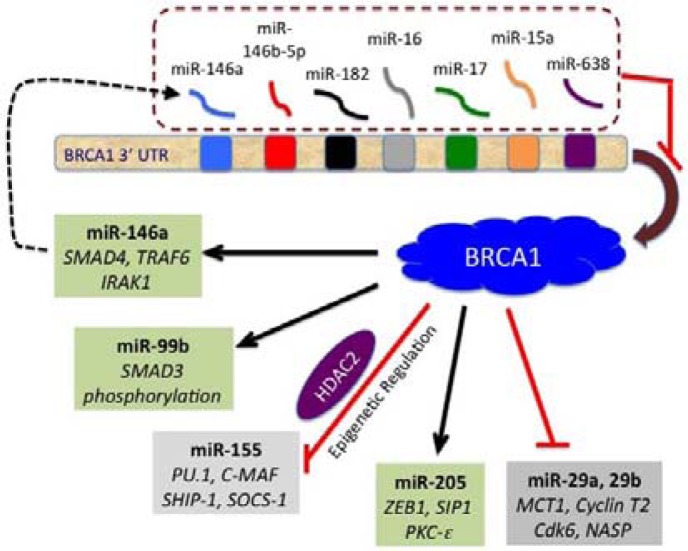
It is estimated that more than 60% of cellular mRNAs are under the control of miRNAs (Friedman et al., 2009; Lewis et al., 2005). BRCA1 is no exception, as several recent studies have shown it to be regulated by miRNAs (summarized in Table 1). The human BRCA1 mRNA has a 1.5-kb 3’UTR that is predicted to bind to 20–100 miRNAs (Griffiths-Jones et al., 2008; John et al., 2004). To date, seven of these miRNAs have been identified and shown to regulate BRCA1 (Fig. 1). Because miRNAs are known to down-regulate the level of the target protein by degrading mRNA or inhibiting its translation (Nelson et al., 2003), the binding of miRNA to the BRCA1 mRNA can have the same effect as a loss-of-function mutation in BRCA1. Indeed, a recent study revealed that the Argonate/miR-182 complex is selectively enriched for the BRCA1 mRNA. Moreover, the overexpression of miR-182 reduces the level of BRCA1 (Moskwa et al., 2011), resulting in hypersensitivity of the cells to γ-irradiation (IR) and defects in homologous recombination, which are the hallmarks of BRCA1 loss. In addition, the authors found that treatment with Poly (ADP-ribose) polymerase 1 (PARP1) inhibitors affects the growth of miR-182-expressing tumors in mice. These results suggest that combining miR-182 with PARP1 inhibitors confers a therapeutic advantage. Currently, PARP1 inhibitors are being used in clinical trials to treat BRCA1- and BRCA2-deficient tumors that are defective in homologous recombination (Fong et al., 2009; Gelmon et al., 2011).
Table 1.
Summary of BRCA1-related miRNAs and related signaling
| Sub group | miRNA | Related pathways | References |
|---|---|---|---|
| Upstream signals | |||
| Targeting BRCA1 | miR-182 | Repressed by IR | Moskwa et al. (2011) |
| miR-146a, miR146-5p | Diflourinated-curcumin (CDF) / EZH2 | Garcia et al. (2011); Shen et al. (2008) | |
| miR-15a, miR-16 | Induced by circumin | Zhu et al. (2009) | |
| miR-638 | Induced by benzo(a)pyrene (BaP) | Li et al. (2012). | |
| miR-17 | Myc, Cyclin D1 | Shen et al. (2009) | |
| Downstream signals | |||
| Targeted by BRCA1 | miR-155 | Immune response, anti-apototic (PU.1, p53INP1…) | Rodriguez et al. (2007); Thai et al. (2007) |
| miR-148, miR-152 | Tumor-suppressor in endometrial cancer (EC) | Tsuruta et al. (2011) | |
| miR-205 | EMT, invasion (ZEB1, ZEB2, CK-epsilon) | Gregory et al. (2008) | |
| miR-99b | NF-κB, MAPK pathway (TRAF2) | Tanic et al. (2011); Zilahi et al. (2012) | |
| miR-146a | NF-κB, MAPK pathway (TRAF2) | Tanic et al. (2011); Zilahi et al. (2012) |
Fig. 1.
BRCA1-related miRNA network. miRNAs that regulate BRCA1 by interacting with its 3’UTR are shown in the upper part (dotted box). In the lower part, the miRNAs that are up- or down-regulated by BRCA1 are shown in green or gray boxes, respectively. Some of the known targets (or effect) of each miRNA are indicated in italics. A possible feedback loop for the miR-146a is indicated by a dotted line with an arrowhead.
In addition to miR-182, miR-146a and miR-146-5p have been shown to bind to the 3′ UTR of BRCA1 (Garcia et al., 2011a; Shen et al., 2008). The reduction in the BRCA1 level caused by these two miRNAs resulted in increased cell proliferation and a reduction in homologous recombination. Moreover, the authors showed that the two miRNAs are up-regulated in the basal-like tumor cell lines and triple-negative breast tumors, suggesting that the up-regulation of these two miRNAs may be the driving force for the basal-like sporadic cancers caused by a reduction in BRCA1 expression. On the other hand, miR-15a and miR-16 are identified as BRCA1-targeting miRNAs in nasopharyngeal carcinoma (NPC) (Zhu et al., 2009). It was reported that six out of eight NPCs exhibited a loss of BRCA1 expression, suggesting the involvement of BRCA1 in Epstein-Barr virus-mediated induction of NPC. Lastly, Li et al. (2012) showed that BRCA1 is also targeted by miR-638, which is induced in human bronchial epithelial cells by benzo(a)pyrene (BaP). As BaP is a known carcinogen, this finding suggests that induced miR-638 might down-regulate BRCA1, impairing DNA damage repair and causing cancer.
