Abstract
Nitric oxide (NO) is known for its role in the activation of plant defense responses. To examine the involvement and mode of action of NO in plant defense responses, we introduced calmodulin-dependent mammalian neuronal nitric oxide synthase (nNOS), which controls the CaMV35S promoter, into wild-type and NahG tobacco plants. Constitutive expression of nNOS led to NO production and triggered spontaneous induction of leaf lesions. Transgenic plants accumulated high amounts of H2O2, with catalase activity lower than that in the wild type. nNOS transgenic plants contained high levels of salicylic acid (SA), and they induced an array of SA-, jasmonic acid (JA)-, and/or ethylene (ET)-related genes. Consequently, NahG co-expression blocked the induction of systemic acquired resistance (SAR)-associated genes in transgenic plants, implying SA is involved in NO-mediated induction of SAR genes. The transgenic plants exhibited enhanced resistance to a spectrum of pathogens, including bacteria, fungi, and viruses. Our results suggest a highly ranked regulatory role for NO in SA-, JA-, and/or ET-dependent pathways that lead to disease resistance.
Keywords: nitric oxide (NO), nitric oxide synthase (NOS), plant defense signaling, reactive oxygen species, salicylic acid
INTRODUCTION
Nitric oxide (NO) is a ubiquitous biological messenger that is involved in a wide range of functions such as regulation of vascular tone, neuronal communication, and immune response to infection in animal cells (Schmidt and Walter, 1994). In 1998, NO was shown to have a role in plants (Delledonne et al., 1998; Durner et al., 1998), followed by detection of nitric oxide synthase (NOS)-like activity in various plants (Barroso et al., 1999; Corpas et al., 2009; Wendehenne et al., 2001). Several studies have shown that NO may also function as an important signaling molecule during various aspects of plant growth and development, such as photomorphogenesis, leaf expansion, root growth, senescence, programmed cell death, and cytokinin signaling (Beligni and Lamattina, 2000; Beligni et al., 2002; Gupta et al., 2011; Leshem, 1996; Leshem et al., 1998; Tun et al., 2001).
Nitric oxide (NO) is a ubiquitous biological messenger that is involved in a wide range of functions such as regulation of vascular tone, neuronal communication, and immune response to infection in animal cells (Schmidt and Walter, 1994). In 1998, NO was shown to have a role in plants (Delledonne et al., 1998; Durner et al., 1998), followed by detection of nitric oxide synthase (NOS)-like activity in various plants (Barroso et al., 1999; Corpas et al., 2009; Wendehenne et al., 2001). Several studies have shown that NO may also function as an important signaling molecule during various aspects of plant growth and development, such as photomorphogenesis, leaf expansion, root growth, senescence, programmed cell death, and cytokinin signaling (Beligni and Lamattina, 2000; Beligni et al., 2002; Gupta et al., 2011; Leshem, 1996; Leshem et al., 1998; Tun et al., 2001).
NO, along with salicylic acid (SA) and reactive oxygen species (ROS), also plays a pivotal role in signaling and initiation of plant defense responses to microbial pathogens (Durner and Klessig, 1999; Van Camp et al., 1998; Wendehenne et al., 2001; Yoshioka et al., 2008; 2011). When NO is exogenously applied to tobacco and soybean suspension cells, it triggers the expression of several defense-related genes and potentiates ROS-induced cell death (Delledonne et al., 1998; 2001; Durner et al., 1998;). Infection with either avirulent bacterial pathogens or elicitors triggers an NO burst in suspension-cultured Arabidopsis and tobacco cells (Clarke et al., 2000; Foissner et al., 2000). A NOS-like enzyme (AtNOS1) has been identified in Arabidopsis thaliana but is not directly involved in NO synthesis; therefore, AtNOS1 was renamed AtNOA1 for NO ASSOCIATED1 (Crawford et al., 2006; Guo et al., 2003; Zemojtel et al., 2006). Recently, AtNOA1 showed GTPase activity and was needed in chloroplast biogenesis (Flores-Pérez et al., 2008; Moreau et al., 2008; Sudhamsu et al., 2008; Van Ree et al., 2011). Although supported by a growing body of evidence that suggests the existence of NOS-like activities in higher plants, no genuine plant NOS gene has been cloned to date.
Plant cells evoke multiple responses to defend themselves against pathogenic infections, such as the hypersensitive response (HR) and systemic acquired resistance (SAR). During plant defense against microbial pathogens, SA and NO play key roles as second messengers. In addition to SA-dependent defense responses, either plant hormones or growth regulators, such as jasmonic acid (JA) and ethylene (ET), may function as alternative signals that induce resistance against necrotrophic pathogens and regulate a subset of pathogenesis-related (PR) genes, including PR-3, PR-4 (also called as Hel) and PDF1.2 (Dong, 1998; Thomma et al., 2001). The reported contribution by NO to disease resistance, against necrotrophic pathogens, might imply that NO functions as a key factor in plant adaptation to a wide spectrum of pathogens (Asai and Yoshioka, 2009).
Relationships between NO, SA, and ROS during the establishment of disease resistance have been studied. Similar to HR, NO acts synergistically with ROS to potentiate cell death in soybean suspension cells (Delledonne et al., 1998). NO also functions independently from ROS to induce defense-related gene expression. NOS inhibitors have been shown to compromise the HR in Arabidopsis and tobacco (Delledonne et al., 1998; 2001; Durner et al., 1998). In addition, NO appears to activate defense responses through an SA-dependent signaling pathway: NO treatment of tobacco leaves led to a significant increase in endogenous SA, as well as in defense-related gene expression, but it failed to increase these same responses in NahG transgenic tobacco plants (Durner et al., 1998). NO-releasing compounds also induce disease resistance against tobacco mosaic virus (TMV) in tobacco, and NO is required for the development of SAR, which is induced by SA in TMV-infected tobacco (Song and Goodman, 2001). NO, therefore, may act synergistically with ROS and SA to transduce plant defense signals. Furthermore, S-nitrosylation is an important route for the transfer of NO bioactivity, and it is significantly involved in plant defense signaling (Feechan et al., 2005; Yu et al., 2012). In S-nitrosylation, a NO moiety is covalently attached to a protein cysteine thiol to form an S-nitrosothiol, which is recently emerging as a key regulatory process during the establishment of plant disease resistance (Spadaro et al., 2010; Tada et al., 2008; Yun et al., 2011).
To understand the involvement of NO in plant defense responses in more detail, we generated transgenic tobacco plants that over-express mammalian NOS cDNA. NOS transgenic tobacco plants exhibited HR-like lesions and accumulated both SA and ROS. In addition to SA-responsive genes, we also observed up-regulation of JA/ET-responsive gene expression, including PIN2, EREBP, and ACC oxidase, in the transgenic tobacco plants. Most importantly, the over-expression of mammalian NOS cDNA conferred broad-spectrum resistance against bacterial, fungal, and viral pathogens. The results suggest that, together with SA and ROS, JA and/or ET participate in NO-mediated plant defense signaling.
MATERIALS AND METHODS
Construction of transgenic plants, plant materials, and growth conditions
For the construction of transgenic plants, rat brain NOS cDNA was first ligated into a plant binary vector (pCAMBIA1300) that harbors hpt as a selection marker, which was placed under the control of the CaMV35S promoter for sense orientation. The recombinant plasmid was introduced into either wild-type tobacco plants (Nicotiana tabacum cv Xanthi-nc) or NahG-10 tobacco plants (gift from J. Ryals, Paradigm Genetics, Research Triangle Park, NC), via the Agrobacterium tumefaciens, (EHA101)-mediated, leaf disc, transformation method (Horsch et al., 1985). Either the neuronal NOS (nNOS) or nNOS/NahG transformants were selected on Murashige-Skoog medium that contained either hygromycin (20 mg L−1) or hygromycin/kanamycin (100 mg L−1), respectively. T2 progeny, of transgenic plants that expressed nNOS, were used for the experiments and maintained at 25/20°C (day/night) in a growth chamber, with a 16-hr photoperiod and 65% relative humidity.
