Abstract
Quantitative RNA analyses of autopsy materials to diagnose the cause and mechanism of death are challenging tasks in the field of forensic molecular pathology. Alterations in mRNA profiles can be induced by cellular stress responses during supravital reactions as well as by lethal insults at the time of death. Here, we demonstrate that several gene transcripts encoding heat shock proteins (HSPs), a gene family primarily responsible for cellular stress responses, can be differentially expressed in the occipital region of postmortem human cerebral cortices with regard to the cause of death. HSPA2 mRNA levels were higher in subjects who died due to mechanical asphyxiation (ASP), compared with those who died by traumatic injury (TI). By contrast, HSPA7 and A13 gene transcripts were much higher in the TI group than in the ASP and sudden cardiac death (SCD) groups. More importantly, relative abundances between such HSP mRNA species exhibit a stronger correlation to, and thus provide more discriminative information on, the death process than does routine normalization to a housekeeping gene. Therefore, the present study proposes alterations in HSP mRNA composition in the occipital lobe as potential forensic biological markers, which may implicate the cause and process of death.
Keywords: cause of death, heat shock protein (HSP), molecular pathology, mRNA, postmortem brain tissue
INTRODUCTION
Postmortem RNA profiling along with the classical examination of morphological, pathological, and laboratory changes has become an emerging field of forensic medical science, supporting advanced molecular autopsy. Compared with other biochemical substances, RNA transcripts have advantages in terms of immediate responsiveness, chemical stability, and technical convenience in quantitative analysis. Extensive studies have currently been carried out to examine gene expression at the time of death to address essential issues in forensic medicine, including the cause and process of death, survival period, postmortem interval (PMI), and pathophysiological conditions of diseases and injuries (Maeda et al., 2010; Vennemann and Koppelkamm, 2010a). For instance, postmortem profiles of several mRNA species, such as immediate early genes, hypoxia-inducible factor-1α, erythropoietin, vascular endothelial growth factor, glucose transporter-1, surfactants, and dual-specificity phosphatase 1, have been proposed to be related to certain causes and circumstances of death (Ikematsu et al., 2005; 2008; Miyazato et al., 2012; Takahashi et al., 2009; Zhao et al., 2006; 2008; 2009). However, in contrast to the identification of body fluids and tissues using nucleic acid biomarkers (Courts and Madea, 2011; Haas et al., 2009; Juusola and Ballantyne, 2003; Zubakov et al., 2008; 2010), the application of molecular pathological examination based on postmortem gene expression profiles to death processes appears to be still challenging. Despite technical advances in resolving the difficulties of sensitivity and standardization (Catts, 2005; Heinrich et al., 2007b; Lee et al., 2005), there remain limitations primarily due to the restricted understanding of molecular physiology at the time of death and, subsequently, the insufficiency of well-established forensic RNA biomarkers.
For this purpose, alterations in postmortem mRNA profiles mediating cellular stress responses are worthwhile to be examined, since agonal responses to lethal insults as well as supravital reactions primarily involve cellular damage. Indeed, both genomic and proteomic analyses have suggested that key cellular pathways involving cellular stress responses, protein biosynthesis, cell cycle regulation, and apoptosis appear to be induced during the acute postmortem period (Bjarnadóttir et al., 2010; Jardine et al., 2011; Sanoudou et al., 2004). In this regard, a family of transcripts encoding heat shock proteins (HSPs) is a good candidate to be evaluated. HSPs play pivotal roles in mediating cellular stress responses to a wide range of cellular stresses, such as increased temperature, radiation, exposure to toxic substances, oxidative stress, and various physiological and pathological stimuli (Calabrese et al., 2006; Romi et al., 2011; Stetler et al., 2010). HSPs can be categorized into several subfamilies with dozens of members according to their molecular weights and structural characteristics, such as HSPAs/HSP70s, HSPBs/small HSPs, HSPCs/HSP90s, HSPD/HSP60, HSPE/HSP10, and HSPHs/HSP105/110. Although HSPs were originally discovered as stress-inducible proteins, it is currently understood that some of them are inducible under the control of heat shock factor-1 (HSF1), whereas others are highly constitutive in their expression to perform housekeeping functions (Fujimoto and Nakai, 2010; Stetler et al., 2010).
Considering the diversity in the modes and mechanisms of transcriptional regulation among HSPs, we hypothesized that differential death processes can affect HSP mRNA profiles in response to cellular stress, which can persist and manifest in forensic autopsy samples. To test this idea, we primarily focus on brain tissues because neuronal cells abundantly express HSPs and are highly vulnerable to a variety of cellular stresses, such as hypoxia/anoxia as well as disturbances in energy homeostasis (Hecker and McGarvey, 2011; Stetler et al., 2010). Furthermore, brain tissues also have a practical advantage in postmortem RNA profiling; it has been demonstrated that postmortem gene transcripts in brain tissues are relatively stable and intact with longer PMI, and are thus suitable for extraction and quantitative analyses of RNA (Heinrich et al., 2007a; 2007b; Leonard et al., 1993; Popova et al., 2008). Therefore, the present study aims to quantitatively analyze a subgroup of brain-enriched HSP mRNA transcripts particularly in the postmortem human occipital lobes, thus gaining useful clues on their differential profiles by the cause of death, such as traumatic injury (TI), mechanical asphyxiation (ASP), or sudden cardiac death (SCD).
MATERIALS AND METHODS
Subjects and tissue preparation
Tissue specimens from the occipital region of the cerebral cortex were collected in selected medicolegal autopsy cases within 5 days postmortem by the help of the Korean National Forensic Service. The occipital lobe was mainly focused on because the cerebral cortical subregion is known to be relatively less influenced by preexisting neurodegenerative diseases and psychiatric disorders than do other cortical lobes such as frontal and temporal lobes (Brun and Gustafson, 1976; Dabir et al., 2004; Forman et al., 2004; Price and Drevets, 2012). Thus, it can be a suitable brain region for evaluating temporal alterations in gene expression potentially by the process and circumstance of death. The causes of death examined in the present study were TI, acute mechanical ASP, including both hanging and strangulation, and SCD, classified by routine morphological, biochemical, and toxicological findings (Table 1). Collected tissues were immediately treated with RNAlater™ solution (QIAGEN GmbH, Germany) according to the manufacturer’s instructions and stored at −70°C until use.
Table 1.