Polymorphism in the miRNA genes may affect BRCA1 expression
It is possible that a mutation in a gene encoding a miRNA can alter its sequence such that gene(s) that may not normally be targeted by this miRNA may become a potential target. Such mutations can have serious consequences especially if the target gene(s) are functionally important, as is the case with BRCA1. This prediction is supported by Shen et al. (2008) who found that a G-to-C polymorphism (rs2910164) in the miR-146a precursor resulted in increased production of mature miR-146a. In addition, the authors found that the C allele miR-146a binds significantly more tightly to the BRCA1 UTR than the common G allele. They also revealed that carriers of this polymorphism have an earlier breast cancer diagnosis, suggesting the downregulation of BRCA1 by the polymorphic miR-146a has a clinical impact. This finding is further confirmed by a recent study conducted in a European population (Lian et al., 2012). However, Garcia et al. (2011b) found no association between the miR-146a SNP and breast cancer risk in a large-scale analysis of breast cancer samples. Clearly, further studies are needed to establish the possible role of a miR-146a polymorphism on BRCA1 regulation. In another study, Shen et al. (2009) found that a rare variant in the miR-17 gene resulted in altered expression of mature miR-17, which could bind to the BRCA1 UTR. These results suggest that the variation in the miRNA gene is one of the mechanisms for the miRNA-mediated downregulation of BRCA1.
Mutation in the BRCA1 UTR may affect miRNA binding
In addition to the polymorphisms in miRNAs, variations in the UTR of BRCA1 may alter the miRNA regulation on BRCA1. Notably, if the mutation creates or enhances a specific miRNA interaction, the effect can be similar to a complete loss of function mutation in BRCA1. Indeed, Nicoloso et al. (2010) reported that the rs799917-BRCA1 SNP is associated with a higher risk of breast cancer and showed that this SNP is more susceptible to regulation by miR-638. In keeping with this finding, we identified, through bioinformatics analysis of BRCA1 UTRs, several SNPs that can potentially generate miRNA binding sites. A number of other reports have also identified BRCA1 UTR variations associated with breast cancer risk. For example, two polymorphic alleles, c.5711+421 (G or T) and the c.5711+1286(C or T), showed a significant association with cancer (Pongsavee et al., 2009). Likewise, rs8176318 on the BRCA1 3’UTR is also shown to be associated with breast cancer risk (Pelletier et al., 2011). Identifying miRNAs that potentially control these UTR variations will provide a better understanding of the clinical impact of these variants.
BRCA1 CONTROLS THE EXPRESSION OF MIRNAS
BRCA1 down-regulates miRNAs
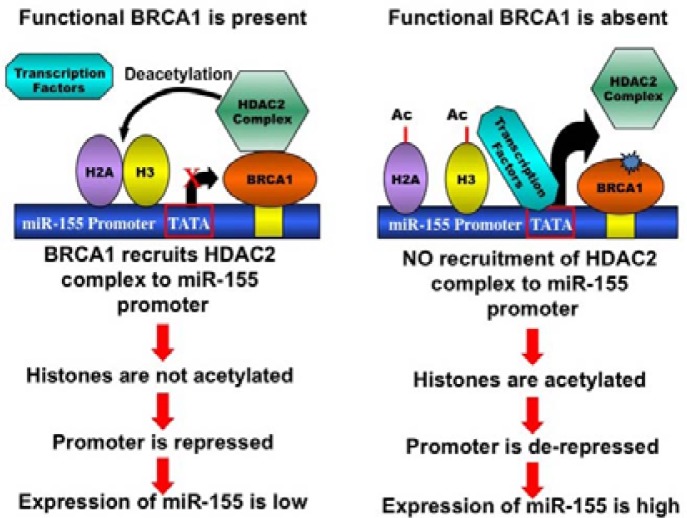
In addition to the DNA damage repair function, BRCA1 exerts its effect on diverse cellular processes such as cell cycle progression, chromatin remodeling, and transcriptional regulation (Huen et al., 2010; Ohta et al., 2011). Because of its role as a transcriptional regulator, BRCA1 has been predicted to regulate the expression of miRNAs. This prediction is supported by the identification of a number of miRNAs that are regulated by BRCA1 (Table 1). Recently, we discovered a novel role of BRCA1 in regulating the expression of an oncogenic miRNA, miR-155 (Chang et al., 2011). Using a miRNA array-based expression analysis in differentiating mouse embryonic stems cells with BRCA1 mutation, we identified miR-155 as one of the up-regulated miRNAs. Further investigation revealed that BRCA1 suppresses the miR-155 promoter epigenetically via its interaction with the HDAC2 complex (Fig. 2). This finding represents an example of how BRCA1 can exert its tumor suppression function through miRNA regulation. We also found miR-652 to be up-regulated in BRCA1 mutant cells. At present, it is not clear whether, like miR-155, miR-652 is directly repressed by BRCA1. Considering that BRCA1 functions as a transcription co-repressor for several genes (Aiyar et al., 2007; Wang et al., 1998; Yu et al., 1998), it will be meaningful to find other oncogenic miRNAs that are directly regulated by BRCA1. Of note, a recent report showed that DNMT1, which encodes a methylation maintenance enzyme, is a transcriptional target of BRCA1 (Shukla et al., 2010). The authors also showed that the loss of BRCA1 resulted in global DNA hypomethylation, suggesting that some miRNAs are indirectly repressed by BRCA1 through the promoter hypomethylation.
Fig. 2.
BRCA1-dependent epigenetic regulation of miR-155 promoter. In the BRCA1 wild-type cells (on left), there is an association of the HDAC2 complex on the miR-155 promoter, mediated by BRCA1. The HDAC2 complex deacetylates H2A and H3, thereby epigenetically repressing the miR-155 promoter. In the BRCA1 mutant cells (on right), the HDAC2 association on the miR-155 promoter is disrupted. As a result, there is more H2A and H3 acetylation, which in turn activates the miR-155 promoter epigenetically.