NO detection by fluorescence analysis
Intracellular NO, in leaves of tobacco transgenic plants, was detected with a fluorescent dye: 4-amino-5-methylamino-2′7′-difluorofluorescein diacetate (DAF-FM-DA, Alexis Biochemicals; Guo et al., 2003). Samples were incubated in a solution with a NO probe (10 μM DAF-FM-DA), for 30 min, washed three times with AT3 medium, and mounted on microscopic slides. Fluorescence was estimated from a confocal laser scanning microscopic image (Nikon PCM2000; excitation wavelength 488 nm; emission wavelength 515 to 530 nm).
Histochemistry and microscopy
The accumulation of autofluorescent materials was detected, using methods described by Dietrich et al. (1994). Briefly, leaves were cleared by boiling them in lactophenol, rinsing them in 50% ethanol, and rinsing them in water. We observed autofluorescence with an ultraviolet epifluorescence microscope. Staining for the presence of H2O2via the diaminobenzidine (DAB) uptake method was performed as described in Thordal-Christensen et al. (1997).
Antioxidant enzyme assays
Activities of catalase (CAT) and peroxidase (POX) were measured following the methods of Rao et al. (1997). Soluble protein (1 g) was extracted from leaves of 4-week-old tobacco plants by grinding tissues in a mortar with 3 ml of 100 mM potassium phosphate buffer (pH 7.5) that contained 2 mM Na2-EDTA, 1% (w/v) polyvinylpyrrolidone-40, and 1 mM phenylmethylsulfonyl fluoride. Extracts were centrifuged for 15 min at 12,000 × g and desalted by gel filtration (PD-10 column). CAT activity was determined after consumption of H2O2 at 240 nm for 5 min in 3 ml reaction mixture that contained 100 mM potassium phosphate buffer (pH 7.5) and 10 ml 30% H2O2. POX activity was determined at 470 nm for 5 min in 3 ml reaction mixture that contained 100 mM potassium phosphate buffer (pH 6.5), 16 mM guaiacol, and 10 ml 10% H2O2. All enzyme assays were performed at 25°C on 20 to 50 mg of protein. All experiments described above were repeated twice, with three replicates per experiment.
Measurement of SA
Plant material (usually 1.2 to 1.5 g young leaf) was grounded up and frozen in liquid nitrogen. Briefly, each tissue was extracted in 6 ml of ice-cold methanol at 4°C for 24 h and added to 3.6 ml ice-cold water and 3 ml chloroform, with 10 mM of a 3,4,5-trimethoxy-trans-cinamic acid as internal standard. After vortexing, the samples were kept at 4°C for 12 h. The combined supernatants were dried in a speed vacuum. The residue was re-suspended in 0.6 ml ice-cold water/methanol (1:1 v/v) and analyzed by high-performance liquid chromatography (HPLC), as described by Meuwly and Métraux (1993).
RNA extraction and RNA blot analysis
Isolation of total RNA and Northern hybridizations were performed as described by Park et al. (2004). The tobacco PR cDNA probes were provided by J. Ryals (Paradigm Genetics, Research Triangle Park, USA) and the phenylalanine ammonialyase cDNA probe was donated by C. Lamb (John Innes Centre, UK). Tobacco cDNA clones, one encoding ACC oxidase and one encoding PIN2, were provided by W.T. Kim (Yonsei University, Korea) and S.H. Lee (Pohang University of Science and Technology, Korea), respectively. Tobacco EREBP cDNA and actin cDNAs were amplified by polymerase chain reaction using specific primers that were based on published sequences. All probes were radiolabeled with [α-32P]dATP, using a random-primed labeling method.
Bioassays of transgenic plants
A bacterial pathogen, Pseudomonas syringae pv tabaci, was grown in King’s B (KB) medium supplemented with 50 mg L−1 rifampicin. The bacterial culture was washed with 10 mM MgCl2 and re-suspended in 10 mM MgCl2. Bacterial density was determined by measuring OD600 nm. The bacteria were diluted to the desired concentrations in 10 mM MgCl2 and inoculated into the leaves of mature tobacco with a needleless 1-ml syringe. Leaf discs (5 mm in diameter) were taken from the infection site at different time points, ground in 10 mM MgCl2, diluted, and plated on a KB agar plate that contained 50 mg L−1 rifampicin. Bacterial growth was monitored by counting colony-forming units (cfus). Measurements for each time point were performed in triplicate. Inoculation of tobacco with P. parasitica was conducted under aseptic conditions. To assay resistance to the infection by the oomycete, wild-type and transgenic tobacco plants were inoculated with agar plugs that contained hyphae of P. parasitica. In addition, fully-expanded young leaves were inoculated with TMV in TET buffer (10 mM Tris-HCl, 1 mM EDTA, 0.1% Triton X-100, pH 7.0) by gently rubbing leaves with both carborundum and 10 mg-per-leaf TMV. Inoculated plants were maintained at 22°C, and the size and the number of HR lesions were monitored at different time points.
RESULTS
Expression of the mammalian nNOS gene in tobacco plants
The initial research that explored the role of NO in the plant defense system indicates that infiltration of mammalian NOS enzyme into plant cells successfully triggers both NO production and activation of plant defense-related gene expression (Durner et al., 1998). Based on those results, we tested whether constitutive expression of the mammalian NOS gene in a plant could activate a plant defense system against pathogens. We did this by expressing rat brain NOS (nNOS) (Bredt et al., 1991) cDNA, which was under the control of the constitutive Cauliflower Mosaic Virus (CaMV) 35S promoter in tobacco plants (Nicotiana tabacum cv. Xanthi-nc). We also generated transgenic tobacco plants that co-expressed nNOS and NahG genes, which encode bacterial salicylate hydroxyalse (named nNOS/NahG), since SA plays crucial roles in NO-mediated defense signaling (Durner et al., 1998; Klessig et al., 2000). The transgenic tobacco T0 lines that expressed nNOS and nNOS/NahG were initially screened by Northern blot analysis (Supplementary Fig. S1). Based on the transcript level of nNOS, we selected two independent transgenic lines that showed strong nNOS expression out of about 15 T0nNOS and nNOS/NahG plants. We further selected homozygous T2 progeny from the T0 nNOS and nNOS/NahG lines and used them for further analyses.
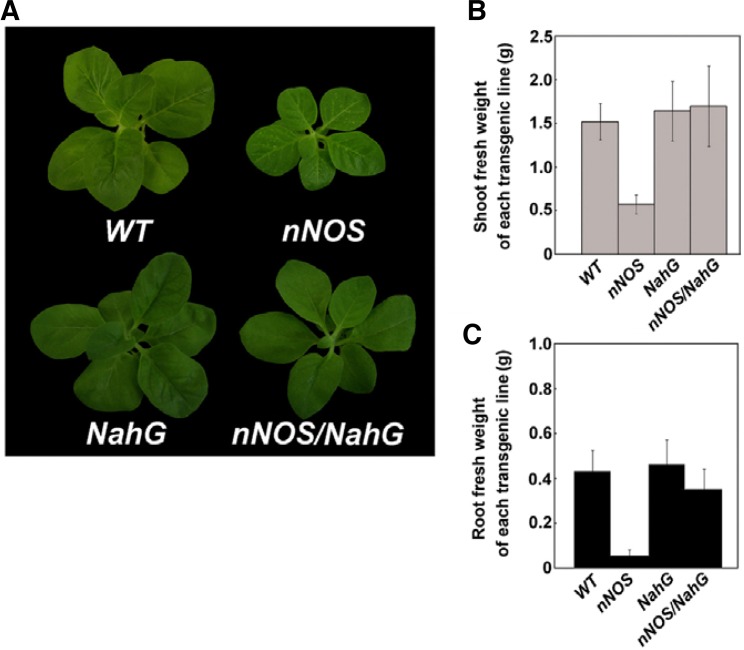
Interestingly, nNOS transgenic plants formed spontaneous necrotic lesions on their leaves (Figs. 1A and 3). The lesions were observed on all plant leaves and at all developmental stages, with the exception of the latest-developing top leaves. Untransformed wild-type and empty, vector-transformed, transgenic, control plants, both grown under identical conditions, did not show these symptoms. In addition, the nNOS transgenic plant showed a smaller-sized, morphological phenotype than that in both wild-type and NahG plants (Fig. 1A). In contrast, nNOS/NahG transgenic plants showed a phenotype similar to wild-type and NahG plants, implying NO involvement in the SA-related, plant defense signaling. We also measured shoot and root fresh weights. The growth of nNOS transgenic plants decreased by 3- and 8-fold in shoot and root, respectively, when compared to the growth of wild-type, NahG, and nNOS/NahG plants (Figs. 1B and 1C).