Case profiles
| Group | Size | Male/female | Age (years)
|
28S:18S ratio (mean ± SE) |
|---|---|---|---|---|
| Range (mean ± SE) | ||||
| TI | n = 19 | 13/6 | 32–64 (48.11 ± 2.20) | 0.932 ± 0.043 |
| ASP | n = 22 | 8/14 | 25–54 (40.77 ± 1.87) | 0.877 ± 0.034 |
| SCD | n = 15 | 15/0 | 40–69 (53.33 ± 1.73) | 0.849 ± 0.026 |
TI, traumatic injury; ASP, asphyxiation; SCD, sudden cardiac death
Microarray
Gene expression profiling by microarray analysis was performed to identify human brain-enriched HSP gene transcripts as described previously with minor modifications (Choe et al., 2011). Briefly, human brain RNA (a mixture with equal amounts of 18 randomly chosen human RNA samples from the occipital lobes) was subjected to microarray analysis. Two hundred ng of total RNA was amplified and labeled. The GeneChip Human Gene 1.0 ST arrays (Affymetrix Inc., USA) were then hybridized in the presence of 5 μg labeled cDNA, washed, stained, and scanned according to the protocol described in the Affymetrix GeneChip Expression Analysis Manual. Normalized hybridization intensities for each HSP gene in comparison to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were analyzed.
RNA isolation and quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Quantitative analyses of mRNA profiles were performed as previously described with modifications (Choe et al., 2011). Total RNA was isolated using the RNeasy mini kit (QIAGEN) from 20–50 mg tissue and then quantified by spectrophotometry. Aliquots of total RNA samples (5 μg) were resolved on 1.2% denaturing agarose gels and stained with ethidium bromide. Signal intensities of 28S and 18S rRNAs were measured with Bio-1D image analysis software (Vilber-Lourmat, Cedex, France). The ratio of 28S to 18S signals were used as indices for RNA integrity (Table 1). For qRT-PCR, 1 μg of each total RNA sample was reverse-transcribed using MMLV reverse transcriptase (Promega, USA) by the random priming method. Then, aliquots of cDNA were subjected to quantitative real-time PCR in the presence of SYBR Green I (Life Technologies Corp., USA). Serial dilutions of reference cDNA samples were utilized to construct a regression curve. HSP mRNA expression levels were normalized to GAPDH. All PCR products were cloned in the pGEM-T vector (Promega), and their sequence identities were confirmed by chain termination sequencing. Primer sequences used for real-time qRT-PCR were as follows: HSPA1A up, 5′-GCC TTT CCA AGA TTG CTG TT-3′; HSPA1A dn, 5′-TCA ACA TTG CAA ACA CAG GA-3′; HSPA1B up, 5′-AGG GTG TTT CGT TCC CTT TA-3′; HSPA1B dn, 5′-CAT TCC CAG CCT TTG TAG TG-3′; HSPA2 up, 5′-TCG AGG TGG CCG TTA GTT G-3′; HSPA2 dn, 5′-AAA GGC GAG CGA CGT TAG G-3′; HSPA4 up, 5′-GCA ACA GCA GCA GAC ACC AGC A-3′; HSPA4 dn, 5′-GCC TTC TTG GCT TGG GGT GGT-3′; HSPA5 up, 5′-TCT TGT TGG TGG CTC GAC TC-3′; HSPA5 dn, 5′-ATC TGG GTT TAT GCC ACG GG-3′; HSPA7 up, 5′-CAA CCT GCT GGG GCG TTT TGA-3′; HSPA7 dn, 5′-CCG GCC CTT GTC ATT GGT GAT CTT-3′; HSPA8 up, 5′-GGC ATA CCT CCT GCA CCC CGA-3′; HSPA8 dn, 5′-GTC TTC CTT GCT CAA ACG GCC C-3′; HSPA9 up, 5′-TGG AAT GCC GGC CAA GCG AC-3′; HSPA9 dn, 5′-GCC TCA ACC CAG GCA TCA CCA-3′; HSPA12A up, 5′-GCA CGC CTT GCG GAA AAG CA-3′; HSPA12A dn, 5′-GGC ATC TGG ACT CAT CCG CAG C-3′; HSPA13 up, 5′-ACC GCA GAA GAG TTG GAG GCT GA-3′; HSPA13 dn, 5′-TCT GGG GAC ACT GTG ATG GTC TCA-3′; HSPD1 up, 5′-TGG CCG TTA CAA TGG GGC CAA-3′; HSPD1 dn, 5′-AGC AGT GGT AGT GCC ATC CCC A-3′; HSPH1 up, 5′-AGG AGT TCC ATA TCC AGA A-3′; HSPH1 dn, 5′-CAG CTC AAC ATT CAC CAC-3′; HSP90AA1 up, 5′-AGC TCA AGC CCT AAG AGA CAA CT-3′; HSP90AA1 dn, 5′-AAG ATG ACC AGA TCC TTC ACA GA-3′; GAPDH up, 5′-GAA ATC CCA TCA CCA TCT TCC AGG-3′; GAPDH dn, 5′-GAG CCC CAG CCT TCT CCA TG-3′.
Statistical analysis
Group differences in HSP mRNA levels were statistically evaluated by one-way analysis of variance (ANOVA) followed by Newman-Keuls test for post-hoc comparison. Statistical significance was set at p < 0.05.
RESULTS
Identification of HSP mRNA species enriched in the human occipital lobe
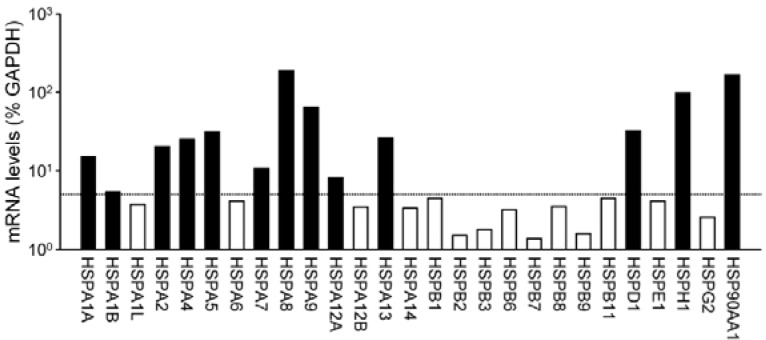
We initially aimed to identify members of the HSP gene family that are significantly detected in postmortem human brain tissues. For this purpose, we carried out microarray analysis of human total RNA samples derived from the cortical tissues of the occipital lobes. Relative signal intensity for each HSP gene relative to that of GAPDH was evaluated and expressed as the percent intensity of GAPDH (Fig. 1). Among tested HSP mRNA species, several members belonging to the HSPA subfamily encoding 70-kDa HSPs (HSP70s) were significantly detected; hybridization signal intensities for HSPA1A, A2, A4, A5, A8, A9, and A13 exhibited more than 10% of the GAPDH signals. HSPA1B, A7 and A12A gene transcripts were also significantly detected at the levels of 5–10% of the GAPDH signals. By contrast, mRNA transcripts for small HSPs belonging to the HSPB subfamily were barely detectable. Their expression levels were comparable with the average signal intensities for negative control probes (< 5% of GAPDH transcript levels). Other members including HSPD1, H1, and 90AA1 were also abundantly expressed in brain tissues. Based on their expression levels, 13 HSPs gene transcripts were chosen and subjected to qRT-PCR analyses.