BRCA1 up-regulates miRNAs
It is well known that BRCA1 is a component of the RNA Pol II holoenzyme complex (Anderson et al., 1998; Scully et al., 1997) and induces a set of target genes, including p21, Gadd45a (Harkin et al., 1999; Somasundaram et al., 1997). A recent genome-wide study identified 44 gene promoters associated with BRCA1 (Gorski et al., 2011). Consistent with these findings, BRCA1 has been found to be involved in the transcriptional activation of miRNAs, as shown in Fig. 1. Tanic et al. showed that miR-146a, miR-99b, and miR-205 are induced in BRCA1-deficient HCC1937 cells after they were reconstituted with BRCA1 (Tanic et al., 2011). Interestingly, miR-146a has also been shown to regulate BRCA1 (Garcia et al., 2011a), suggesting a possible negative feedback loop. Our study also showed that several miRNAs, including miR-148, miR-152, and miR-744 (Chang et al., 2011), are down-regulated in mutant BRCA1 cells. Interestingly, miR-152 is identified as a tumor-suppressor miRNA in endometrial cancer (Tsuruta et al., 2011). Thus, it is possible that BRCA1 exerts its tumor-suppressor function by up-regulating tumor-suppressor miRNAs. Furthermore, hypermethylation was shown in the promoters of mir-148 and mir-152 in 34–86% of 71 primary human breast cancer specimens (Lehmann et al., 2007), suggesting the functional impact of the repression of these miRNAs. At this point, it is not clear whether these miRNAs are regulated directly or indirectly by BRCA1. Further studies, including the examination of methylation control of the promoters of the miRNA genes and the identification of transcriptional activators, will reveal the role of BRCA1 in the control of these miRNAs.
SIGNALING PATHWAYS AFFECTING THE MIRNAS THAT TARGET BRCA1
miR-146a
A recent report revealed that diflourinated-curcumin (CDF) treatment on pancreatic cancer cells reduces EZH2 expression (Bao et al., 2012). Several tumor-suppressor miRNAs are induced by EZH2, including let-7a, b, c, d, miR-26a, miR-101, miR-146a, and miR-200b and c. Because EZH2 is a histone methyltransferase involved in epigenetic regulation of many genes (Simon and Lange, 2008), the effect of EZH2 on miR-146a expression suggests an epigenetic regulation of this miRNA. Once CDF reduces the EZH2 level, the induction of miR-146a will be compromised by the increase in methylated histones on the promoter. Conversely, the increased level of EZH2 is likely to result in more miR-146a, which in turn will down-regulate BRCA1. Consistent with this expectation, knockdown of EZH2 was shown to increase BRCA1 (Gonzalez et al., 2009). Interestingly, a report showed EZH2 is up-regulated in BRCA1-deficient tumors and tumor cell lines (Puppe et al., 2009), suggesting a positive feedback loop that down-regulates BRCA1 by the EZH2-miR-146a pathway.
miR-15a and miR-16
miR-15a has been proposed to be a tumor-suppressor miRNA because it induces apoptosis and reduces tumorigenicity in chronic lymphocytic leukemia through the regulation of antiapoptotic Bcl-2 expression (Cimmino et al., 2005). This suggestion is further supported by Yang et al., who showed curcumin, an anti-cancer agent, up-regulates miR-15a and miR-16, which can down-regulate Bcl-2 in MCF7 cells (Yang et al., 2010). Since miR-15a and miR-16 were shown to target BRCA1, these miRNAs appear to have a dual role. Further studies will reveal how these two functions are regulated in different cellular contexts. Interestingly, Cittelly et al. (2010) showed that the HER2Δ16, which is an oncogenic isoform of HER2, suppresses miR-15a and miR-16. This study also showed that decreased miR-15a and miR-16 confer tamoxifen resistance in ER-α positive cells by up-regulating Bcl-2. It is not clear whether the increased miR-15a and miR-16 can target Bcl-2 in NPC, where BRCA1 was identified as a target of these miRNAs. Further studies are needed to understand the effect of oncogenic HER2Δ 16 on the regulation of BRCA1 through miR-15a and miR-16.
miR-182
miR-182 was recently identified as a miRNA that is downregulated in response to IR in undifferentiated, proliferating progenitor K562 cells (Moskwa et al., 2011). It was reasoned that decreased expression of miRNA may allow more production of the DNA damage repair proteins that will be used to repair the IR-induced DNA damage in undifferentiated cells than in terminally differentiated cells. As mentioned above, miR-182 was shown to regulate the expression of BRCA1. A recent study showed miR-182 to be regulated by RNA helicase DDX5 in basal breast cancer cells (Wang et al., 2012). Interestingly, another study found that p53 interacts with the Drosha processing complex through its association with DDX5, which facilitates the processing of primary miRNAs, including miR-16-1 and miR-143 (Suzuki et al., 2009). Therefore, it will be interesting to examine whether the DDX5/p53/Drosha complex can also control the processing of miR-182, especially after IR.
miR-17
miR-17 is transcribed from a miRNA cluster gene, miR-17-92. A study by Castellano et al. (2009) revealed that the miR-17-92 is transactivated by Myc in an ER-alpha-dependent manner. Because miR-17 targets BRCA1, this regulation suggests a mechanism by which the oncogene Myc may repress tumor-suppressor BRCA1. On the other hand, a study by Yu et al. (2008) showed that Cyclin D1 induces miR-17-92, and miR-17/20a represses Cyclin D1 levels, forming a negative feedback loop. Because BRCA1 is dynamically regulated during cell cycle progression, the feedback loop suggests a possible mechanism of cell cycle-associated change in the BRCA1 level.
