Abstract
Secreted proteins are known to have multiple roles in plant development, metabolism, and stress response. In a previous study to understand the roles of secreted proteins, Capsicum annuum secreted proteins (CaS) were isolated by yeast secretion trap. Among the secreted proteins, we further characterized Capsicum annuum senescence-delaying 1 (CaSD1), a gene encoding a novel secreted protein that is present only in the genus Capsicum. The deduced CaSD1 contains multiple repeats of the amino acid sequence KPPIHNHKPTDYDRS. Interestingly, the number of repeats varied among cultivars and species in the Capsicum genus. CaSD1 is constitutively expressed in roots, and Agrobacterium-mediated transient overexpression of CaSD1 in Nicotiana benthamiana leaves resulted in delayed senescence with a dramatically increased number of trichomes and enlarged epidermal cells. Furthermore, senescence- and cell division-related genes were differentially regulated by CaSD1-overexpressing plants. These observations imply that the pepper-specific cell wall protein CaSD1 plays roles in plant growth and development by regulating cell division and differentiation.
Keywords: cell wall protein, Nicotiana benthamiana, pepper (Capsicum annuum L.), senescence, trichome
INTRODUCTION
Plants have developed unique structures and mechanisms to control growth and development or to overcome biotic or abiotic stresses. One of the unique features of plant cells is the presence of a cell wall, which is a dynamic structure and varies according to developmental stage and environmental conditions. The plant cell wall is primarily composed of polysaccharides, such as cellulose, hemicelluloses, and pectin, which together make up to 90% of the dry weight of the plant (Cosgrove, 2005; Lerouxel et al., 2006). However, the plant cell wall also contains proteins that often play crucial roles in maintaining the structure and function of the cell wall (Agrawal et al., 2010; Showalter, 1993).
The group of proteins exported through the secretory pathway and localized in the cell wall or extracellular space is called the secretome. Members of plant secretomes have various roles in plant cell expansion, differentiation, cell-to-cell communication, and defense against pathogens (Humphrey et al., 2007; Lee et al., 2004). Structural proteins, such as hydroxyproline-rich glycoproteins, proline-rich proteins, glycine-rich proteins, and arabinogalactan proteins, act on polysaccharides and are primarily involved in plant growth and development. For example, LRR/extensin-1 (LRX1) is a hydroxyproline-rich glycoprotein that is linked to root hair morphogenesis and elongation (Baumberger et al., 2001). In addition, expansins, xyloglucan endotransglucosylase/hydrolases, and endo-(1,4)-β-D-glucanases are wall-associated proteins that play roles in cell extensibility and differentiation. In particular, expansins have been extensively studied and are known to control leaf initiation and shape (Cho and Cosgrove, 2000; Fleming et al., 1997; Pien et al., 2001). Other secreted proteins are known to be involved in cell wall strengthening and signal transmission in the defense against pathogens (Hematy et al., 2009; Huckelhoven, 2007; Oh et al., 2005; Yeom et al., 2012). However, the roles of a large portion of the plant secretome remain to be elucidated.
Senescence is the final stage of growth and development for all living organisms. In plants, the degradation of chlorophyll, proteins, and nucleic acids is the main symptom in senescing leaves (Lim et al., 2007; Quirino et al., 2000). To date, genetic and molecular approaches have been performed to elucidate the senescence mechanism. Using T-DNA insertion or chemical-induced mutagenesis, a number of mutants showing delayed senescence were identified. For example, mutation of ORE9, which contains an F-box motif and 18 leucine-rich repeats, delayed overall senescence symptoms in Arabidopsis (Woo et al., 2001). A mutant produced by T-DNA insertion into the AtATE gene encoding arginyl-tRNA:protein arginyltransferase also delayed age-dependent and dark-induced senescence (Yoshida et al., 2002). In a molecular approach, DNA microarray analysis was used to profile the expression of genes in senescing leaves (Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006). Over 800 genes were upregulated more than 3-fold during senescence and were referred to as senescence-associated genes (SAGs).