Fig. 1.
Relative mRNA levels of various HSP genes in postmortem human occipital lobes of cerebral cortices. mRNA levels of HSPs obtained from microarray analyses were normalized to those of GAPDH. Signal intensities from two independent replicates were averaged and expressed as % intensity of GAPDH in a log scale. Based on negative control probes, mRNA levels of HSPs higher than 5% of GAPDH were considered to be significantly detected.
HSPs mRNA profiles in subjects with different causes of death
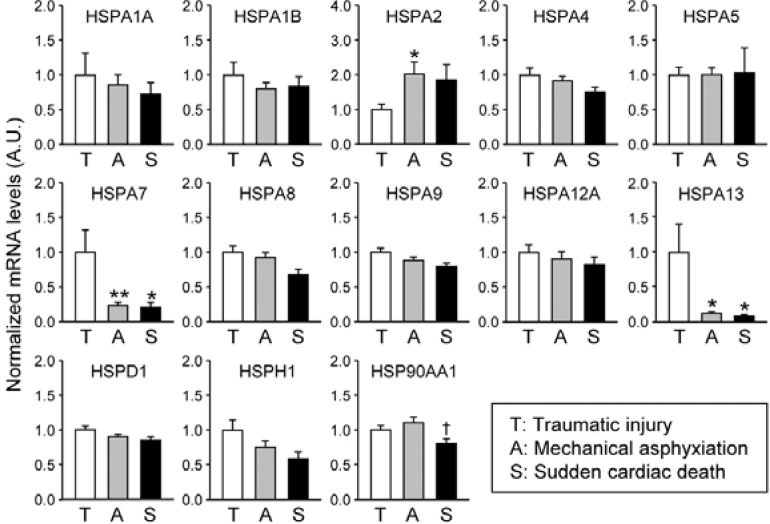
In the next set of experiments, we compared mRNA profiles of the selected HSP genes in subjects with three different causes of death, including TI, mechanical ASP, and SCD (Fig. 2 and Table 2). Among tested HSP gene transcripts, HSPA2 mRNA levels were significantly higher in the ASP subjects compared to the TI subjects by approximately 2-fold; it also shows a higher tendency in the SCD subjects than in the TI group although the difference is not significant by Newman-Keuls post-hoc comparison (ASP: 201.73 ± 34.82% of the mean value of the TI group; SCD: 184.83 ± 45.12). By contrast, HSPA7 and A13 mRNA levels were significantly lower in both ASP and SCD samples (ASP: 23.70 ± 4.35%; SCD: 21.08 ± 6.85%). Of interest, HSP90AA1 mRNA, which is known as a housekeeping gene, showed modestly but significantly lower levels in the SCD group than in those of the ASP samples.
Fig. 2.
HSP mRNA levels in the occipital lobes influenced by different causes of death. Occipital lobe-enriched HSP mRNA species were quantitatively analyzed by real-time qRT-PCR in subjects who died by traumatic injury (T), mechanical asphyxiation (A), or sudden cardiac death (S). Data were normalized to GAPDH mRNA levels and expressed the mean ± SE in arbitrary units (A.U.), where the mean values of the T group were set as 1. The group differences were statistically evaluated by one-way ANOVA followed by Newman-Keuls test (*p < 0.05 and **p < 0.01 vs. T; †p < 0.05 vs. A).
Table 2.
Statistical evaluation of group differences in HSP mRNA levels
| Gene name | F(2, 53) | P value |
|---|---|---|
| HSPA1A | 0.349 | 0.707 |
| HSPA1B | 0.610 | 0.547 |
| HSPA2 | 3.333 | 0.043* |
| HSPA4 | 2.145 | 0.127 |
| HSPA5 | 0.010 | 0.990 |
| HSPA7 | 5.325 | 0.008** |
| HSPA8 | 2.549 | 0.088 |
| HSPA9 | 2.951 | 0.061 |
| HSPA12A | 0.524 | 0.596 |
| HSPA13 | 4.582 | 0.015* |
| HSPD1 | 2.630 | 0.081 |
| HSPH1 | 2.836 | 0.068 |
| HSP90AA1 | 4.466 | 0.016* |
p < 0.05 and
p < 0.01 by one-way ANOVA
In quantitative RNA profiling of human autopsy materials, various factors related to RNA quality as well as premortem heterogeneity can be essential issues in the interpretation of results. We therefore evaluated our standardization method based on a housekeeping gene such as GAPDH. For this purpose, we plotted the cycle threshold (Ct) values of each HSP mRNA in relation to those of GAPDH, and the linearity of the correlations between two factors was then examined (Fig. 3 and Table 3). Most HSP mRNA species with higher basal expression levels, regardless of stressful stimuli, such as HSPA4, A8, A9, H1, and 90AA1 mRNA transcripts, showed a strong correlation to GAPDH in their mRNA abundances, with R2 values usually higher than 0.6. However, mRNA levels of three differentially expressed HSPs (i.e., HSPA2, A7, and A13) as well as known inducible HSPAs (i.e., HSPA1A and A1B), whose expression levels are highly induced during the acute postmortem period (Daugaard et al., 2007; Jardine et al., 2011) showed a less linear correlation to GAPDH. These results indicate that normalization to GAPDH mRNA levels is applicable for quantitative analyses on forensic RNA samples, which are degraded to variable extents. Furthermore, it can be postulated that HSP mRNA species with poor correlations to GAPDH mRNA might be altered in their expression levels either at the time of death or during supravital reactions.
Fig. 3.

Correlation of individual HSP mRNA levels to GAPDH. The Ct values for HSPA2 and A8 were plotted with those of GAPDH, and linear regression was carried out for each group (TI, open; ASP, gray; SCD, closed circles). Slopes and linear correlation coefficients expressed as R2 are summarized in Table 2.