TARGET MRNAS AND SIGNALING PATHWAYS AFFECTED BY THE BRCA1-DEPENDENT MIRNAS
miR-155
The role of miR-155 has been extensively studied in the immune system. The importance of miR-155 in the immune system was evident by the phenotype of the miR-155 knockout mice (Rodriguez et al., 2007; Thai et al., 2007; Tili et al., 2009), which were immunodeficient (Turner and Vigorito, 2008). Mice lacking miR-155 are viable and fertile but are deficient in lymphocyte development and generation of B- and T-cell responses after B-cell receptor or T-cell receptor activation. Also, dendritic cells in miR-155-deficient mice have been shown to have an impaired antigen-presenting function (Martinez-Nunez et al., 2009), supporting the importance of miR-155 in the immune cells. Many known targets of miR-155 (more than 40 so far), such as PU.1 (Martinez-Nunez et al., 2009), c-MAF (Thai et al., 2007), SHIP-1 (Kurowska-Stolarska et al., 2011), and SOCS-1, have been shown to affect cell differentiation and function (Tili et al., 2009). Interestingly, SOCS-1 is also reported to be a target of miR-155 in breast cancer cells (Jiang et al., 2010). Jiang et al. showed that SOCS-1 knockdown has a proliferation effect similar to that of the miR-155 expression, whereas restoration of SOCS-1 expression can block the oncogenic potential of miR-155 in MDA-MB-231 cells. On the other hand, FOXO3a, which affects cell survival and chemosensitivity, is also reported to be a target of miR-155 (Kong et al., 2010). Lastly, a SILAC analysis to identify the targets of miR-155 revealed 46 putative miR-155 target proteins in HEK293 cells (Lossner et al., 2011). The authors identified CKAP5 as a novel target of miR-155 and suggested the role of miR-155 in the cell cycle regulation. These results suggest that the repression of multiple targets by miR-155 may contribute to breast tumor formation.
miR-146a and miR-99b
miR-146a and -99b negatively regulate NF-kappa B activity through the TRAF6 and IRAK1 (Tanic et al., 2011; Zilahi et al., 2012). It has been reported that miR-146a can inhibit the invasion and migration potential of highly metastatic human breast cancer cell line MDA-MB-231 (Bhaumik et al., 2008). Likewise, a study in androgen-independent cell lines revealed that miR-146a inhibits cell proliferation and migration as well as in vivo tumor growth and angiogenesis by repressing EGFR and MMP2 (Xu et al., 2011). Additionally, a recent report showed miR-146a also controls SMAD4 (Liu et al., 2012; Xiao et al., 2012). These data demonstrate the tumor-suppressive role of miR-146a. Because miR-146a not only represses BRCA1, but it can also be up-regulated by BRCA1, suggesting that this feedback loop may contribute to fine tuning the level of expression of BRCA1 and/or miR-146a (Fig. 1). Importantly, another study using a knockout mouse model confirmed IRAK1 and TRAF6 to be targets of miR-146a (Zhao et al., 2011). These are key molecules in proinflammatory responses and adaptor/scaffold proteins in the IL-1 as well as in the Toll-like receptor signaling pathway. These pathways are known to positively regulate NF-kappa B activity (Andreakos et al., 2005). Collectively, these studies suggest that miR-146a negatively regulates NF-kappa B signaling and plays tumor-suppressive as well as anti-inflammatory roles.
miR-205
Along with the miR-200 family, the miR-205 was shown to control the expression of E-cadherin transcriptional repressors ZEB1 (also known as deltaEF1) and SIP1 (also known as ZEB2) (Gregory et al., 2008). Gregory et al. found that these miRNAs are down-regulated during epithelial-mesenchymal transition (EMT) and subsequently confirmed this finding in invasive cancer cell lines with a mesenchymal phenotype. Moreover, Wu et al. (2009) showed suppression of cell growth and invasion caused by the ectopic expression of miR-205. They further showed that Protein Kinase C-epsilon is the target of miR-205 in this process. These data demonstrate that miR-205 has a tumor-suppression function, which may be enhanced by its upregulation via BRCA1.
MIRNAS IN BRCA1-MEDIATED TUMOR SUPPRESSION
miR-335
Overexpression of miR-335 has been shown to up-regulate BRCA1 through activators such as ERα, IGF1R, and SP1 (Heyn et al., 2011), resulting in decreased cell viability and increased apoptosis. It has been reported that miR-335 acts as a metastasis suppressor in human breast cancer (Tavazoie et al., 2008), suggesting that this miRNA has a tumor-suppressive function by up-regulating BRCA1 expression.
THERAPEUTIC POTENTIAL OF TARGETING THE BRCA1-RELATED MIRNAS
Along with the rigorous research to reveal the role of miRNA in human diseases, the therapeutic potential of anti-miRNAs or miRNA mimics has also been tested extensively. So far, the only miRNA-targeting agent in clinical trials is anti-miR-122, which was shown to regulate hepatitis C virus (HCV) (Sarasin-Filipo-wicz et al., 2009). However, many other miRNA-targeting agents (antagomir or mimic) have been tested in various mouse models and shown to have therapeutic potential. Some of the BRCA1-associated miRNAs were also targeted, and the results are summarized in Table 2. Interestingly, many of these antagomirs or miRNA mimics have been tested in the treatment of various diseases or conditions other than cancer, providing the potential for using the same agent for the treatment of BRCA1-associated breast cancer. For example, anti-miR-182 was successfully used to inhibit helper T-lymphocyte expansion (Stittrich et al., 2010), and the introduction of double-stranded miR-146a resulted in the suppression of cartilage and bone destruction (Nakasa et al., 2011). Likewise, the inhibition of miR-17 was shown to improve heart and lung function in pulmonary hypertension (Pullamsetti et al., 2012). All of these miRNAs were shown to target BRCA1 mRNA, as described above. Therefore, the inhibition of these miRNAs may increase the expression of BRCA1, leading to an enhanced tumor-suppressive network.
Table 2.