In previous research, 101 unique proteins in the secretome that may be related to plant cell defense and development were isolated from peppers using the yeast secretion trap (YST) system (Yeom et al., 2011). One of the members of the secretome, Capsicum annuum senescence-delaying 1 (CaSD1), has a novel feature and was selected for in-depth study. Here, we functionally characterize the CaSD1 gene, which is present only in the genus Capsicum and has a unique repeat region with variable length among Capsicum species. Gain-of-function studies using ectopic transient overexpression in Nicotiana benthamiana revealed that CaSD1 plays roles in the delay of senescence and trichome formation.
MATERIALS AND METHODS
Plant materials
Pepper plants were used for cloning, gene expression, and loss-of-function analysis. Capsicum annuum L. cv. ‘Bukang’ was used to clone the full-length CaSD1 cDNA and to evaluate organ-specific expression. Four cultivars or germplasms of each of five different Capsicum species were randomly selected from RDA-Genebank information center, and genomic DNAs of each germplasm were also provided for polymerase chain reaction (PCR) amplification of the repeat region (Supplementary Table 1). Nicotiana benthamiana plants were used for subcellular localization and transient overexpression (TOE) of CaSD1. All plants were grown in a walk-in chamber maintained at 22–25°C and subjected to a 16-h photoperiod for 4–6 weeks.
Cloning and DNA sequence analysis
The full-length sequence of CaSD1 was isolated from a bacterial artificial chromosome (BAC) 529L23 sequence of C. annuum ‘CM334’ (Yoo et al., 2003). Both the gene-coding region and the 3′ untranslated region were predicted using the FGENESH program (http://linux1.softberry.com/berry.phtml). Gene-specific primers were designed for PCR amplification of CaSD1 based on the BAC sequence (Supplementary Table 2). PCR was performed to amplify CaSD1 using ‘Bukang’ cDNA, which was cloned into the pJET vector using the pJET™ PCR Cloning Kit (Fermentas Canada Inc., Canada). The CaSD1 sequence was confirmed by DNA sequencing using the ABI 3730 XL (Applied Biosystems Inc, USA) at the National Instrumentation Center for Environmental Management (NICEM, Korea). The sequence analyses were performed using the ExPasy translation tool (http://us.expasy.org/tools/dna.html), National Center for Biotechnology Information Blast search (Altschul et al., 1997), SignalP signal peptide prediction (http://www.cbs.dtu.dk/services/SignalP/), and the TCoffee multi-alignment tool (http://www.ebi.ac.uk/Tools/msa/tcoffee/).
RNA extraction and gene expression analysis
For gene expression analysis, total RNA was extracted from frozen pepper and N. benthamiana leaves using the TRIzol reagent (Molecular Research Center, USA). First-strand cDNA was synthesized from 5 μg total RNA using Oligo (dT) and SuperScript II reverse transcriptase (Invitrogen, USA). Quantitative or semi-quantitative RT-PCR was performed using gene-specific primers designed using Primer3 software (http://frodo.wi.mit.edu/primer3, Supplementary Table 2). For quantitative gene expression analysis, quantitative reverse transcription (RT)-PCR was performed with a Rotor-Gene 2000 (Qiagen, USA) using Syto 9 (Invitrogen). Fluorescence was measured at 72°C for 60 cycles, and expression levels of each sample were normalized to those of actin. Semi-quantitative RT-PCR was carried out using a MyCycler (Biorad, USA), and PCR products were electrophoresed in 1% agarose gels, stained with ethidium bromide, and photographed under ultraviolet light.
Subcellular localization of CaSD1-soluble modified form of green fluorescent protein (smGFP) or smGFP
Both pMBP1:CaSD1-smGFP (pBI121-Modified) and pMBP1: smGFP were constructed using the ligation-independent cloning method (Oh et al., 2010) and transformed into Agrobacterium tumefaciens strain GV2260. The transformed cells were cultured overnight in YEP medium at 30°C with shaking (200 rpm), centrifuged, and resuspended in infiltration buffer containing 10 mM MES (pH 5.5)/10 mM MgCl2. The cell suspension (O.D600 = 0.5) was incubated with 200 μM acetosyringone for 3 h at room temperature. The cell suspension was then pressure-infiltrated into the backsides of N. benthamiana leaves using a needleless syringe. One day after infiltration, the abaxial epidermal cell layer was peeled off, and GFP fluorescence was observed by confocal laser scanning microscopy (LSM510, Carl Zeiss, Germany).