Table 3.
Correlation between HSP and GAPDH mRNA levels
| Gene name (vs. GAPDH) | Slope (R2)
|
||
|---|---|---|---|
| TI | ASP | SCD | |
| HSPA1A | 0.2191 (0.0184) | 0.4981 (0.1748) | 0.6595 (0.2105) |
| HSPA1B | 0.3977 (0.0602) | 0.8076 (0.3977) | 0.7671 (0.3392) |
| HSPA2 | 0.6440 (0.2532) | 0.4944 (0.1100) | 0.7468 (0.2596) |
| HSPA7 | 0.0672 (0.0016) | 0.0285 (0.0031) | −0.0956 (0.0052) |
| HSPA13 | 0.0669 (0.0019) | 0.1465 (0.0193) | −0.0814 (0.0087) |
| HSPA4 | 0.9944 (0.7868) | 0.9781 (0.8245) | 0.8459 (0.6770) |
| HSPA5 | 1.2393 (0.6999) | 1.2014 (0.6469) | 1.1153 (0.6135) |
| HSPA8 | 1.4790 (0.9336) | 1.2918 (0.8583) | 1.3844 (0.9003) |
| HSPA9 | 1.1677 (0.9513) | 1.1678 (0.8827) | 1.1603 (0.8491) |
| HSPA12A | 1.3192 (0.7621) | 1.4035 (0.6470) | 0.9991 (0.5962) |
| HSPD1 | 0.7583 (0.5742) | 0.8311 (0.7421) | 0.8386 (0.8277) |
| HSPH1 | 1.8179 (0.8058) | 1.6483 (0.8318) | 1.8931 (0.9326) |
| HSP90AA1 | 0.8901 (0.5900) | 0.8507 (0.6250) | 0.9569 (0.6496) |
GAPDH, glyceraldehyde 3-phosphate dehydrogenase; TI, traumatic injury; ASP, asphyxiation; SCD, sudden cardiac death
Ratios of HSPA2 mRNA levels relative to those of HSPA7 or A13 as potential forensic biomarkers
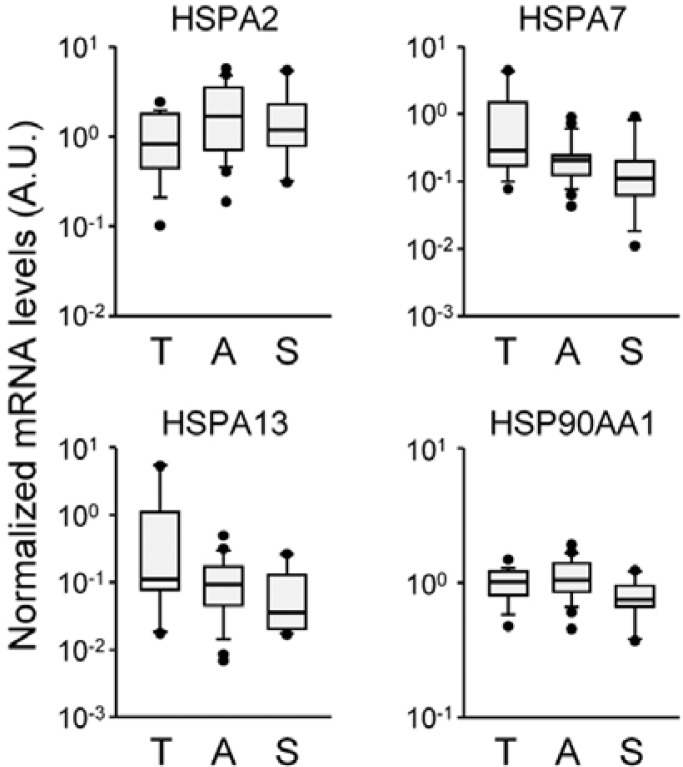
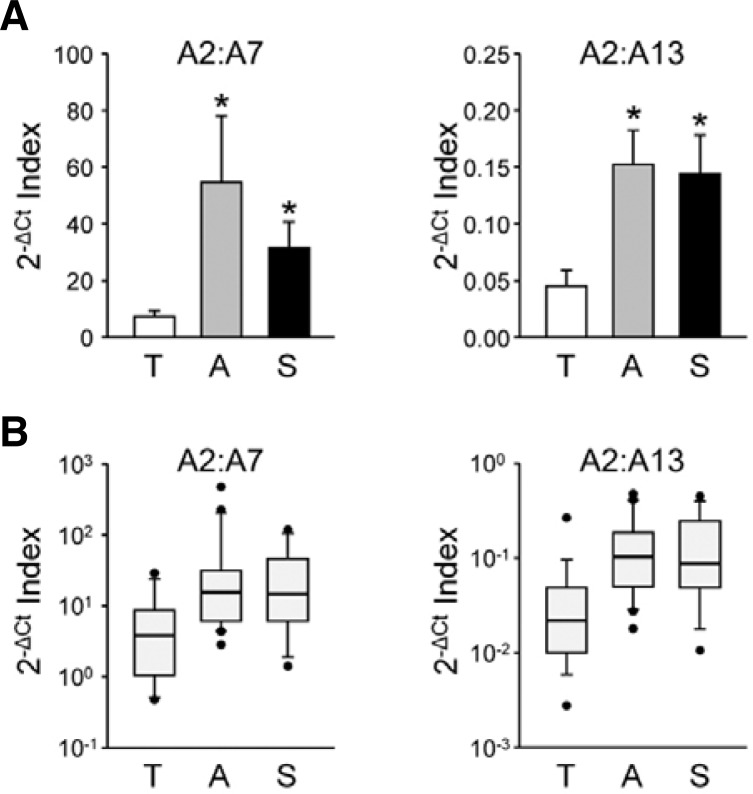
Although several HSP mRNA species exhibit significant group differences by cause of death, large variations within the group are found in all selected HSP genes. Box-and-whisker diagrams for differentially expressed HSP mRNA transcripts indicate that more than one-half of measured values in a group overlap with those of others, indicating poor discriminating power to be applied to molecular autopsy (Fig. 4). These results suggest that mRNA abundance for a certain HSP gene relative to a housekeeping mRNA may not provide practical information by itself. However, it is plausible that alterations in the composition of brain HSP gene transcripts are valuable indices with better resolution. To test this idea, we compared ratios of the relative abundances of HSPA2 to A7 or A13 showing larger group differences in opposite directions by evaluating the differences in their Ct values. As shown in Fig. 5A, 2−ΔCt indices for the HSPA2:A7/A13 ratios were higher in ASP and SCD subjects compared to TI subjects (for the HSPA2:A7 ratio, ASP: 728.07 ± 308.59% and SCD: 421.64 ± 45.12% of the mean value of TI group, F(2, 53) = 4.218, p < 0 .05 by one-way ANOVA; for the HSPA2:A13 ratio, ASP: 309.82 ± 65.51% and SCD: 318.37 ± 75.89%, F(2, 53) = 4.779, p < 0.05). More importantly, box-and-whisker diagrams for these ratios show better resolution between TI and the other groups (Fig. 5B). In both cases, overlapping sections were reduced by approximately 25%.