Fig. 1.
Phenotypes of transgenic tobacco plants that constitutively express mammalian neuronal nitric oxide synthase (nNOS). (A) Formation of spontaneous disease, lesion-like, necrotic regions in the NOS transgenic plants. The whole plant morphology of a 6-week-old representative of a NOS transgenic plant (nNOS) showed that lesions only appeared in the older leaves. Wild-type plant (WT), NahG transgenic plant (NahG), and nNOS/NahG transgenic plant (nNOS/NahG) of similar ages are shown. (B) Shoot fresh weight of each transgenic line. The fresh weight was calculated by shoot fresh weight/plant. Error bars represent standard deviations (n = 12). (C) Root fresh weight of each transgenic line. The fresh weight was calculated by root fresh weight/ plant. Error bars represent standard deviations (n = 12).
Identification of NO in the nNOS transgenic tobacco plants
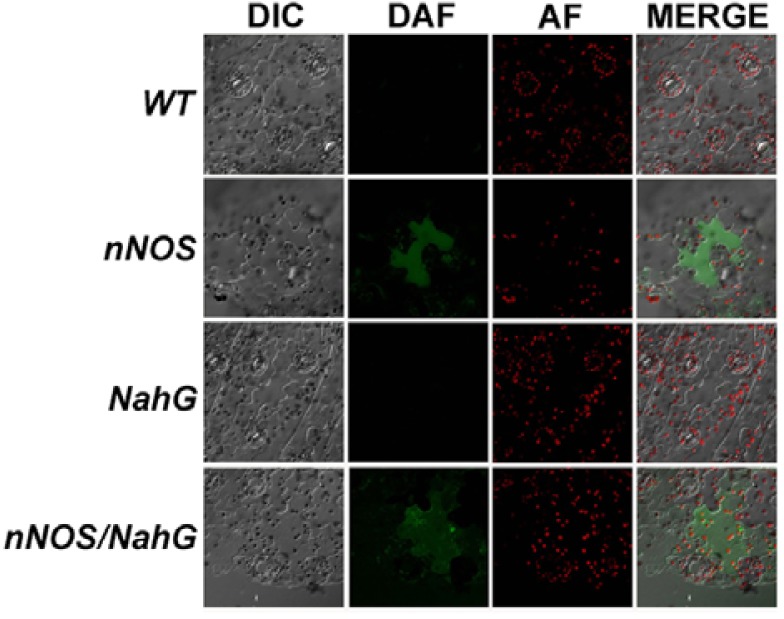
We first tested if expression of mammalian nNOS led to the production of NO in tobacco transgenic plants by using DAF-FM-DA. DAF-FM DA is cell-permeable and reacts with the more oxidized NO forms (e.g., NO+ and N2O3), and it has been widely used to detect NO levels in plants (Guo et al., 2003; Lozano-Juste and Leon, 2010; Planchet and Kaiser, 2006; Zottini et al., 2007). Since epidermal peels of transgenic tobacco plants can be effectively loaded with DAF-FM-DA, we did so and analyzed the peels with confocal laser-scanning microscopy. After treatment, strong green fluorescence was observed in a number of leaf epidermal cells, indicating NO production in the leaves of both nNOS and nNOS/NahG transgenic tobacco plants, but not in those of wild-type and NahG plants (Fig. 2). The results indicated that constitutive expression of mammalian NOS effectively triggered NO production in plant cells. In addition, integration of NO production with the previously observed NahG phenotype tentatively showed that SA is in a position downstream of nNOS-mediated NO production.
Fig. 2.
NOS activity in nNOS transgenic tobacco plant. NOS activities were measured, using fluorescence microscopy and intracellular DAF-FM-DA signals (DAF), on epidermal cells from wild-type (WT), nNOS transgenic (nNOS), NahG transgenic (NahG), and nNOS/NahG transgenic (nNOS/NahG) plants. The epidermis of transgenic tobacco leaves was loaded with 10 μM DAF-FM-DA. The images were obtained with a confocal laser scanning microscope (Nikon PCM2000). DIC, differential interference contrast; DAF, 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate (DAF-FM-DA) treatments; AF, autofluorescent materials; MERGE, overlapping images of DIC, DAF, and AF.
In animals, NO is primarily generated by a NOS enzyme that converts L-arginine to both L-citrulline and NO (Marletta, 1994). Furthermore, injection of recombinant mammalian NOS enzymes into tobacco plants induces the expression of defense-related genes, such as PR-1 and PAL; and, like a mammalian system, NOS activity in tobacco plants is also dependent on both a NOS substrate (arginine) and cofactors (Durner et al., 1998). Based on previous reports, we speculate that expression of rat nNOS gene in transgenic tobacco plants may produce NO through the same mechanism known in animals.
HR-like lesion formation and H2O2 accumulation in nNOS transgenic tobacco plants
The nNOS plants showed reduced shoot growth (Fig. 1B), reduced root growth (Fig. 1C), and reduced fertility (data not shown). The most obvious phenotype of nNOS transgenic plants was the development of spontaneous lesions on their leaves (Figs. 1 and 3). The cell death phenotype was more obvious on the lower (older) leaves than on the upper (younger) leaves of nNOS transgenic tobacco plants. The phenotypes of nNOS/NahG transgenic tobacco plants, however, were similar to those of wild-type plants, indicating important roles for SA in both the formation of lesions and the developmental defects caused by constitutive expression of nNOS.
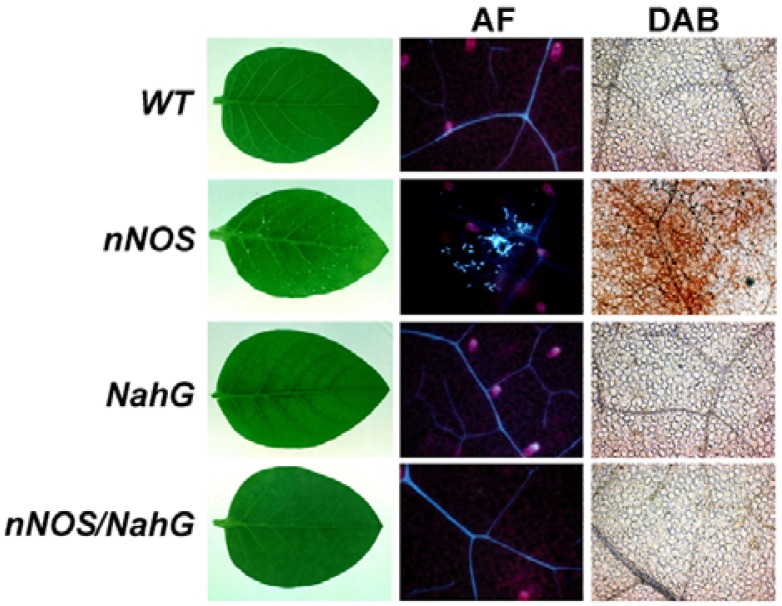
To determine whether the lesions mimicked a pathogen-induced HR, we tested for the presence of cellular and biochemical markers associated with HR (Hammond-Kosack and Jones, 1996). Substantial accumulations of autofluorescent material and H2O2 production (revealed by brown color development after staining with DAB) were detected on the leaves of nNOS transgenic tobacco plants. In contrast, nNOS/NahG transgenic plants did not exhibit spontaneous cell death, autofluorescence, or H2O2 accumulation in their leaves (Fig. 3). These results suggest that the expression of mammalian nNOS in tobacco plants generates a signal that leads to HR-like cell death and the commonly associated oxidative burst.