Transient overexpression of CaSD1 in N. benthamiana
The CaSD1 open reading frame (ORF) containing the 3′ untranslated region was amplified with primers containing BamHI and SacI recognition sequences at the 5′ and 3′ ends, respectively. The PCR product was cloned into a pMBP1 vector treated with BamHI and SacI (Suh et al., 1998). The cloning product was transformed into Agrobacterium strain GV2260. Agrobacterium transformed with pMBP1:CaSD1 and the pMBP1 control were cultured overnight in YEP medium at 30°C with shaking (200 rpm). The cells were centrifuged and resuspended in infiltration buffer containing 10 mM MES (pH 5.5)/10 mM MgCl2. The resuspended bacterial suspension (O.D600 = 0.5) was incubated with 200 μM acetosyringone for 3 h at room temperature. After incubation, the bacterial suspension was pressure-infiltrated into the backsides of N. benthamiana leaves using a needleless syringe. The infiltrated leaves were used for RNA extraction and microscopic observation at the indicated time points.
Light and field emission scanning electronic microscopy
The surfaces of N. benthamiana leaves transiently overexpressing CaSD1 were monitored and photographed 10 days after TOE by light microscopy (Siwon Optical Technology, Korea) to observe the trichomes. For analysis of trichome density and cell size, field emission scanning electron microscopy (SUPRA 55VP, Carl Zeiss, Wetzlar, Germany, NICEM at Seoul National University) was performed 2, 7, and 10 days after TOE without processing samples in low-vacuum mode. The samples were monitored with a 15-kV accelerating voltage, and pictures were digitally captured.
RESULTS
CaSD1 is a novel protein present only in species of the Capsicum genus
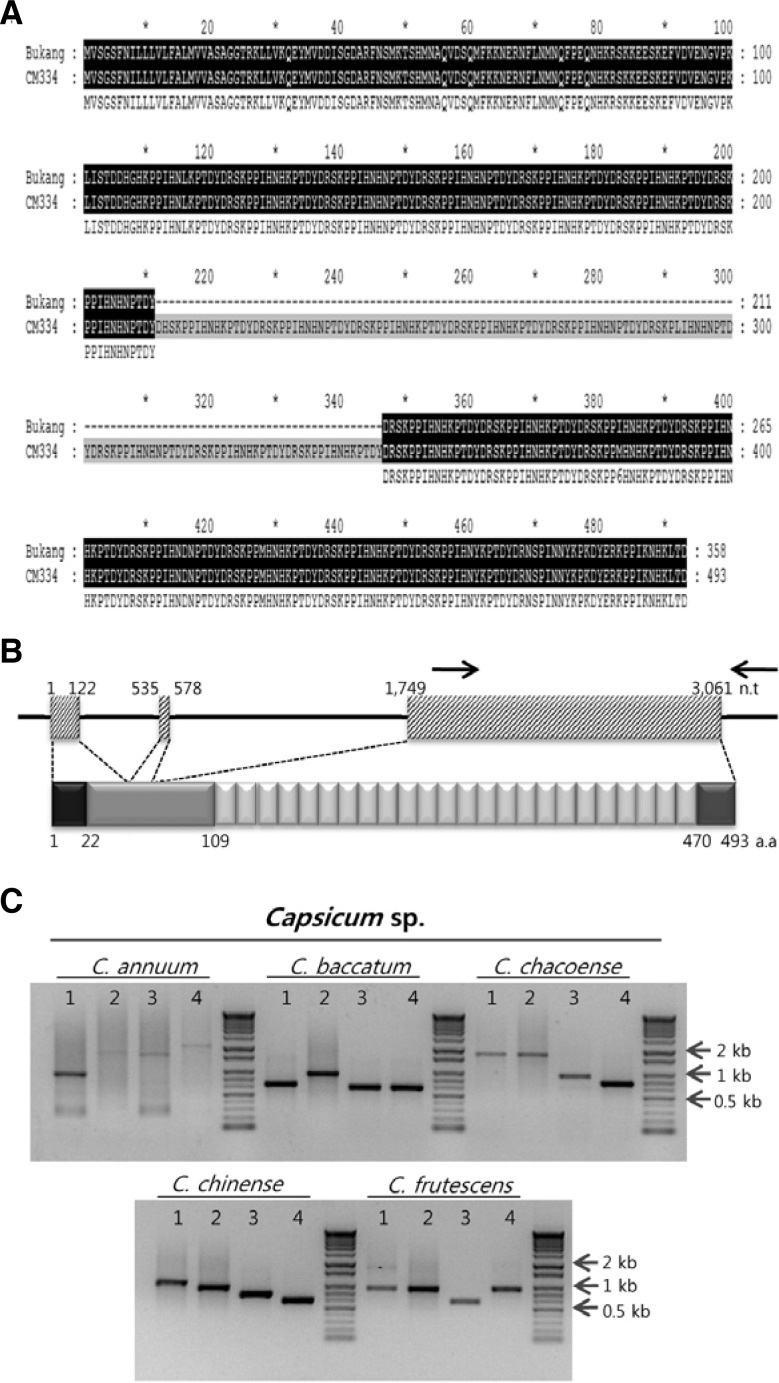
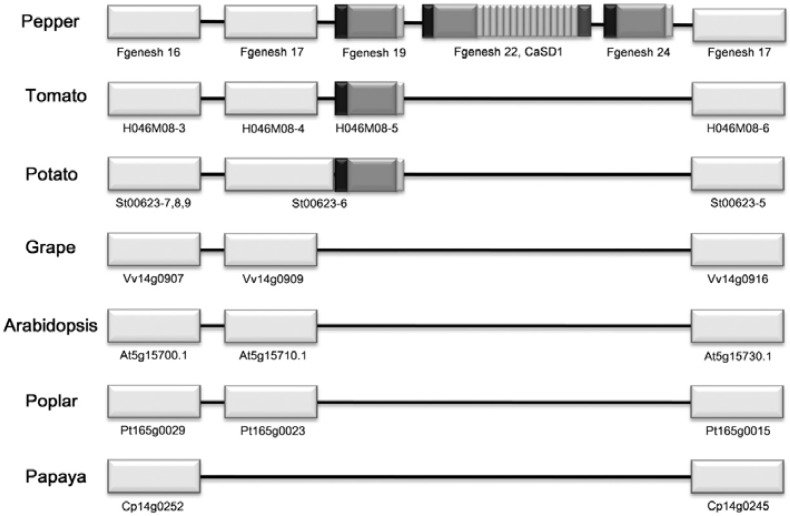