Fig. 4.
Variations of selected HSP mRNA levels within the group. Relative mRNA levels of the differentially regulated HSPs were expressed by box-and-whisker diagrams in log scales. The median values are expressed as lines in the middle of the boxes.
Fig. 5.
Relative mRNA abundances between two differentially expressed HSP gene transcripts. Relative mRNA abundances between HSPA2 and A7 (left) or A13 (right) were examined by use of 2−ΔCt indices of two mRNA transcripts (ΔCt = CtHSPA2 − CtHSPA7/A13) and then plotted using standard bar charts indicating the mean ± SE (A; *p < 0.05 vs. T by Newman-Keuls test as post-hoc comparison) or box-and-whisker diagrams (B).
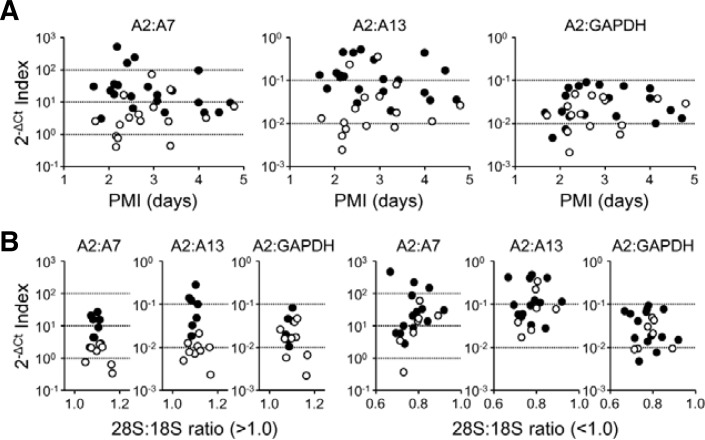
Next, we further dissected our results in relation to PMI or RNA quality, particularly between the TI and ASP subjects, as the PMIs of our SCD cases were quite uncertain. When we compared the individual results of the two groups of subjects in relation to PMI, the 2−ΔCt indices for HSPA2:A7 or HSPA2:A13 ratios were more clearly split than those of HSPA2:GAPDH by cause of death. More importantly, the split patterns appear to be influenced to a lesser extent by PMI (Fig. 6A). Rather, RNA integrity is likely to be a crucial determinant. For cases with relatively intact RNA (>1.0 for 28S:18S rRNA ratio), the 2−ΔCt indices for HSPA2:A7 or HSPA2:A13 were clearly separated between the TI and ASP groups, but the 2−ΔCt indices for A2:GAPDH failed to exhibit such discrimination (Fig. 5B, left panels). However, when severe RNA degradation (< 1.0 for 28S:18S rRNA ratio) is observed, no indices could clearly distinguish the cause of death (Fig. 6B, right panels). Taken together, our findings strongly suggest that alterations in the compositions of the HSP isoforms may be better forensic molecular indices for cause of death than relative mRNA levels normalized to housekeeping genes with a higher resolution.
Fig. 6.
Individual profiles of the TI and ASP subjects in relation to PMI and RNA integrity. (A) HSPA2 mRNA abundance relative to A7 (left), A13 (middle), or GAPDH (right) of each subject were plotted in relation to PMI (TI, open; ASP, closed symbols). (B) Subjects from the TI and ASP groups were categorized into two subgroups according to the 28S:18S rRNA ratio, and HSPA2 mRNA abundance relative to A7 (left), A13 (middle), or GAPDH (right) of a subject were plotted in relation to the 28S:18S rRNA ratio (TI, open; ASP, closed symbols).
DISCUSSION
We intended to examine whether cause and process of death could differentially affect cellular stress responses, leading to altered HSP mRNA profiles that can persist and become detectable in postmortem brain tissues. For this purpose, we compared the HSP mRNA profiles in the occipital lobes from subjects with three different causes of death: TI, ASP, and SCD. Although previous studies suggested that the expression of inducible HSPs may increase in various postmortem tissues after somatic death as a cellular stress response (Bjarnadóttir et al., 2010; Jardine et al., 2011; Mash et al., 2009; Sanoudou et al., 2004), the present study dissects the differential profiles of brain HSP mRNA expression according to the cause of death. The key finding of the present study is that mRNA profiles of several HSPs, such as HSPA2, A7, and A13, are distinct in the ASP and SCD groups compared with the TI group. It should be noted that mechanical ASP and SCD can evoke an immediate anoxic/hypoxic state and subsequently disrupt metabolic/energy homeostasis in the brain tissues, leading to acute somatic death. By contrast, TI is usually accompanied by blood loss and anemia, thereby causing the gradual deterioration of vital functions with longer survival times in many cases (Maeda et al., 2011; Zhao et al., 2009). Therefore, differences in mRNA profiles among HSPA/HSP70 members are most likely attributable to differences in conditions of death, particularly in terms of severity and robustness of anoxic/hypoxic damage to the brain and survival periods after lethal insults.