Therapeutic Potential of BRCA1 related miRNAs
| miRNA | Type | Disease models | References |
|---|---|---|---|
| miR-182 | AntagomiR | Melanoma -Liver metastasis | Segura et al. (2009) |
| AntagomiR | Helper T cell mediated immune response | Stittrich et al. (2010) | |
| miR-146a | Mimic | Rheumatoid arthritis | Nakasa et al. (2011) |
| miR-15a / miR-16a | AntagomiR | Prostate Cancer | Bandi et al. (2009) |
| miR-17 | AntagomiR | Pulmonary hypertension | Pullamsetti et al. (2012) |
| miR-155 | Mimic | Tumor associated dendritic cell-Ovarian cancer | Cubillos-Ruiz et al. (2012) |
| miR-205 | Mimic | Renal cancer | Majid et al. (2011) |
| Mimic | Breast cancer (Targeting HER3) | Iorio et al. (2009) | |
| miR-99a/99b | AntagomiR | Breast Cancer (EMT) | Turcatel et al. (2012). |
A study by Bandi et al. (2009) revealed that the expression of miR-16 and miR-15a induced growth arrest and apoptosis that resulted in the inhibition of prostate tumor proliferation in xenografts. On the other hand, inhibition of miR-182 was shown to impede the metastasis of melanoma cells (Segura et al., 2009). Also, Iorio et al. showed that miR-205 targets HER3, which inhibits downstream Akt in breast cancer cells (Iorio et al., 2009). Additionally, the expression of miR-205 was recently shown to inhibit renal tumor cell growth in vivo (Majid et al., 2011), suggesting that the same miRNA mimic has a tumor-suppressive effect in other types of cancer. A recent report provided a good example of how to take advantage of the spontaneous, enhanced endocytic activity of ovarian cancer-associated dendritic cells (Cubillos-Ruiz et al., 2012). The authors used an immune stimulatory miR-155 mimic to successfully activate dendritic cells that abrogated the progression of ovarian cancer. Lastly, the inhibition of miR-99a and -99b was recently shown to induce the EMT of breast epithelial cells (Turcatel et al., 2012). Since miR-99b is positively regulated by BRCA1, the use of a miR-99b mimic might have the potential to inhibit the metastasis of BRCA1-deficient tumors.
CONCLUSIONS
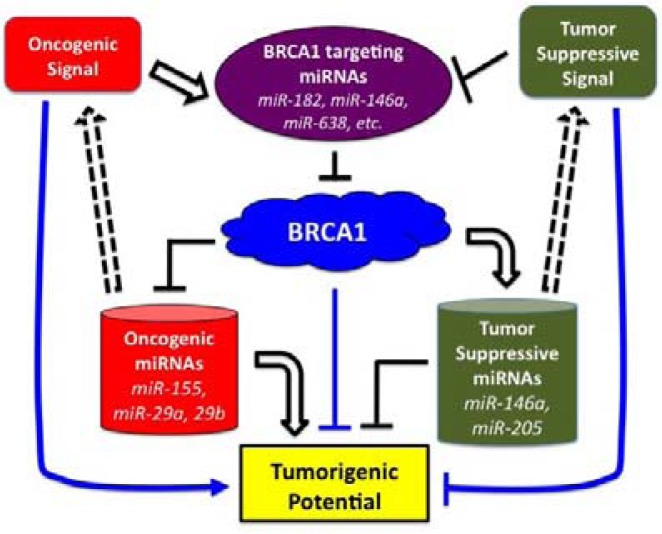
As summarized in Table 1, the number of miRNAs either shown to regulate the expression of BRCA1 or identified as a transcriptional target of BRCA1 is rapidly increasing. Considering the tumor-suppressive role of BRCA1, any perturbation in these regulatory functions is likely to have an effect on BRCA1-mediated tumorigenesis. For example, up-regulation of BRCA1-targeting miRNAs may result in a reduction in levels of BRCA1, which will cause genomic instability due to defects in DNA damage repair. On the other hand, loss of BRCA1 can up-regu-late oncogenic miRNAs, such as miR-155, or down-regulate tumor-suppressive miRNAs, such as miR-205. It is important to note that the change in the expression of these miRNAs will affect the protein level of multiple target proteins, which, in turn, triggers changes in downstream signaling and thereby perturbing diverse cellular processes (Fig. 3). Similarly, there are upstream signaling pathways that affect the level of miRNAs that regulate BRCA1. Integrating these signaling pathways and understanding their interactions is challenging, but will certainly be useful for the development of therapeutic agents targeting these networks. For example, the use of miRNA agents, such as a miR-155 antagomir or a miRNA mimic of miR-205, in BRCA1-associated breast cancer would be of great interest. Finally, combining these miRNA agents with other therapeutic drugs might be a useful strategy for treating BRCA1-associated human cancers.
Fig. 3.
Schematic diagram showing the concept of the BRCA1-associated miRNA network. There are oncogenic and tumor-suppressive signals that control the level of BRCA1-targeting miRNAs. The BRCA1, on the other hand, exerts its function by up-regulating tumor-suppressive miRNAs (right barrel) or down-regulating oncogenic miRNAs (left barrel). Additionally, these BRCA1-dependent miRNAs can alter oncogenic or tumor-suppressive signaling through their target regulation (Table 1). The integrated output of this miRNA-BRCA1 network determines the tumorigenic potential of a cell (box at the bottom).
Acknowledgments
This work is supported by the Center for Cancer Research Intramural Program, National Cancer Institute, National Institutes of Health, a part of the U.S. Department of Health and Human Services.