To identify secreted proteins involved in growth, development, and plant-pathogen interactions, C. annuum secretome (CaS) genes were isolated from pepper roots following P. capsici infection using YST (Yeom et al., 2011). Among them, CaS113 was selected for further study. Using the partial sequence of CaS113, a putative full-length cDNA was identified from C. annuum ‘CM334’ BAC sequences (Yoo et al., 2003). The predicted gene encodes a 493-amino acid protein with a molecular mass of 58 kDa composed of a signal peptide (SP), N-terminal non-repeat region, multiple repeat region, and C-terminal non-repeat region (Fig. 1B). The gene was thereafter referred to as C. annuum senescence-delaying gene 1 (CaSD1). In the multiple repeat regions, each repeat unit was 15 amino acids in length (KPPIHNHKPTDYDRS), often with one or two amino acids differing among the repeat units. CaSD1 from the pepper cultivar ‘Bukang’ was cloned, and the number of repeats was fewer than that of the ‘CM334’ cultivar. Other regions, except for the repeat number, were identical (Fig. 1A). Further investigation using PCR amplification of genomic DNA and sequence analysis revealed that the number of repeat units was variable among species and cultivars in the Capsicum genus (Fig. 1C). We cloned the full-length CaSD1 homologs in two Capsicum chinense cultivars, ‘Jolokia’ and ‘Numex Suave Orange,’ in which the SPs and non-repeat regions of the deduced proteins showed 92% sequence similarity with CaSD1. However, these homologs had 9 and 12 repeat units, respectively (Supplementary Fig. 1). Additionally, two genes predicted from the BAC sequence around CaSD1 had conserved SPs and non-repeat regions but no repeat regions (Fig. 2). Comparative genomic analysis revealed that genes lacking a repeat region exist in the Solanacea family but that CaSD1 homologs with a repeat region are only present in the Capsicum genus (Fig. 2). Additional searches in the GenBank database and Interpro-Scan could not retrieve any significantly matched genes or domains except for the signal peptide (data not shown). These results suggest that CaSD1 is a novel Capsicum-specific gene.