The HSPA/HSP70 family of molecular chaperones is presently the most widely studied group, comprised of more than 13 highly related members that are either constitutively expressed or induced by cellular stress (Daugaard et al., 2007). HSPA1A and A1B are known as major inducible HSP70s. For instance, global ischemia can dramatically increase both mRNA and protein levels of HSPA1 (Aoki et al., 1993; Muranyi et al., 2005; Sharp et al., 1993). HSPA1 protein levels were also reported to be increased to a greater extent in the brain stem after drowning compared with other causes of ASP (Gotohda et al., 2000). However, we could not find any significant differences in HSPA1 mRNA levels in the occipital lobes of the cerebral cortex by three different causes of death (Fig. 2). Rather, some of less-inducible members, such as HSPA2, A7, and A13, exhibit significant differences. Among tested HSPs, only HSPA2 mRNA levels were significantly higher, whereas HSPA7 and A13 mRNA levels were lower in the ASP and SCD groups. Since HSPA7 and A13 are also known as inducible members of HSPs (Leung et al., 1992; Otterson et al., 1994), it can be postulated that these HSPs are not induced in subjects with ASP or acute heart failure as robustly as in those with TI. Rapid gene transcription of inducible HSP mRNAs primarily lies under the control of HSF1, acting on downstream target genes bearing heat shock elements in their promoter region (Chang et al., 2012; Fujimoto and Nakai, 2010), other cis-elements in the promoters often contribute to differential modes of transcriptional induction in different cellular contexts as well as by exposure to different types and severities of stressful stimuli (Diller, 2006; Stetler et al., 2010). Therefore, it is reasonable that circumstances of death can differentially influence transcriptional induction of each member of the inducible HSPs, leading to alterations in postmortem mRNA profiles.
It is also noteworthy that ubiquitously expressed HSP90AA1 mRNA levels were significantly different between the ASP and SCD groups. HSP90AA1 belongs to the HSP90 subfamily that accounts for 1–2% of all cellular proteins in most cells, even under non-stress conditions (Csermely et al., 1998). Although HSP90AA1 gene expression can be induced by severe cellular stress, it is transcriptionally active under normal physiological conditions. This strongly suggests that its expression can be largely affected by preexisting pathologies and genetic diversities of the subjects. Furthermore, modest group differences in their mRNA levels as well as their higher correlations to GAPDH mRNA profiles (see Fig. 4 and Table 3) also suggest that HSP90AA1 is inappropriate as forensic biological markers for cause of death.
It is fundamentally difficult to standardize RNA profiling results in forensic medicine. This is primarily due to the fact that postmortem human tissues are heterogeneous. Postmortem intervals and conditions as well as antemortem factors such as age, gender, body mass, and the physiological/pathological conditions of the agonal phase can profoundly affect expression profiles and half-lives of certain gene transcripts in postmortem tissues, obscuring the employment of molecular biomarkers to investigate the cause and process of death (Durrenberger et al., 2010; Preece and Cairns, 2003; Tomita et al., 2004). Therefore, data normalization and RNA integrity have been essential issues in the field of forensic molecular pathology (Vennemann and Koppelkamm, 2010b). Various normalization methods using the total amount of tissue, DNA, or RNA, and endogenous reference genes have been proposed (Huggett et al., 2005; Talaat et al., 2002; Vandesompele et al., 2002).
In this regard, it should be noted that there are large variations among RNA species in susceptibility to postmortem degradation. A subgroup of mRNA transcripts carrying the 3′-UTR AUUUA motif was shown to be particularly susceptible to PMI-related degradation in animal models (Catts et al., 2005). In addition, a systemic analysis of postmortem brain tissues also demonstrated that RNA quality is a more critical determinant in analyzing a broad spectrum of mRNA transcripts, even more so than agony factors (Popova et al., 2008). Thus, data normalization to endogenous housekeeping reference genes without regard to RNA quality effects could be misleading. As shown in Fig. 6, individual HSPA2 mRNA profiles normalized to GAPDH cannot clearly distinguish between TI and ASP subjects, even when total RNA samples are relatively intact. On the other hand, alterations in mRNA compositions among related members belonging to a large family such as HSPs can be a better strategy in postmortem analyses, because they usually show higher homologies in their sequences and, at least in part, share posttranscriptional regulatory mechanisms. For example, comparison of human HSPA2 (NM_021979) and A7 (NR_024151) mRNA sequences revealed 77% homology in the coding region. The indices based on the abundance of HSPA2 mRNA relative to HSPA7 or A13 discriminated more than 70% of examined subjects belonging to the TI and ASP groups. More importantly, our indices appear to be less affected by PMI and better applicable when RNA samples are less degraded (Fig. 6).
In conclusion, the present study suggests that composition of a subset of HSP mRNA species in postmortem occipital lobes can be affected by different death processes, thereby implying cause of death. Although gene expression alterations in certain classes of genes cannot be used as the sole convincing evidence, accumulation of data that enables combined analyses on a wide spectrum of gene transcripts would provide essential information on the cause and process of death, supporting the classical approaches based on morphological, pathological, and toxicological features.
Acknowledgments
This work was supported by the Korean government through the National Forensic Service, Ministry of Public Administration and Security (Grant #: 1315000312, NFS-NG-2011-01) and the Ministry of Education, Science and Technology (Grant #: 2009-0088886 and 2011-0019232), and by a grant to Dr. J.-J. Hwang from the Glami Co., Ltd., Seoul, Korea. Microarray experiments were carried out by the help of Neurogene Bank Service in the Korea University College of Medicine supported by the 21stC Frontier R&D Program in Neuroscience. BioScience Writers edited the manuscript prior to submission.