REFERENCES
- Aiyar S.E., Cho H., Lee J., Li R. Concerted transcriptional regulation by BRCA1 and COBRA1 in breast cancer cells. Int. J. Biol. Sci. 2007;3:486–492. doi: 10.7150/ijbs.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S.F., Schlegel B.P., Nakajima T., Wolpin E.S., Parvin J.D. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat. Genet. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- Andreakos E., Sacre S., Foxwell B.M., Feldmann M. The toll-like receptor-nuclear factor kappaB pathway in rheumatoid arthritis. Front. Biosci. 2005;10:2478–2488. doi: 10.2741/1712. [DOI] [PubMed] [Google Scholar]
- Bandi N., Zbinden S., Gugger M., Arnold M., Kocher V., Hasan L., Kappeler A., Brunner T., Vassella E. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–5559. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- Bao B., Ali S., Banerjee S., Wang Z., Logna F., Azmi A.S., Kong D., Ahmad A., Li Y., Padhye S., et al. Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer Res. 2012;72:335–345. doi: 10.1158/0008-5472.CAN-11-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik D., Scott G.K., Schokrpur S., Patil C.K., Campisi J., Benz C.C. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27:5643–5647. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano L., Giamas G., Jacob J., Coombes R.C., Lucchesi W., Thiruchelvam P., Barton G., Jiao L.R., Wait R., Waxman J., et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc. Natl. Acad. Sci. USA. 2009;106:15732–15737. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Wang R.H., Akagi K., Kim K.A., Martin B.K., Cavallone L., Haines D.C., Basik M., Mai P., Poggi E., et al. Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nat. Med. 2011;17:1275–1282. doi: 10.1038/nm.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A., Calin G.A., Fabbri M., Iorio M.V., Ferracin M., Shimizu M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M., et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cittelly D.M., Das P.M., Salvo V.A., Fonseca J.P., Burow M.E., Jones F.E. Oncogenic HER2{Delta}16 suppresses miR-15a/16 and deregulates BCL-2 to promote endocrine resistance of breast tumors. Carcinogenesis. 2010;31:2049–2057. doi: 10.1093/carcin/bgq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz J.R., Baird J.R., Tesone A.J., Rutkowski M.R., Scarlett U.K., Camposeco-Jacobs A.L., Anadon-Arnillas J., Harwood N.M., Korc M., Fiering S.N., et al. Reprogramming tumor-associated dendritic cells in vivo using miRNA mimetics triggers protective immunity against ovarian cancer. Cancer Res. 2012;72:1683–1693. doi: 10.1158/0008-5472.CAN-11-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton D.F., Ford D., Bishop D.T. Breast and ovarian cancer incidence in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Am. J. Hum. Genet. 1995;56:265–271. [PMC free article] [PubMed] [Google Scholar]
- Fackenthal J.D., Olopade O.I. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat. Rev. Cancer. 2007;7:937–948. doi: 10.1038/nrc2054. [DOI] [PubMed] [Google Scholar]
- Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O’Connor M.J., et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A.I., Buisson M., Bertrand P., Rimokh R., Rouleau E., Lopez B.S., Lidereau R., Mikaelian I., Mazoyer S. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol. Med. 2011a;3:279–290. doi: 10.1002/emmm.201100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A.I., Cox D.G., Barjhoux L., Verny-Pierre C., Barnes D., Antoniou A.C., Stoppa-Lyonnet D., Sinilnikova O.M., Mazoyer S. The rs2910164:G>C SNP in the MIR146A gene is not associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Hum. Mutat. 2011b;32:1004–1007. doi: 10.1002/humu.21539. [DOI] [PubMed] [Google Scholar]
- Gelmon K.A., Tischkowitz M., Mackay H., Swenerton K., Robidoux A., Tonkin K., Hirte H., Huntsman D., Clemons M., Gilks B., et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- Gonzalez M.E., Li X., Toy K., DuPrie M., Ventura A.C., Banerjee M., Ljungman M., Merajver S.D., Kleer C.G. Down-regulation of EZH2 decreases growth of estrogen receptor-negative invasive breast carcinoma and requires BRCA1. Oncogene. 2009;28:843–853. doi: 10.1038/onc.2008.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski J.J., Savage K.I., Mulligan J.M., McDade S.S., Blayney J.K., Ge Z., Harkin D.P. Profiling of the BRCA1 transcriptome through microarray and ChIP-chip analysis. Nucleic Acids Res. 2011;39:9536–9548. doi: 10.1093/nar/gkr679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., Goodall G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkin D.P., Bean J.M., Miklos D., Song Y.H., Truong V.B., Englert C., Christians F.C., Ellisen L.W., Maheswaran S., Oliner J.D., et al. Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1. Cell. 1999;97:575–586. doi: 10.1016/s0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- Heyn H., Engelmann M., Schreek S., Ahrens P., Lehmann U., Kreipe H., Schlegelberger B., Beger C. MicroRNA miR-335 is crucial for the BRCA1 regulatory cascade in breast cancer development. Int. J. Cancer. 2011;129:2797–2806. doi: 10.1002/ijc.25962. [DOI] [PubMed] [Google Scholar]
- Huen M.S., Sy S.M., Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell Biol. 2010;11:138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio M.V., Casalini P., Piovan C., Di Leva G., Merlo A., Triulzi T., Menard S., Croce C.M., Tagliabue E. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69:2195–2200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
- Jiang S., Zhang H.W., Lu M.H., He X.H., Li Y., Gu H., Liu M.F., Wang E.D. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–3127. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K.L., Buzdar A.U. A review of adjuvant hormonal therapy in breast cancer. Endocr. Relat. Cancer. 2004;11:391–406. doi: 10.1677/erc.1.00594. [DOI] [PubMed] [Google Scholar]