Fig. 1.
Sequence analysis of CaSD1. (A) Sequence alignment of CaSD1 in C. annuum L. cv. ‘Bukang’ and ‘CM334.’ Black box denotes sequence match between ‘Bukang’ and ‘CM334.’ (B) Schematic structure of CaSD1 genomic DNA region and amino acid sequence of CaSD1. The black line indicates the 5′ and 3′ untranslated regions and introns. The box with diagonal line indicates exons of CaSD1. Black arrow designates primers for PCR amplification of the repeat region. Black box: signal peptide, light gray: non-repeat region, white: repeat region, dark gray: another non-repeat region. n.t, nucleotide. a.a, amino acid. (C) Four genomic DNA samples from each Capsicum species were used for PCR amplification. PCR products were electrophoresed in 1% agarose gels, stained with ethidium bromide, and photographed under ultraviolet light.
Fig. 2.
Comparative genomic analysis of syntenic segments of known plant genomes. Syntenic segments including CaSD1 were compared to those of other known plant genomes. The ORFs of pepper BAC 529L23 were predicted using the FGENESH program, and each ORF was presented according to the order of FGENESH number. White boxes represent other proteins in syntenic regions. Proteins with sequence similarity were placed at the same vertical position.
Organ-specific expression and subcellular localization of CaSD1
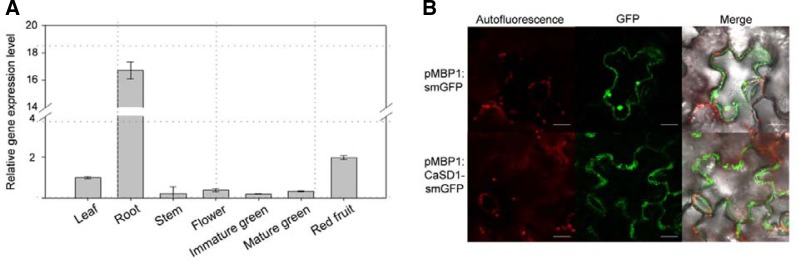
To investigate the organ-specific expression of CaSD1, real-time RT-PCR was performed using RNA samples from different pepper organs. CaSD1 was constitutively expressed in roots, where the transcript levels were 16.7-fold higher than those of leaves (Fig. 3A). To determine the cellular localization of CaSD1, targeting experiments in plants were performed by TOE using Agrobacterium carrying CaSD1 fused to a smGFP as a fluorescent marker (Davis and Vierstra, 1998). Agrobacterium suspensions carrying pMBP1:CaSD1-smGFP or pMBP1:smGFP were pressure-infiltrated into N. benthamiana leaves. One day after TOE, abaxial epidermal cell layers of the leaves were observed by confocal laser scanning microscopy. GFP fluorescence of pMBP1:CaSD1-smGFP was observed in the cell wall and extracellular matrix, while pMBP1:smGFP was localized throughout the cells, including the nucleus (Fig. 3B). These observations indicate that CaSD1 is secreted and localized to the extracellular space, including the plasma membrane or cell wall, in N. benthamiana plants.
Fig. 3.