REFERENCES
- Aoki M., Abe K., Kawagoe J., Sato S., Nakamura S., Kogure K. Temporal profile of the induction of heat shock protein 70 and heat shock cognate protein 70 mRNAs after transient ischemia in gerbil brain. Brain Res. 1993;601:185–192. doi: 10.1016/0006-8993(93)91709-2. [DOI] [PubMed] [Google Scholar]
- Bjarnadóttir S.G., Hollung K., Faergestad E.M., Veiseth-Kent E. Proteome changes in bovine longissimus thoracis muscle during the first 48 h postmortem shifts in energy status and myofibrillar stability. J. Agric. Food Chem. 2010;58:7408–7414. doi: 10.1021/jf100697h. [DOI] [PubMed] [Google Scholar]
- Brun A., Gustafson L. Distribution of cerebral degeneration in Alzheimer’s disease. A clinico-pathological study. Arch. Psychiatr. Nervenkr. 1976;223:15–33. doi: 10.1007/BF00367450. [DOI] [PubMed] [Google Scholar]
- Calabrese V., Butterfield D.A., Scapagnini G., Stella A.M., Maines M.D. Redox regulation of heat shock protein expression by signaling involving nitric oxide and carbon monoxide: relevance to brain aging, neurodegenerative disorders and longevity. Antioxid. Redox. Signal. 2006;8:444–477. doi: 10.1089/ars.2006.8.444. [DOI] [PubMed] [Google Scholar]
- Catts V.S., Catts S.V., Fernandez H.R., Taylor J.M., Coulson E. J., Lutze-Mann L.H. A microarray study of postmortem mRNA degradation in mouse brain tissue. Brain Res. Mol. Brain Res. 2005;138:164–177. doi: 10.1016/j.molbrainres.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Chang Z., Lu M., Park S.M., Park H.K., Kang H.S., Pak Y., Park J.S. Functional HSF1 requires aromatic-participant interactions in protecting mouse embryonic fibroblasts against apoptosis via G2 cell cycle arrest. Mol. Cells. 2012;33:465–470. doi: 10.1007/s10059-012-2246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe H.K., Son G.H., Chung S., Kim M., Sun W., Kim H., Geum D., Kim K. Maternal stress retards fetal development in mice with transcriptome-wide impact on gene expression profiles of the limb. Stress. 2011;14:194–204. doi: 10.3109/10253890.2010.529972. [DOI] [PubMed] [Google Scholar]
- Courts C., Madea B. Specific micro-RNA signatures for the detection of saliva and blood in forensic body-fluid identification. J. Forensic Sci. 2011;56:1464–1470. doi: 10.1111/j.1556-4029.2011.01894.x. [DOI] [PubMed] [Google Scholar]
- Csermely P., Schnaider T., Soti C., Prohászka Z., Nardai G. The 90-kDa molecular chaperone family: structure, function and clinical applications. A comprehensive review. Pharmacol. Ther. 1998;79:129–168. doi: 10.1016/s0163-7258(98)00013-8. [DOI] [PubMed] [Google Scholar]
- Dabir D.V., Trojanowski J.Q., Richter-Landsberg C., Lee V.M., Forman M.S. Expression of the small heat-shock protein alphaB-crystallin in tauopathies with glial pathology. Am. J. Pathol. 2004;164:155–166. doi: 10.1016/s0002-9440(10)63106-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugaard M., Rohde M., Jäättelä M. The heat shock protein 70 family Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- Diller K.R. Stress protein expression kinetics. Annu. Rev. Biomed. Eng. 2006;8:403–424. doi: 10.1146/annurev.bioeng.7.060804.100449. [DOI] [PubMed] [Google Scholar]
- Durrenberger P.F., Fernando S., Kashefi S.N., Ferrer I., Hauw J.J., Seilhean D., Smith C., Walker R., Al-Sarraj S., Troakes C., et al. Effects of antemortem and postmortem variables on human brain mRNA quality a BrainNet Europe study. J. Neuropathol. Exp. Neurol. 2010;69:70–81. doi: 10.1097/NEN.0b013e3181c7e32f. [DOI] [PubMed] [Google Scholar]
- Forman M.S., Trojanowski J.Q., Lee V.M. Neurodegenerative diseases a decade of discoveries paves the way for therapeutic breakthroughs. Nat. Med. 2004;10:1055–1063. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- Fujimoto M., Nakai A. The heat shock factor family and adaptation to proteotoxic stress. FEBS J. 2010;277:4112–4125. doi: 10.1111/j.1742-4658.2010.07827.x. [DOI] [PubMed] [Google Scholar]
- Gotohda T., Kubo S., Kitamura O., Tokunaga I., Eguchi A., Orihara Y., Tsuda R., Ikematsu K., Nakasono I. HSP70 and c-Fos expression of brain stem hypoglossal nucleus in drowning. J. Med. Invest. 2000;47:76–79. [PubMed] [Google Scholar]
- Haas C., Klesser B., Maake C., Bär W., Kratzer A. mRNA profiling for body fluid identification by reverse transcription endpoint PCR and realtime PCR. Forensic Sci. Int. Genet. 2009;3:80–88. doi: 10.1016/j.fsigen.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Hecker J.G., McGarvey M. Heat shock proteins as biomarkers for the rapid detection of brain and spinal cord ischemia: a review and comparison to other methods of detection in thoracic aneurysm repair. Cell Stress Chaperones. 2011;16:119–131. doi: 10.1007/s12192-010-0224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich M., Lutz-Bonengel S., Matt K., Schmidt U. Real-time PCR detection of five different “endogenous control gene” transcripts in forensic autopsy material. Forensic Sci. Int. Genet. 2007a;1:163–169. doi: 10.1016/j.fsigen.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Heinrich M., Matt K., Lutz-Bonengel S., Schmidt U. Successful RNA extraction from various human postmortem tissues. Int. J. Legal Med. 2007b;121:136–142. doi: 10.1007/s00414-006-0131-9. [DOI] [PubMed] [Google Scholar]
- Huggett J., Dheda K., Bustin S., Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- Ikematsu K., Tsuda R., Nakasono I. Gene response of mouse skin to pressure injury in the neck region. Leg. Med (Tokyo) 2005;8:128–131. doi: 10.1016/j.legalmed.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Ikematsu K., Takahashi H., Kondo T., Tsuda R., Nakasono I. Temporal expression of immediate early gene mRNA during the supravital reaction in mouse brain and lung after mechanical asphyxiation. Forensic Sci. Int. 2008;179:152–156. doi: 10.1016/j.forsciint.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Jardine D., Cornel L., Emond M. Gene expression analysis characterizes antemortem stress and has implications for establishing cause of death. Physiol. Genomics. 2011;43:974–980. doi: 10.1152/physiolgenomics.00062.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juusola J., Ballantyne J. Messenger RNA profiling a prototype method to supplant conventional methods for body fluid identification. Forensic Sci. Int. 2003;135:85–96. doi: 10.1016/s0379-0738(03)00197-x. [DOI] [PubMed] [Google Scholar]
- Lee J., Hever A., Willhite D., Zlotnik A., Hevezi P. Effects of RNA degradation on gene expression analysis of human postmortem tissues. FASEB J. 2005;19:1356–1358. doi: 10.1096/fj.04-3552fje. [DOI] [PubMed] [Google Scholar]