- Kong W., He L., Coppola M., Guo J., Esposito N.N., Coppola D., Cheng J.Q. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J. Biol. Chem. 2010;285:17869–17879. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kurowska-Stolarska M., Alivernini S., Ballantine L.E., Asquith D.L., Millar N.L., Gilchrist D.S., Reilly J., Ierna M., Fraser A.R., Stolarski B., et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc. Natl. Acad. Sci. USA. 2011;108:11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann U., Hasemeier B., Romermann D., Muller M., Langer F., Kreipe H. Epigenetic inactivation of microRNA genes in mammary carcinoma. Verh. Dtsch. Ges. Pathol. 2007;91:214–220. [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li D., Wang Q., Liu C., Duan H., Zeng X., Zhang B., Li X., Zhao J., Tang S., Li Z., et al. Aberrant expression of miR-638 contributes to benzo(a) pyrene-induced human cell transformation. Toxicol. Sci. 2012;125:382–391. doi: 10.1093/toxsci/kfr299. [DOI] [PubMed] [Google Scholar]
- Lian H., Wang L., Zhang J. Increased risk of breast cancer associated with CC genotype of Has-miR-146a Rs29 10164 polymorphism in Europeans. PLoS One. 2012;7:e31615. doi: 10.1371/journal.pone.0031615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Lu C.L., Cui L.P., Hu Y.L., Yu Q., Jiang Y., Ma T., Jiao D.K., Wang D., Jia C.Y. MicroRNA-146a modulates TGF-beta1-induced phenotypic differentiation in human dermal fibroblasts by targeting SMAD4. Arch. Dermatol. Res. 2012;304:195–202. doi: 10.1007/s00403-011-1178-0. [DOI] [PubMed] [Google Scholar]
- Lossner C., Meier J., Warnken U., Rogers M.A., Lichter P., Pscherer A., Schnolzer M. Quantitative proteomics identify novel miR-155 target proteins. PLoS One. 2011;6:e22146. doi: 10.1371/journal.pone.0022146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid S., Saini S., Dar A.A., Hirata H., Shahryari V., Tanaka Y., Yamamura S., Ueno K., Zaman M.S., Singh K., et al. MicroRNA-205 inhibits Src-mediated oncogenic pathways in renal cancer. Cancer Res. 2011;71:2611–2621. doi: 10.1158/0008-5472.CAN-10-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Nunez R.T., Louafi F., Friedmann P.S., Sanchez-Elsner T. MicroRNA-155 modulates the pathogen binding ability of dendritic cells (DCs) by down-regulation of DC-specific intercellular adhesion molecule-3 grabbing non-integrin (DC-SIGN) J. Biol. Chem. 2009;284:16334–16342. doi: 10.1074/jbc.M109.011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y., Swensen J., Shattuck-Eidens D., Futreal P.A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L.M., Ding W., et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- Moskwa P., Buffa F.M., Pan Y., Panchakshari R., Gottipati P., Muschel R.J., Beech J., Kulshrestha R., Abdelmohsen K., Weinstock D.M., et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell. 2011;41:210–220. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison E.P., Hannon G.J. miRNAs on the move miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 2004;16:223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Nakasa T., Shibuya H., Nagata Y., Niimoto T., Ochi M. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum. 2011;63:1582–1590. doi: 10.1002/art.30321. [DOI] [PubMed] [Google Scholar]
- Nelson P., Kiriakidou M., Sharma A., Maniataki E., Mourelatos Z. The microRNA world small is mighty. Trends Biochem. Sci. 2003;28:534–540. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Nicoloso M.S., Sun H., Spizzo R., Kim H., Wickramasinghe P., Shimizu M., Wojcik S.E., Ferdin J., Kunej T., Xiao L., et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70:2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., Sato K., Wu W. The BRCA1 ubiquitin ligase and homologous recombination repair. FEBS Lett. 2011;585:2836–2844. doi: 10.1016/j.febslet.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Pelletier C., Speed W.C., Paranjape T., Keane K., Blitzblau R., Hollestelle A., Safavi K., van den Ouweland A., Zelterman D., Slack F.J., et al. Rare BRCA1 haplotypes including 3’UTR SNPs associated with breast cancer risk. Cell Cycle. 2011;10:90–99. doi: 10.4161/cc.10.1.14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongsavee M., Yamkamon V., Dakeng S., O-charoenrat P., Smith D.R., Saunders G.F., Patmasiriwat P. The BRCA1 3′-UTR: 5711+421T/T_5711+1286T/T genotype is a possible breast and ovarian cancer risk factor. Genet. Test Mol. Biomarkers. 2009;13:307–317. doi: 10.1089/gtmb.2008.0127. [DOI] [PubMed] [Google Scholar]
- Pullamsetti S.S., Doebele C., Fischer A., Savai R., Kojonazarov B., Dahal B.K., Ghofrani H.A., Weissmann N., Grimminger F., Bonauer A., et al. Inhibition of microRNA-17 improves lung and heart function in experimental pulmonary hypertension. Am. J. Respir. Crit. Care Med. 2012;185:409–419. doi: 10.1164/rccm.201106-1093OC. [DOI] [PubMed] [Google Scholar]
- Puppe J., Drost R., Liu X., Joosse S.A., Evers B., Cornelissen-Steijger P., Nederlof P., Yu Q., Jonkers J., van Lohuizen M., et al. BRCA1-deficient mammary tumor cells are dependent on EZH2 expression and sensitive to polycomb repressive complex 2-inhibitor 3-deazaneplanocin A. Breast Cancer Res. 2009;11:R63. doi: 10.1186/bcr2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramus S.J., Gayther S.A. The contribution of BRCA1 and BRCA2 to ovarian cancer. Mol. Oncol. 2009;3:138–150. doi: 10.1016/j.molonc.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Vigorito E., Clare S., Warren M.V., Couttet P., Soond D.R., van Dongen S., Grocock R.J., Das P.P., Miska E.A., et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M., Krol J., Markiewicz I., Heim M.H., Filipowicz W. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat. Med. 2009;15:31–33. doi: 10.1038/nm.1902. [DOI] [PubMed] [Google Scholar]