Organ-specific expression of CaSD1 and localization of CaSD1 in the cell wall. (A) Organ-specific expression of CaSD1 in pepper plants. Real-time PCR was performed with CaSD1-specific primers, and the values were normalized to CaActin and calculated relative to expression levels in leaves. Bars represent standard deviation of four replicates. (B) Agrobacterium containing pMBP1: smGFP or pMBP1: CaSD1-smGFP was infiltrated into N. bentha-miana leaves. Pictures were taken 1 day after infiltration by confocal laser scanning microscopy. White bars, 20 μm.
Transient overexpression of CaSD1 causes delay of senescence
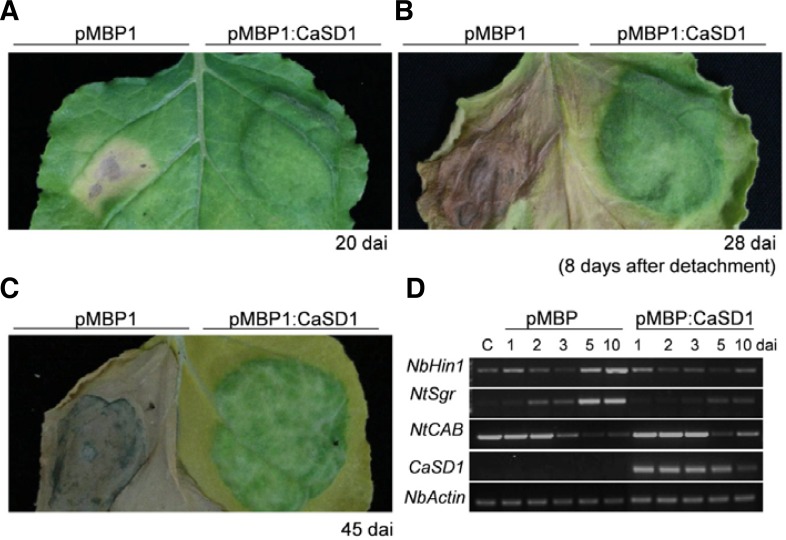
To investigate the function of CaSD1 in plants, we adopted loss-of-function and gain-of-function approaches. Virus-induced gene silencing (VIGS) in pepper was conducted for loss-of-function study. However, CaSD1-silenced pepper plants showed no significant difference compared to those of control plants. To investigate gain-of-function of CaSD1 in plants, Agrobacterium-mediated TOE of pMBP1 or pMBP1:CaSD1 was performed in N. benthamiana leaves. Significant differences between CaSD1-and control vector-expressing plants were observed about 5–7 days after inoculation (dai). CaSD1-overexpressing regions showed delayed senescence, while senescence was accelerated in empty vector pMBP1-expressing regions (Fig. 4A). Furthermore, the senescence-delayed region remained as a green island even after the leaf was detached (Fig. 4B). The CaSD1-expressing region of intact plants remained green until the pMBP1-control region withered away (Fig. 4C). To better understand the effects of CaSD1 on senescence, gene profiling experiments were conducted. Sgr (Staygreen) is a typical senescence-associated (SAG) gene which regulates chlorophyll degradation and sgr mutant rice show senescence-delayed phenotype (Park et al., 2007). Also, Harpin-induced 1 (Hin1) is a marker gene of senescence as well as cell death in plants (Pontier et al., 1999). Expression levels of Hin1 and Sgr were downregulated in CaSD1-overexpressing plants compared to control plants (Fig. 4D). On the other hand, the transcript levels of CAB (Chlorophyll a/b binding protein gene), which is related to the progression of photosynthesis and a representative control for leaf senescence (Lohman et al., 1994; Weaver et al., 1998), were higher in CaSD1-overexpressing plants than in vector control-expressing plants during senescence. These reults indicate that overexpression of CaSD1 suppressed senescence by affecting the expression levels of senescence-related genes and chlorophyll-associated genes.
Fig. 4.
Transient overexpression of CaSD1 delays senescence and regulates expression of senescence-related genes. Agrobacterium-mediated overexpression of pMBP1 or pMBP1:CaSD1 in N. benthamiana leaves. Photographs were taken of the same leaf (A) 20 days after inoculation (dai) and (B) 8 days after detachment. (C) Photograph was taken 45 dai. Similar results were obtained more than three times of independent experiments. (D) Total RNA was isolated from leaves at the indicated time points after TOE, and semi-quantitative RT-PCR was performed using gene-specific primer sets. NbActin gene was the control. C, control plants. NbHin1, N. benthamiana harpin-induced 1; NtSgr, N. tabaccum Staygreen; NtCAB, N. tabaccum chlorophyll a/b binding protein gene.