- Leonard S., Logel J., Luthman D., Casanova M., Kirch D., Freedman R. Biological stability of mRNA isolated from human postmortem brain collections. Biol. Psychiatry. 1993;33:456–466. doi: 10.1016/0006-3223(93)90174-c. [DOI] [PubMed] [Google Scholar]
- Leung T.K., Hall C., Rajendran M., Spurr N.K., Lim L. The human heat-shock genes HSPA6 and HSPA7 are both expressed and localize to chromosome 1. Genomics. 1992;12:74–79. doi: 10.1016/0888-7543(92)90409-l. [DOI] [PubMed] [Google Scholar]
- Maeda H., Zhu B.L., Ishikawa T., Michiue T. Forensic molecular pathology of violent deaths. Forensic Sci. Int. 2010;203:83–92. doi: 10.1016/j.forsciint.2010.07.024. [DOI] [PubMed] [Google Scholar]
- Maeda H., Ishikawa T., Michiue T. Forensic biochemistry for functional investigation of death: concept and practical application. Leg. Med (Tokyo) 2011;13:55–67. doi: 10.1016/j.legalmed.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Mash D.C., Duque L., Pablo J., Qin Y., Adi N., Hearn W.L., Hyma B.A., Karch S.B., Druid H., Wetli C.V. Brain biomarkers for identifying excited delirium as a cause of sudden death. Forensic Sci. Int. 2009;190:e13–19. doi: 10.1016/j.forsciint.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Miyazato T., Ishikawa T., Michiue T., Maeda H. Molecular pathology of pulmonary surfactants and cytokines in drowning compared with other asphyxiation and fatal hypothermia. Int. J. Legal Med. 2012;126:581–587. doi: 10.1007/s00414-012-0698-2. [DOI] [PubMed] [Google Scholar]
- Muranyi M., He Q.P., Fong K.S., Li P.A. Induction of heat shock proteins by hyperglycemic cerebral ischemia. Brain Res. Mol. Brain Res. 2005;139:80–87. doi: 10.1016/j.molbrainres.2005.05.023. [DOI] [PubMed] [Google Scholar]
- Otterson G.A., Flynn G.C., Kratzke R.A., Coxon A., Johnston P.G., Kaye F.J. Stch encodes the ‘ATPase core’ of a microsomal stress 70 protein. EMBO J. 1994;13:1216–1225. doi: 10.1002/j.1460-2075.1994.tb06371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova T., Mennerich D., Weith A., Quast K. Effect of RNA quality on transcript intensity levels in microarray analysis of human post-mortem brain tissues. BMC Genomics. 2008;9:91. doi: 10.1186/1471-2164-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preece P., Cairns N.J. Quantifying mRNA in postmortem human brain: influence of gender, age at death, postmortem interval, brain pH, agonal state and inter-lobe mRNA variance. Brain Res. Mol. Brain Res. 2003;118:60–71. doi: 10.1016/s0169-328x(03)00337-1. [DOI] [PubMed] [Google Scholar]
- Price J.L., Drevets W.C. Neural circuits underlying the pathophysiology of mood disorders. Trends Cogn. Sci. 2012;16:61–71. doi: 10.1016/j.tics.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Romi F., Helgeland G., Gilhus N.E. Heat-shock proteins in clinical neurology. Eur. Neurol. 2011;66:65–69. doi: 10.1159/000329373. [DOI] [PubMed] [Google Scholar]
- Sanoudou D., Kang P.B., Haslett J.N., Han M., Kunkel L.M., Beggs A.H. Transcriptional profile of postmortem skeletal muscle. Physiol. Genomics. 2004;16:222–228. doi: 10.1152/physiolgenomics.00137.2003. [DOI] [PubMed] [Google Scholar]
- Sharp F.R., Kinouchi H., Koistinaho J., Chan P.H., Sagar S.M. HSP70 heat shock gene regulation during ischemia. Stroke. 1993;24:172–175. [PubMed] [Google Scholar]
- Stetler R.A., Gan Y., Zhang W., Liou A.K., Gao Y., Cao G., Chen J. Heat shock proteins cellular and molecular mechanisms in the central nervous system. Prog. Neurobiol. 2010;92:184–211. doi: 10.1016/j.pneurobio.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Ikematsu K., Tsuda R., Nakasono I. Increase in dual specificity phosphatase 1, TGF-beta stimulated gene 22, domain family protein 3 and Luc7 homolog (S. cerevisiae)-like messenger RNA after mechanical asphyxiation in the mouse lung. Leg. Med (Tokyo) 2009;11:181–185. doi: 10.1016/j.legalmed.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Talaat A.M., Howard S.T., Hale W., Lyons R., Garner H., Johnston S.A. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 2002;30:e104. doi: 10.1093/nar/gnf103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita H., Vawter M.P., Walsh D.M., Evans S.J., Choudary P.V., Li J., Overman K.M., Atz M.E., Myers R.M., Jones E.G., et al. Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol. Psychiatry. 2004;55:346–352. doi: 10.1016/j.biopsych.2003.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennemann M., Koppelkamm A. mRNA profiling in forensic genetics I Possibilities and limitations. Forensic Sci. Int. 2010a;203:71–75. doi: 10.1016/j.forsciint.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Vennemann M., Koppelkamm A. Postmortem mRNA profiling II Practical considerations. Forensic Sci. Int. 2010b;203:76–82. doi: 10.1016/j.forsciint.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Zhao D., Zhu B.L., Ishikawa T., Li D.R., Michiue T., Maeda H. Quantitative RT-PCR assays of hypoxia-inducible factor-1alpha, erythropoietin and vascular endothelial growth factor mRNA transcripts in the kidneys with regard to the cause of death in medicolegal autopsy. Leg. Med (Tokyo) 2006;8:258–263. doi: 10.1016/j.legalmed.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Zhao D., Ishikawa T., Quan L., Li D.R., Michiue T., Yoshida C., Komatu A., Chen J.H., Zhu B.L., Maeda H. Tissue-specific differences in mRNA quantification of glucose transporter 1 and vascular endothelial growth factor with special regard to death investigations of fatal injuries. Forensic Sci. Int. 2008;177:176–183. doi: 10.1016/j.forsciint.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Zhao D., Ishikawa T., Quan L., Michiue T., Yoshida C., Komatu A., Chen J.H., Wang Q., Zhu B.L., Maeda H. Evaluation of pulmonary GLUT1 and VEGF mRNA levels in relation to lung weight in medicolegal autopsy cases. Leg. Med (Tokyo) Suppl. 2009;1:S290–293. doi: 10.1016/j.legalmed.2009.01.094. [DOI] [PubMed] [Google Scholar]
- Zubakov D., Hanekamp E., Kokshoorn M., van Ijcken W., Kayser M. Stable RNA markers for identification of blood and saliva stains revealed from whole genome expression analysis of time-wise degraded samples. Int. J. Legal Med. 2008;122:135–142. doi: 10.1007/s00414-007-0182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubakov D., Boersma A.W., Choi Y., van Kuijk P.F., Wiemer E.A., Kayser M. MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int. J. Legal Med. 2010;124:217–226. doi: 10.1007/s00414-009-0402-3. [DOI] [PMC free article] [PubMed] [Google Scholar]