- Scully R., Anderson S.F., Chao D.M., Wei W., Ye L., Young R.A., Livingston D.M., Parvin J.D. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA. 1997;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura M.F., Hanniford D., Menendez S., Reavie L., Zou X., Alvarez-Diaz S., Zakrzewski J., Blochin E., Rose A., Bogunovic D., et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc. Natl. Acad. Sci. USA. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Ambrosone C.B., DiCioccio R.A., Odunsi K., Lele S.B., Zhao H. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–1966. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- Shen J., Ambrosone C.B., Zhao H. Novel genetic variants in microRNA genes and familial breast cancer. Int. J. Cancer. 2009;124:1178–1182. doi: 10.1002/ijc.24008. [DOI] [PubMed] [Google Scholar]
- Shukla V., Coumoul X., Lahusen T., Wang R.H., Xu X., Vassilopoulos A., Xiao C., Lee M.H., Man Y.G., Ouchi M., et al. BRCA1 affects global DNA methylation through regulation of DNMT1. Cell Res. 2010;20:1201–1215. doi: 10.1038/cr.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J.A., Lange C.A. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Somasundaram K., Zhang H., Zeng Y.X., Houvras Y., Peng Y., Wu G.S., Licht J.D., Weber B.L., El-Deiry W.S. Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21WAF1/CiP1. Nature. 1997;389:187–190. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- Stittrich A.B., Haftmann C., Sgouroudis E., Kuhl A.A., Hegazy A.N., Panse I., Riedel R., Flossdorf M., Dong J., Fuhrmann F., et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat. Immunol. 2010;11:1057–1062. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- Suzuki H.I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- Tanic M., Zajac M., Gomez-Lopez G., Benitez J., Martinez-Delgado B. Integration of BRCA1-mediated miRNA and mRNA profiles reveals microRNA regulation of TRAF2 and NFkappaB pathway. Breast Cancer Res. Treat. 2011. s10549-011-1905-4. [DOI] [PubMed]
- Tavazoie S.F., Alarcon C., Oskarsson T., Padua D., Wang Q., Bos P.D., Gerald W.L., Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai T.H., Calado D.P., Casola S., Ansel K.M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J.L., et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Tili E., Croce C.M., Michaille J.J. miR-155 on the crosstalk between inflammation and cancer. Int. Rev. Immunol. 2009;28:264–284. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- Tsuruta T., Kozaki K., Uesugi A., Furuta M., Hirasawa A., Imoto I., Susumu N., Aoki D., Inazawa J. miR-152 is a tumor suppressor microRNA that is silenced by DNA hyper-methylation in endometrial cancer. Cancer Res. 2011;71:6450–6462. doi: 10.1158/0008-5472.CAN-11-0364. [DOI] [PubMed] [Google Scholar]
- Turcatel G., Rubin N., El-Hashash A., Warburton D. MIR-99a and MIR-99b modulate TGF-beta induced epithelial to mesenchymal plasticity in normal murine mammary gland cells. PLoS One. 2012;7:e31032. doi: 10.1371/journal.pone.0031032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M., Vigorito E. Regulation of B- and T-cell differentiation by a single microRNA. Biochem. Soc. Trans. 2008;36:531–533. doi: 10.1042/BST0360531. [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhang H., Kajino K., Greene M.I. BRCA1 binds c-Myc and inhibits its transcriptional and transforming activity in cells. Oncogene. 1998;17:1939–1948. doi: 10.1038/sj.onc.1202403. [DOI] [PubMed] [Google Scholar]
- Wang D., Huang J., Hu Z. RNA helicase DDX5 regulates microRNA expression and contributes to cytoskeletal reorganization in basal breast cancer cells. Mol. Cell. Proteomics. 2012;11:M111. doi: 10.1074/mcp.M111.011932. 011932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Zhu S., Mo Y.Y. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B., Zhu E.D., Li N., Lu D.S., Li W., Li B.S., Zhao Y.L., Mao X.H., Guo G., Yu P.W., et al. Increased miR-146a in gastric cancer directly targets SMAD4 and is involved in modulating cell proliferation and apoptosis. Oncol. Rep. 2012;27:559–566. doi: 10.3892/or.2011.1514. [DOI] [PubMed] [Google Scholar]
- Xu B., Wang N., Wang X., Tong N., Shao N., Tao J., Li P., Niu X., Feng N., Zhang L., et al. MiR-146a suppresses tumor growth and progression by targeting EGFR pathway and in a p-ERK-dependent manner in castration-resistant prostate cancer. Prostate. 2011;72:1171–1178. doi: 10.1002/pros.22466. [DOI] [PubMed] [Google Scholar]
- Yang J., Cao Y., Sun J., Zhang Y. Curcumin reduces the expression of Bcl-2 by upregulating miR-15a and miR-16 in MCF-7 cells. Med. Oncol. 2010;27:1114–1118. doi: 10.1007/s12032-009-9344-3. [DOI] [PubMed] [Google Scholar]
- Yu X., Wu L.C., Bowcock A.M., Aronheim A., Baer R. The C-terminal (BRCT). domains of BRCA1 interact in vivo with CtIP a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- Yu Z., Wang C., Wang M., Li Z., Casimiro M.C., Liu M., Wu K., Whittle J., Ju X., Hyslop T., et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J. Cell Biol. 2008;182:509–517. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.L., Rao D.S., Boldin M.P., Taganov K.D., O’Connell R.M., Baltimore D. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl. Acad. Sci. USA. 2011;108:9184–9189. doi: 10.1073/pnas.1105398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.Y., Pfuhl T., Motsch N., Barth S., Nicholls J., Grasser F., Meister G. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J. Virol. 2009;83:3333–3341. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilahi E., Tarr T., Papp G., Griger Z., Sipka S., Zeher M. Increased microRNA-146a/b, TRAF6 gene and decreased IRAK1 gene expressions in the peripheral mononuclear cells of patients with Sjogren’s syndrome. Immunol. Lett. 2012;141:165–168. doi: 10.1016/j.imlet.2011.09.006. [DOI] [PubMed] [Google Scholar]