Transient overexpression of CaSD1 accelerates trichome formation with enlarged cell size
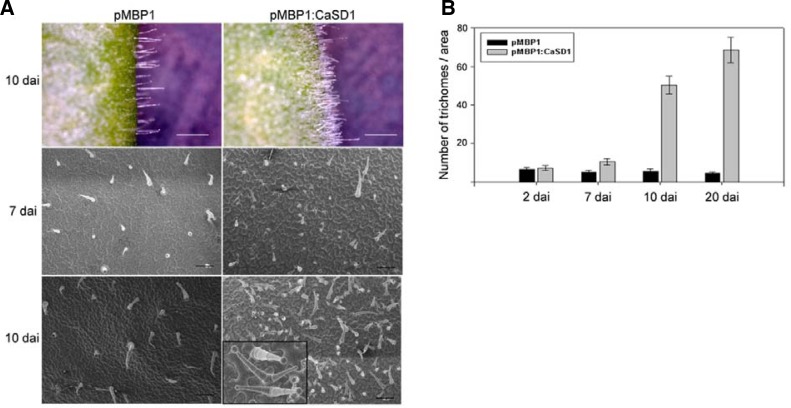
Another peculiar phenotype of CaSD1-overexpressing plants is the induction of trichome formation (Fig. 5). Trichomes are specialized epidermal cells that are regularly distributed on plant leaves (Hulskamp et al., 1994). Similar to the kinetics of CaSD1-induced senescence alterations, a significant difference was observed about 7 days after TOE. Trichome initiation sites (TIS) were significantly increased, and approximately 10-fold more trichomes were observed 10 days after CaSD1 overexpression in N. benthamiana leaves compared to those of control plants. In contrast to a single trichome emerging from one cell on control leaves, CaSD1-overexpressing leaves showed more than one trichome per TIS, forming clusters of adjacent trichomes (Fig. 5A).
Fig. 5.
Increase in trichome density in CaSD1-overexpressing N. benthamiana. (A) Surfaces of pMBP1 vector control (left) or CaSD1-overexpressing (right) N. benthamiana leaves were observed at the indicated time points after TOE by light microscopy or field-emission scanning electron microscopy. White bars, 1 mm. The bottom right hand black box is magnified for clusters of adjacent trichomes. Black bars, 200 μm. (B) pMBP1 vector control (Black) or CaSD1-overexpressing leaves (grey) were comparted into 2 mm × 1.4 mm regions, and trichomes in the fixed regions were counted at the indicated time points after TOE. Bars represent standard deviation (n = 5).