Fig. 3.
Analysis of cell death phenotype in nNOS transgenic tobacco plants. Autofluorescent materials (AFs) were visualized with an ultraviolet epifluorescence microscope. H2O2 accumulation was revealed by DAB staining (DAB). WT, wild-type tobacco plant; nNOS, nNOS transgenic plant; NahG, NahG transgenic plant; nNOS/NahG, nNOS/NahG transgenic plant.
Differential regulation of antioxidant enzymes in nNOS transgenic tobacco plants
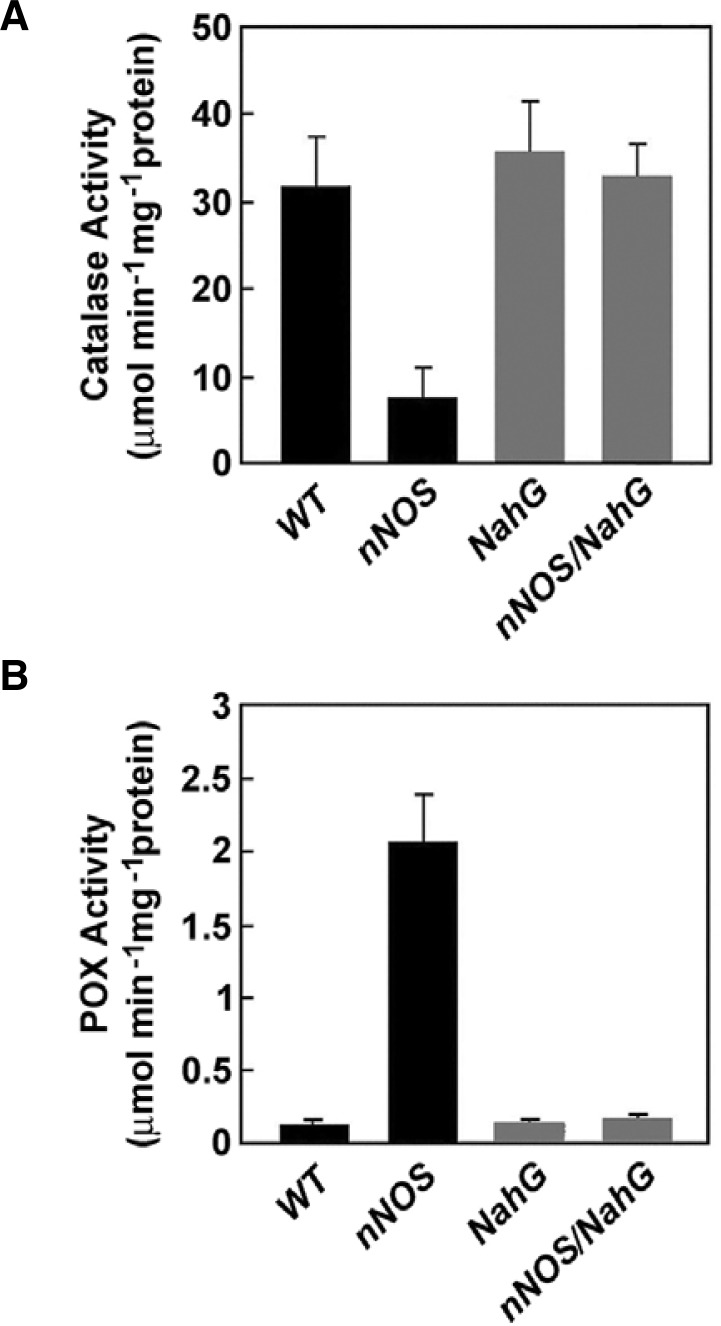
Previous reports had shown transiently stimulated NO production, which was achieved by either infiltration of the mammalian NOS enzyme or the application of NO donors into plant cells, induces both SA accumulation and the expression of defense-related genes. Moreover, activated cell death, concurrent with H2O2, has been demonstrated (Delledonne et al., 1998; Durner et al., 1998). To further understand the role(s) of SA in NOS-mediated cell death and oxidative burst, we measured the activities of antioxidant enzymes in nNOS transgenic plants. Previous reports suggested that SA inhibits CAT and ascorbate peroxidase (APX) activities, leading to an amplification of the oxidative burst (Chen and Klessig, 1991; Durner and Klessig, 1995; 1996). Indeed, the nNOS transgenic tobacco plants exhibited markedly decreased catalase activity, whereas this did not occur in nNOS/NahG transgenic plants (Fig. 4A). This result indicates that elevated levels of SA, induced by nNOS activity, exert an inhibitory effect on endogenous catalase. As a result, H2O2 accumulated in nNOS transgenic plants; however, no significant difference in APX activity was observed to distinguish wild-type from transgenic plants (data not shown). In contrast, the activity of guaiacol POX, which is not affected by SA (Durner and Klessig, 1995), increased by approximately 10-fold in nNOS transgenic plants (Fig. 4B). Upregulation of POX activity in nNOS transgenic plants is therefore caused by SA-dependent H2O2 accumulation. Based on these results, we propose that elevated levels of SA mediate H2O2 accumulation at least partly via the inhibition of CAT activity. Subsequently, increased H2O2 level can act synergistically with NO to trigger HR-like cell death in nNOS transgenic tobacco plants.
Fig. 4.
Differential regulation of antioxidant enzyme activities in nNOS transgenic tobacco plants. The specific activities of catalase (CAT) (A) and guaiacol peroxidase (POX) (B) were measured in soluble crude proteins extracted from leaves of 4-week-old plants, as described in “Materials and Methods”. All experiments were repeated twice, with three replicates per experiment, and error bars represent standard deviations.
Accumulation of SA, and constitutive expression of defense-related genes, in nNOS transgenic tobacco plants
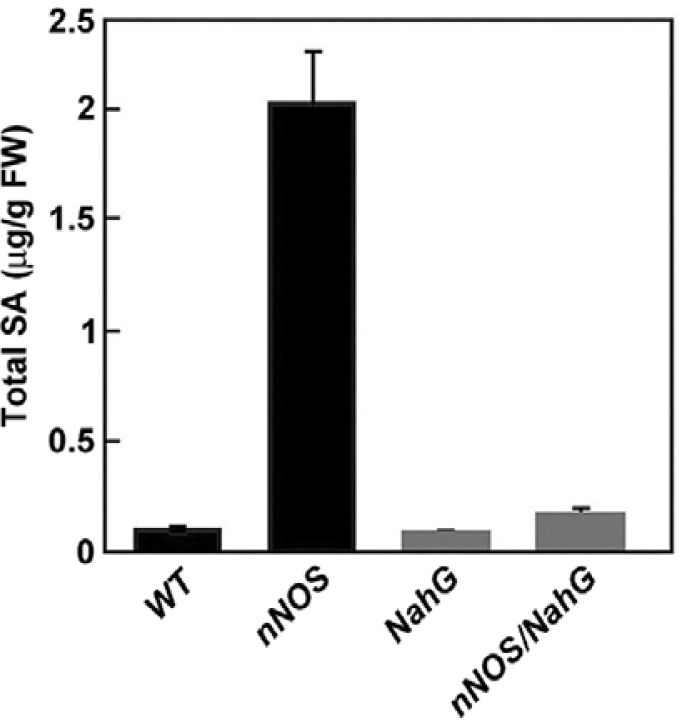
SA is a critical signal for activation of SA-dependent plant defense responses (Durner et al., 1997; Ryals et al., 1996). In several studies of transgenic plants and mutants with spontaneous lesion-mimic phenotypes, elevated levels of endogenous SA were observed and the plants frequently exhibited induction of defense genes (Alvarez, 2000; Dangl et al., 1996; Ryals et al., 1996). We analyzed the endogenous levels of total SA (free and glucose-conjugated SA forms) in nNOS transgenic tobacco plants. Total SA levels in nNOS transgenic plants were approximately 2 μg per gram fresh weight and were 23-fold higher than in wild-type plants (Fig. 5). Co-expression of the bacterial NahG gene was similar to that in wild-type plants that had nNOS-triggered SA accumulation.
Fig. 5.
SA levels in wild-type plant (WT), nNOS transgenic plant (nNOS), NahG transgenic plant (NahG), and nNOS/NahG transgenic plant (nNOS/NahG). Total SA levels (free SA plus glucose-conjugated SA) were determined in transgenic tobacco plants and analyzed in leaves from 4-week-old plants using HPLC. Values represent the mean and standard deviations obtained from three replicates per sample.
Next, we determined whether spontaneous lesion formation and high accumulation of SA in nNOS transgenic plants is accompanied by transcriptional activation of defense-related genes. Total RNA was isolated from the leaves of axenically-grown, nNOS, transgenic plants; and then we looked for the expression of defense-related genes that encode PAL and PR proteins. We found that the transcription of five PR genes, which are known to be enhanced during SAR in tobacco (Ryals et al., 1996), increased in response to nNOS expression, in a SA-dependent manner (Fig. 6). The PAL gene was also induced in nNOS transgenic plants, but in contrast to PR genes, appeared to be SA-independent. These results suggest that constitutive expression of mammalian nNOS is both effective and sufficient for the induction of plant defense signaling pathways.
Fig. 6.
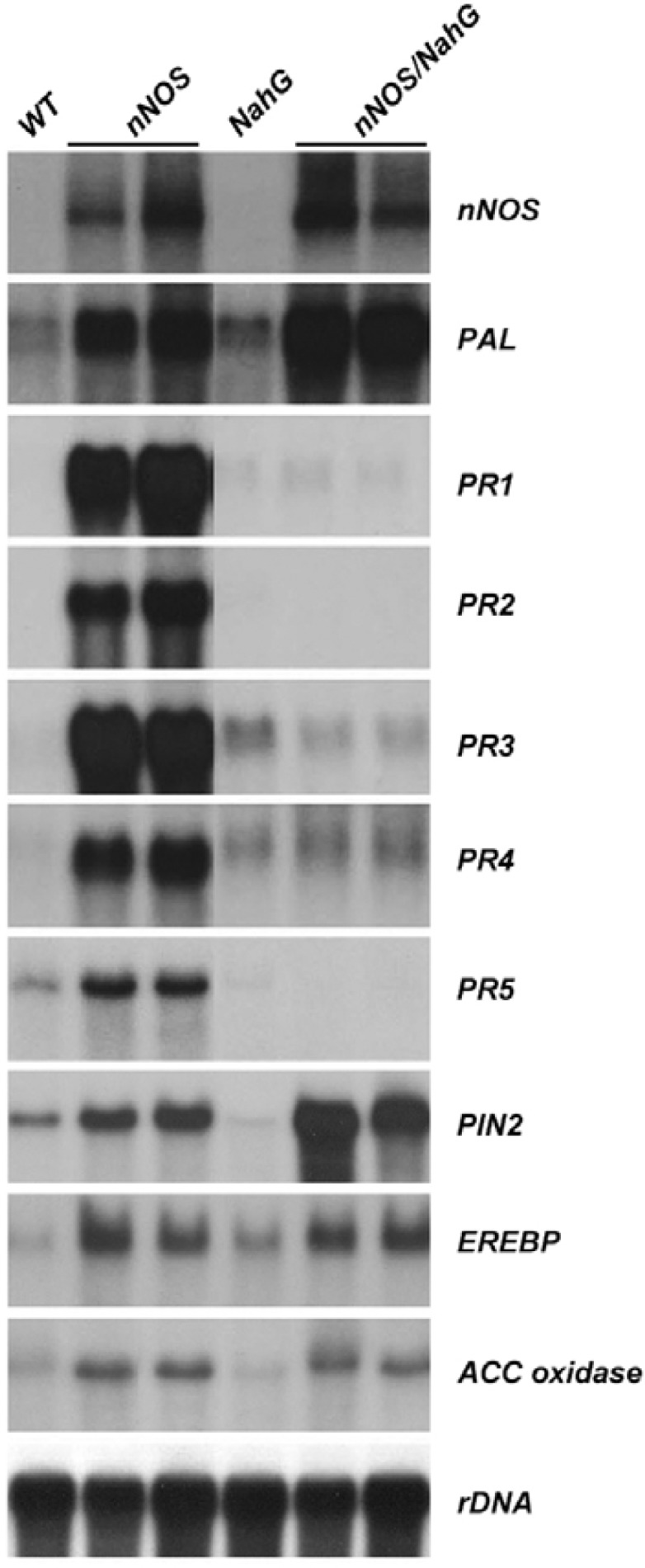
Overexpression of mammalian nNOS in tobacco plants increased defense-related gene expressions. Total RNA was isolated from axenically-grown wild-type (WT), nNOS transgenic (nNOS), NahG transgenic (NahG), and nNOS/NahG transgenic (nNOS/NahG) plants. The endogenous levels of defense-related gene expression were analyzed by Northern blot analysis. The rDNA transcript was used as a loading control.
Plants use multi-faceted signaling cascades such as SA- or JA/ET-dependent pathways to promote resistance against pathogens (Dong, 1998). To determine whether over-expression of mammalian nNOS triggers SA- and JA/ET-dependent plant defense pathways, we analyzed the expression of known, JA/ET-responsive, defense genes in nNOS transgenic tobacco plants. Interestingly, the expression of JA/ET-responsive defense genes, including EREBP (Ohme-Takagi and Shinshi, 1995), ACC oxidase (Kim et al., 1998), and Pin2, was induced in transgenic tobacco plants. The expression of these genes also increased significantly in nNOS/NahG plants, in which SA accumulation was abolished (Fig. 6). Taken together, these results provide biological evidence that JA and/or ET, along with SA, function as signaling molecules in NO-mediated plant defense responses.
Overexpression of mammalian nNOS confers resistance to a broad spectrum of pathogens
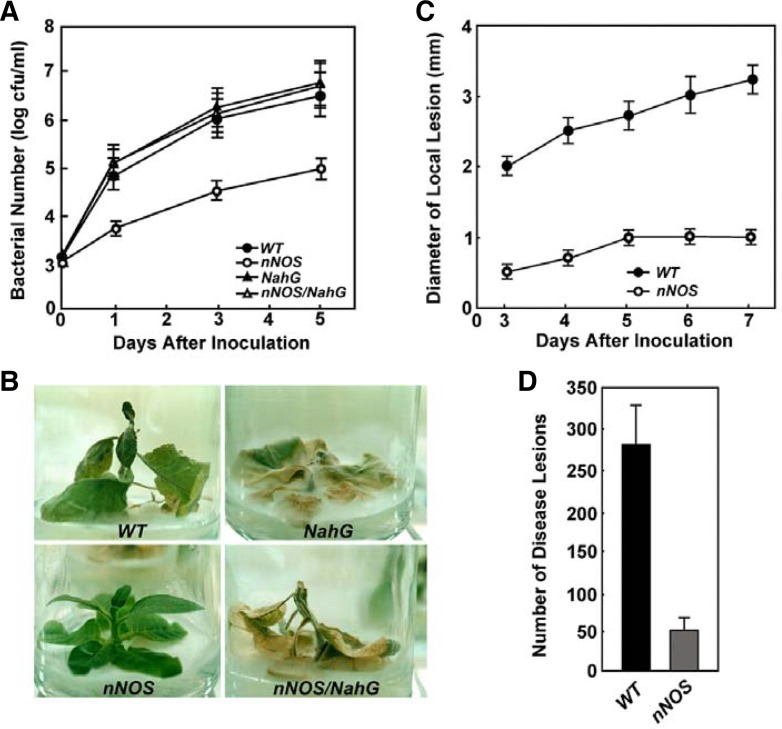
Spontaneous cell death, and accumulation of SA, H2O2, and PR gene transcripts, in nNOS transgenic plants, prompted us to test the resistance of these plants to pathogens. We first infiltrated nNOS transgenic plants with a virulent bacterial pathogen, Pseudomonas syringe pv. tabaci (Pst). Transgenic nNOS plants exhibited a significantly elevated level of resistance to Pst. Inhibition of pathogen growth in nNOS transgenic plants was observed as early as 24 h and for five days after inoculation. Bacterial growth in these plants was inhibited 50-fold more than wild-type plants (Fig. 7A).
Fig. 7.
Broad-spectrum disease resistance of transgenic tobacco plants that constitutively expressed the mammalian neuronal nitric oxide synthase (nNOS). (A) In planta bacterial growth. The bacterial pathogen, Pseudomonas syringae pv tabaci, was inoculated onto leaves of wild-type (WT), nNOS, NahG, and nNOS/NahG transgenic tobacco plants, at 105 cfu/ml, and bacterial growth was monitored over 5 days. Measurements for each time point were performed in triplicate. (B) Disease response to a virulent fungal pathogen, Phytophthora parasitica var. nicotianae. Two agar plugs, containing hyphae of P. parasitica, were placed on agar media and incubated in a culture bottle, together with 1-month-old wild-type (WT), nNOS, NahG, or nNOS/NahG transgenic tobacco plants, for 12 days. (C, D) Enhanced resistance of nNOS transgenic plants to tobacco mosaic virus (TMV) infection. Fully-expanded young leaves of wild-type (WT) and transgenic (nNOS) plants were inoculated with TMV. Lesion size was measured over 7 days (C), and lesion number was counted 5 days after inoculation (D).
We then tested the resistance of these transgenic plants to Phytophthora parasitica var. nicotianae, an oomycete pathogen that causes black shank disease in tobacco. nNOS transgenic and wild-type tobacco plants were inoculated with agar plugs that contained hyphae of P. parasitica and we monitored the development of disease symptoms. Five days post-inoculation, disease symptoms appeared on the wild-type plants but not on transgenic plants. By 8 days post-inoculation, wild-type plants showed severe leaf wilting and stem rot symptoms. They eventually died at 10 days post-inoculation (Fig. 7B); however, nNOS transgenic plants remained healthy without appreciable disease symptoms (Fig. 7B). We also tested for the resistance of nNOS/NahG transgenic plants to bacterial and fungal pathogens, but nNOS/NahG transgenic plants did not show resistance to the pathogens, indicating that SA plays a critical role in NO-mediated defense signaling in plants (Figs. 7A and 7B). In addition, transgenic nNOS plants exhibited enhanced resistance against an avirulent viral pathogen, TMV (Figs. 7C and 7D). The transgenic plants developed TMV-induced HR lesions approximately 12 h earlier than wild-type plants. The TMV-induced average lesion size in the transgenic plants was on average 70% smaller than in wild-type plants (Fig. 7C). Moreover, the transgenic plants showed fewer (∼5-fold less) TMV-induced disease events (Fig. 7D). These two characteristics that reduce lesion size and lesion number are therefore associated with enhanced SAR in response to TMV.
DISCUSSION
In plants, NO both signals and regulates many physiological processes, including germination, flowering, seed dormancy, pollen tube growth, root development, leaf senescence, and stomatal movements. In addition, the NO signals help orchestrate responses to a wide array of stresses, such as temperature extremes, salinity, drought, heavy metals, and pathogens (Baudoin, 2011). Despite the growing body of evidence implicating NO, SA, and ROS in plant-pathogen interactions, including PR gene expression and HR, detailed mechanisms of NO function in plant defense responses are still missing. This might be either partially or entirely due to methodological restrictions. Because plant genes that encode a genuine NO-synthase have yet to be found, only indirect approaches have been used to study NO-mediated, plant, defense signaling, such as infiltration of mammalian NOS protein directly into plant tissues and pharmacological approaches that employ either NO-releasing compounds or NOS inhibitors (Delledonne et al., 1998; Durner et al., 1998). In this study, by analyzing stable plant transformants that expressed mammalian nNOS, we demonstrated that NO communicates with JA and/or ET pathways (and SA and ROS pathways) during defense responses to pathogen attacks.
NO potentiates induction of hypersensitive cell death and defense-related gene expression by acting synergistically with ROS in both soybean cells and Arabidopsis (Delledonne et al., 1998). In addition, application of either recombinant mammalian NOS or NO donors to both tobacco plants and suspension cells triggers SA accumulation and the expression of defense-related genes, such as PR1 and PAL, in an SA-dependent manner (Durner et al., 1998). These findings imply that NO, SA, and ROS act cooperatively to activate plant defense signaling. Here, transgenic tobacco plants that express mammalian nNOS cDNA showed HR-like spontaneous necrotic lesion formation on their leaves, accumulation of H2O2 and SA, and elevated PR gene expression (Figs. 3 and 5). Interestingly, these NO-triggered events were completely abolished in NahG plants that expressed an SA-degrading enzyme. These results suggest that elevated SA levels triggered by NO can potentiate both the NO-mediated oxidative burst and the associated hypersensitive cell death in NOS transgenic tobacco plants. We propose that increased NO induced by over-expression of mammalian nNOS triggers SA biosynthesis through a cGMP and cADP ribose-dependent pathway (Durner et al., 1998). In turn, SA elevates H2O2 accumulation by inhibiting antioxidant enzymes, such as CAT. Finally, NO and H2O2 synergistically participate in hypersensitive cell death of NOS transgenic tobacco plants.
To confirm our hypothesis on H2O2 accumulation, we measured the activities of the endogenous antioxidant enzymes, CAT, APX, and POX. In nNOS transgenic plants (but not in nNOS/NahG transgenic plants), endogenous CAT activity decreased approximately 3-4x, as compared to wild-type tobacco plants (Fig. 4A), thereby supporting our hypothesis. We could not, however, observe any significant differences in APX activity. One possible explanation for this discrepancy may be the existence of different Kd values for the SA binding to CAT and APX, which have been previously reported as 14 mM and 78 mM, respectively (Chen and Klessig, 1991; Durner and Klessig, 1995). Clark et al. (2000) suggest that NO binds directly to CAT and APX, and inhibits their activities. However, SA more likely plays a more critical role in NO-triggered ROS generation because we did not observe inhibition of CAT activity in nNOS/NahG transgenic plants. In contrast to CAT, the activity of SA-insensitive POX increased in nNOS transgenic plants, but did not increase in wild-type and nNOS/NahG transgenic plants (Fig. 4B). POX activity may have increased as a means to scavenge accumulated H2O2 in nNOS transgenic plants.
Plants use multiple signaling cascades to evoke broad-spectrum disease resistance; therefore, induction of both SA-dependent and SA-independent (or JA/ET-dependent) defense response pathways is required (Dong, 1998). To test whether the over-expression of nNOS can effectively induce the JA/ET-dependent pathway, in addition to the SA-dependent pathway, we monitored the expression levels of SA-responsive and JA/ET-responsive genes in nNOS transgenic tobacco plants. We observed an induction of the PAL gene in a NO-dependent fashion; an induction of SA-responsive genes, such as PR1, PR2 and PR5; and elevated expressions of JA/ET-responsive genes, such as PIN2, EREBP, and ACC oxidase (Fig. 6). Interestingly, nNOS/NahG transgenic plants also showed elevated expression of these JA/ET-responsive genes. The latter result is in agreement with data that NO treatment of NahG plants results in activation of JA-responsive genes (Huang et al., 2004). These results suggest that overexpression of nNOS may sufficiently induce both the JA/ET-dependent pathway and the SA-dependent pathway.
To summarize, our results strongly suggest that the crucial participants in animal NO signaling also function in plants. In addition, NO production seems to always be tightly connected to various signals within plant cells. We also provide evidence for the critical involvement of NO in plant defense responses, both by interacting with SA and ROS pathways, and interacting with pathways that involve JA and ET. Identification of either NOS or other NO producers in higher plants may provide unique opportunities to understand the regulation of various metabolic pathways for which NO is the signaling molecule. Further investigations into the regulation of NO production and its downstream effects in plants should involve studies on both the biochemical analysis of NO production and the NO scavenging pathways of plant defense signaling. It will be important to study how other enzymes, such as those involved in nitrate reduction, can be recruited to provide NO signaling. Such information will be crucial for our understanding of NO signaling pathways that occur during development and stress responses. Such knowledge will also provide a better understanding of the roles that NO-related mechanisms play during plant defense responses.
Supplementary Material
Acknowledgments
We thank Dr. Hans J. Bohnert for critical reading and insightful comments. We extend our sincere thanks to Dr. S. H. Snyder for generously providing rat neuronal NOS cDNA. We thank Drs. C. Lamb, J. Ryals, W. T. Kim, and S. H. Lee for tobacco PR protein cDNAs, PAL, ACC oxidase, and PIN2 cDNAs, respectively. We also thank Dr. J. Ryals for providing the NahG plant seeds. This work was supported by grants from the Next-Generation BioGreen21 Program (SSAC, grant#: PJ008025) of the Rural Development Administration, World Class University Program (R32-10148), funded by the Ministry of Education, Science and Technology; and Basic Science Research Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0009092), Republic of Korea.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Alvarez M.E. Salicylic acid in the machinery of hypersensitive cell death and disease resistance. Plant Mol. Biol. 2000;44:429–442. doi: 10.1023/a:1026561029533. [DOI] [PubMed] [Google Scholar]
- Asai S., Yoshioka H. Nitric oxide as a partner of reactive oxygen species participates in disease resistance to necrotrophic pathogen Botrytis cinerea in Nicotiana benthamiana. Mol. Plant-Microbe Interact. 2009;22:619–629. doi: 10.1094/MPMI-22-6-0619. [DOI] [PubMed] [Google Scholar]
- Barroso J.B., Corpas F.J., Carreras A., Sandalio L.M., Valderrama R., Palma J.M., Lupiáñez J.A., del Río L.A. Localization of Nitric-oxide synthase in plant peroxisomes. J. Biol. Chem. 1999;274:36729–36733. doi: 10.1074/jbc.274.51.36729. [DOI] [PubMed] [Google Scholar]
- Baudoin E. The language of nitric oxide signaling. Plant Biol. 2011;13:233–242. doi: 10.1111/j.1438-8677.2010.00403.x. [DOI] [PubMed] [Google Scholar]
- Beligni M.V., Lamattina L. Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta. 2000;210:215–221. doi: 10.1007/PL00008128. [DOI] [PubMed] [Google Scholar]
- Beligni M.V., Fath A., Bethke P.C., Lamattina L., Jones R.L. Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiol. 2002;129:1642–1650. doi: 10.1104/pp.002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredt D.S., Hwang P.M., Glatt C.E., Lowenstein C., Reed R.R., Snyder S.H. Cloned and expressed nitric oxide synthase structurally resembles cytochrome P-450 reductase. Nature. 1991;351:714–718. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- Chen Z., Klessig D.F. Identification of soluble salicylic acid-binding protein that may function in signal transduction in the plant disease-resistance response. Proc. Natl. Acad. Sci USA. 1991;88:8179–8183. doi: 10.1073/pnas.88.18.8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D., Durner J., Navarre D.A., Klessig D.F. Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Mol. Plant-Microbe Interact. 2000;13:1380–1384. doi: 10.1094/MPMI.2000.13.12.1380. [DOI] [PubMed] [Google Scholar]
- Corpas F.J., Palma J.M., del Río L.A., Barroso J.B. Evidence supporting the existence of L-arginine-dependent nitric oxide synthase activity in plants. New Phytol. 2009;184:9–14. doi: 10.1111/j.1469-8137.2009.02989.x. [DOI] [PubMed] [Google Scholar]
- Crawford N.M., Galli M., Tischner R., Heimer Y.M., Okamoto M., Mack A. Response to zemojtel et al: Plant nitric oxide synthase: back to square one. Trends Plant Sci. 2006;11:526–527. [Google Scholar]
- Dangl J.L., Dietrich R.A., Richberg M.H. Death don’t have no mercy: cell death programs in plant-microbe interaction. Plant Cell. 1996;8:1793–1807. doi: 10.1105/tpc.8.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M., Xia Y., Dixon R.A., Lamb C. Nitric oxide signal functions in plant disease resistance. Nature. 1998;394:585–588. doi: 10.1038/29087. [DOI] [PubMed] [Google Scholar]
- Delledonne M., Zeier J., Marocco A., Lamb C. Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci USA. 2001;98:13454–13459. doi: 10.1073/pnas.231178298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich R.A., Delaney T.P., Uknes S.J., Ward E.R., Ryals J.A., Dangl J.L. Arabidopsis mutants simulating disease resistance response. Cell. 1994;77:565–577. doi: 10.1016/0092-8674(94)90218-6. [DOI] [PubMed] [Google Scholar]
- Dong X. SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1998;1:316–323. doi: 10.1016/1369-5266(88)80053-0. [DOI] [PubMed] [Google Scholar]
- Durner J., Klessig D.F. Inhibition of ascorbate peroxidase by salicylic acid and 2,6-dichloroisonicotinic acid, two inducers of plant defense responses. Proc. Natl. Acad. Sci USA. 1995;92:11312–11316. doi: 10.1073/pnas.92.24.11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J., Klessig D.F. Salicylic acid is a modulator of tobacco and mammalian catalases. J. Biol. Chem. 1996;271:28492–28501. doi: 10.1074/jbc.271.45.28492. [DOI] [PubMed] [Google Scholar]
- Durner J., Klessig D.F. Nitric oxide as signal in plants. Curr. Opin. Plant Biol. 1999;2:369–374. doi: 10.1016/s1369-5266(99)00007-2. [DOI] [PubMed] [Google Scholar]
- Durner J., Shah J., Klessig D.F. Salicylic acid and disease resistance in plants. Trends Plant Sci. 1997;2:266–274. [Google Scholar]
- Durner J., Wendehenne D., Klessig D.F. Defense gene induction in tobacco by nitric oxide, cyclic GMP and cyclic ADP ribose. Proc. Natl. Acad. Sci USA. 1998;95:10328–10333. doi: 10.1073/pnas.95.17.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan A., Kwon E., Yun B.W., Wang Y., Pallas J.A., Loake G.J. A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci USA. 2005;102:8054–8059. doi: 10.1073/pnas.0501456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Pérez U., Sauret-Güeto S., Gas E., Jarvis P., Rodríguez-Concepción M. A mutant impaired in the production of plastome-encoded proteins uncovers a mechanism for the homeostasis of isoprenoid biosynthetic enzymes in Arabidopsis plastids. Plant Cell. 2008;20:1303–1325. doi: 10.1105/tpc.108.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissner I., Wendehenne D., Langebartels C., Durner J. In vivo imaging of an elicitor-induced nitric oxide burst in tobacco. Plant J. 2000;23:817–824. doi: 10.1046/j.1365-313x.2000.00835.x. [DOI] [PubMed] [Google Scholar]
- Guo F.-Q., Okamoto M., Crawford N.M. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science. 2003;302:100–103. doi: 10.1126/science.1086770. [DOI] [PubMed] [Google Scholar]
- Gupta K.J., Fernie A.R., Kaiser W.M., van Dongen J.T. On the origins of nitric oxide. Trends Plant Sci. 2011;16:160–168. doi: 10.1016/j.tplants.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack K.E., Jones J.D. Resistance gene-dependent plant defense responses. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch R.B., Fry J.E., Hoffmann N.L., Eichholfz D., Rogers S.G., Fraley R.T. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Huang X., Stettmaier K., Michel C., Hutzler P., Mueller M.J., Durner J. Nitric oxide is induced by wounding and influences jasmonic acid signaling in Arabidopsis thaliana. Planta. 2004;218:938–946. doi: 10.1007/s00425-003-1178-1. [DOI] [PubMed] [Google Scholar]
- Klessig D.F., Durner J., Noad R., Navarre D.A., Wendehenne D., Kumar D., Zhou J.M., Shah J., Zhang S., Kachroo P., et al. Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci USA. 2000;97:8849–8855. doi: 10.1073/pnas.97.16.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Choi D., Lee M.M., Lee S.H., Kim W.T. Biotic and abiotic stress-related expression of 1-aminocyclopropane-1-carboxylate oxidase gene family in Nicotiana glutinosa L. Plant Cell Physiol. 1998;39:565–573. doi: 10.1093/oxfordjournals.pcp.a029406. [DOI] [PubMed] [Google Scholar]
- Leshem Y.Y. Nitric oxide in biological systems. Plant Growth Regul. 1996;18:155–159. [Google Scholar]
- Leshem Y.Y., Wills R.B.H., Ku V.V.V. Evidence for the function of the free radical gas, nitric oxide (NO), as an endogenous maturation and senescence regulating factor in higher plants. Plant Physiol. Biochem. 1998;36:825–833. [Google Scholar]
- Lozano-Juste J., Leon J. Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR-and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiol. 2010;152:891–903. doi: 10.1104/pp.109.148023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marletta M.A. Nitric oxide synthase: aspects concerning structure and catalysis. Cell. 1994;78:927–930. doi: 10.1016/0092-8674(94)90268-2. [DOI] [PubMed] [Google Scholar]
- Meuwly P., Métraux J.P. Ortho-anisic acid as internal standard for the simultaneous quantitation of salicylic acid and its putative biosynthetic precursors in cucumber leaves. Anal. Biochem. 1993;214:500–505. doi: 10.1006/abio.1993.1529. [DOI] [PubMed] [Google Scholar]
- Moreau M., Lee G.I., Wang Y., Crane B.R., Klessig D.F. AtNOS/A1 is a functional Arabidopsis thaliana cGTPase and not a nitric oxide synthase. J. Biol. Chem. 2008;283:32957–32967. doi: 10.1074/jbc.M804838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M., Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.Y., Heo W.D., Yoo J.H., Lee J.H., Kim M.C., Chun H.J., Moon B.C., Kim I.H., Park H.C., Choi M.S., et al. Pathogenesis-related gene expression by specific calmodulin isoforms is dependent on NIM1, a key regulator of systemic acquired resistance. Mol Cells. 2004;18:207–213. [PubMed] [Google Scholar]
- Planchet E., Kaiser W.M. Nitric oxide production in plants. Plant Signal. Behav. 2006;1:46–51. doi: 10.4161/psb.1.2.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M.V., Paliyath G., Ormrod D.P., Murr D.P., Watkins C.B. Influence of salicylic acid on H2O2 production, oxidative stress, and H2O2-metabolizing enzymes. Plant Physiol. 1997;115:137–149. doi: 10.1104/pp.115.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals J.A., Neuenschwander U.H., Willits M.G., Molina A., Stener H.Y., Hunt M.D. Systemic acquired resistance. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H.H., Walter U. NO at work. Cell. 1994;78:919–925. doi: 10.1016/0092-8674(94)90267-4. [DOI] [PubMed] [Google Scholar]
- Song F., Goodman R.M. Activity of nitric oxide is dependent on, but is partially required for function of, salicylic acid in the signaling pathway in tobacco systemic acquired resistance. Mol. Plant-Microbe Interact. 2001;14:1458–1462. doi: 10.1094/MPMI.2001.14.12.1458. [DOI] [PubMed] [Google Scholar]
- Spadaro D., Yun B.W., Spoel S.H., Chu C.C., Wang Y.Q., Loake G.J. The redox switch: dynamic regulation of protein function by cysteine modifications. Physiol Plant. 2010;138:360–371. doi: 10.1111/j.1399-3054.2009.01307.x. [DOI] [PubMed] [Google Scholar]
- Sudhamsu J., Lee G.I., Klessig D.F., Crane B.R. The structure of YqeH: an AtNOS1/AtNOA1 ortholog that couples GTP hydrolysis to molecular recognition. J. Biol. Chem. 2008;283:32968–32978. doi: 10.1074/jbc.M804837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y., Spoel S.H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., Zuo J., Dong X. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science. 2008;321:952–956. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B.P.H.J., Penninckx I.A.M.A., Broekaert W.F., Cammue B.P.A. The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 2001;13:63–68. doi: 10.1016/s0952-7915(00)00183-7. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H., Zhang Z., Wei Y., Collinge D.B. Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley powdery mildew interaction. Plant J. 1997;11:1187–1194. [Google Scholar]
- Tun N.N., Holk A., Scherer G.F.E. Rapid increase of NO release in plant cell cultures induced by cytokinin. FEBS Lett. 2001;509:174–176. doi: 10.1016/s0014-5793(01)03164-7. [DOI] [PubMed] [Google Scholar]
- Van Camp W., Van Montagu M., Inzé D. H2O2 and NO: redox signals in disease resistance. Trends Plant Sci. 1998;3:330–334. [Google Scholar]
- Van Ree K., Gehl B., Chehab E.W., Tsai Y.C., Braam J. Nitric oxide accumulation in Arabidopsis is independent of NOA1 in the presence of sucrose. Plant J. 2011;68:225–233. doi: 10.1111/j.1365-313X.2011.04680.x. [DOI] [PubMed] [Google Scholar]
- Wendehenne D., Pugin A., Klessig D.F., Durner J. Nitric oxide: comparative synthesis and signaling in animal and plant cells. Trends Plant Sci. 2001;6:177–183. doi: 10.1016/s1360-1385(01)01893-3. [DOI] [PubMed] [Google Scholar]
- Yoshioka H., Bouteau F., Kawano T. Discovery of oxidative burst in the field of plant immunity. Plant Signal. Behav. 2008;3:153–155. doi: 10.4161/psb.3.3.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H., Mase K., Yoshioka M., Kobayashi M., Asai S. Regulatory mechanisms of nitric oxide and reactive oxygen species generation and their role in plant immunity. Nitric Oxide. 2011;25:216–221. doi: 10.1016/j.niox.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Yu M., Yun B.W., Spoel S.H., Loake G.J. A sleigh ride through the SNO: regulation of plant immune function by protein S-nitrosylation. Curr. Opin. Plant Biol. 2012;15:1–7. doi: 10.1016/j.pbi.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Yun B.W., Spoel S.H., Loake G.J. Synthesis of and signaling by small, redox active molecules in the plant immune response. Biochim. Biophys Acta. 2012;1820:770–776. doi: 10.1016/j.bbagen.2011.06.015. [DOI] [PubMed] [Google Scholar]
- Zemojtel T., Frohlich A., Palmieri M.C., Kolanczyk M., Mikula I., Wyrwicz L.S., Wanker E.E., Mundlos S., Vingron M., Martasek P., Durner J. Plant nitric oxide synthase: A never-ending story? Trends Plant Sci. 2006;11:524–525. doi: 10.1016/j.tplants.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Zottini M., Costa A., De Michele R., Ruzzene M., Carimi F., Lo Schiavo F. Salicylic acid activates nitric oxide synthesis in Arabidopsis. J. Exp. Bot. 2007;58:1397–1405. doi: 10.1093/jxb/erm001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.