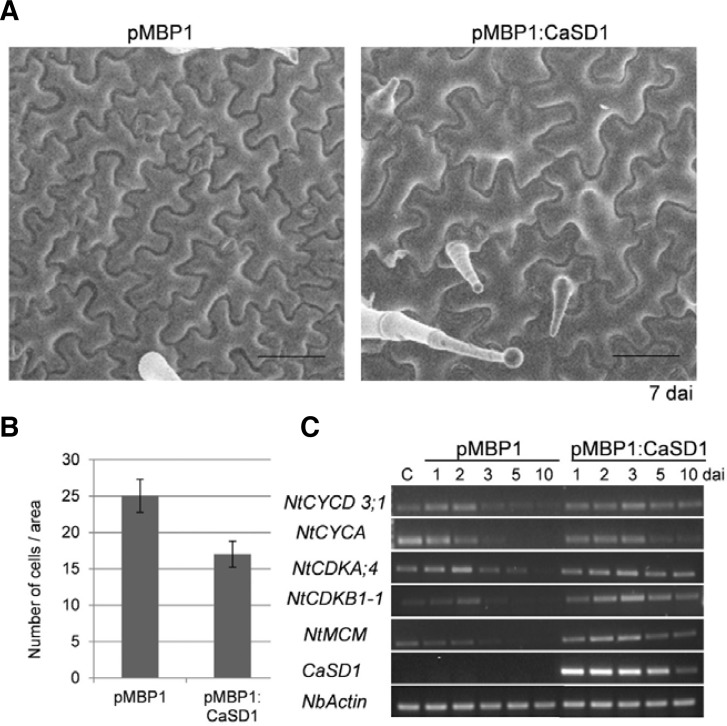
Furthermore, the CaSD1-overexpressing region bulged out, so we speculated that CaSD1 overexpression also affects cell size or number. The CaSD1-overexpressing N. benthamiana leaves were monitored by field-emission scanning electron microscopy. The size of epidermal cells in CaSD1-overexpressing plants was significantly increased compared to the size of those in control plants before trichome cells developed (Fig. 6A). We counted the number of cells in fixed areas and found that there were 30% fewer cells in CaSD1-overexpressing regions than in control regions (Fig. 6B). Trichome development and cell size are related to cell cycle regulation, including endoreduplication (Dewitte et al., 2007; Schnittger et al., 2002; Sugimoto-Shirasu and Roberts, 2003). To elucidate the molecular mechanism of cell cycle regulation in CaSD1-overexpressing plants, semi-quantitative RT-PCR of cell cycle-related genes, such as cyclins, cyclin-dependent kinases, and mini chromosome maintenance (MCM) members, was conducted. The expression levels of those genes were down-regulated during senescence in pMBP1-control plants, but these genes were still expressed in CaSD1-overexpressing plants (Fig. 6C). These results suggest that CaSD1 may play roles in cell fate determination and regulation of cell division.
Fig. 6.
Cell size and expression levels of cell division-related genes in CaSD1-overexpressing N. benthamiana. (A) Surfaces of pMBP1 vector control or CaSD1-overexpressing leaves were observed using field-emission scanning electron microscopy 7 days after inoculation. Black bars, 100 μm. (B) pMBP1 vector control or CaSD1-overexpressing leaves were comparted into 500 μm × 500 μm regions, and cells in the fixed regions were counted. Bars represent standard deviation (n = 40). (C) Expression patterns of cell division-related genes in CaSD1-overexpressing plants. NtCYCD, N. tabaccum D-type cyclin; NtCYCA, N. tabaccum cyclin A; NtCDKA, N. tabaccum cyclin-dependent kinase A; NtCDKB, N. tabaccum cyclin-dependent kinase B; NtMCM, N. tabaccum mini chromosome maintenance.
DISCUSSION
In previous work, we performed YST to isolate secreted proteins related to interactions between C. annuum CM334 and P. capsici (Yeom et al., 2011). Using YST, we isolated a novel gene, CaSD1. We further investigated the biological function of the CaSD1 gene by expression analysis and TOE in N. benthamiana. The predicted CaSD1 protein has repeat sequences that are variable among Capsicum species and cultivars (Figs. 1A and 1C). Indeed, two CaSD1 homologs in C. chinenese have shorter repeat regions than C. annuum ‘CM334’ or ‘Bukang.’ In addition, genes that had a partially conserved sequence with CaSD1 were predicted around the CaSD1 genomic region and in the syntenic segments of tomato and potato but lack repeat regions (Fig. 2). In peppers, duplication of the entire genome did not occur during evolution (Wu et al., 2009). Therefore, CaSD1 may be formed by partial duplication and may thus acquire additional functions.
The other interesting feature of CaSD1 is that there are no known conserved domains or motifs except the signal peptide. However, the repeat unit of CaSD1 shares some amino acid homology with root growth factor 1 (RGF1) from Arabidopsis. RGF1 recovers root meristem activity in short-root mutants as a small signaling peptide that is cleaved from pre-RGF (Matsuzaki et al., 2010). The pre-RGF sequence has no similarity with CaSD1, but the small peptide sequence is similar to a repeat unit of CaSD1. Accordingly, it is possible that CaSD1 has a function during growth and development like RGF1, and the repeat region may play a crucial role in these processes. Indeed, TOE of the CaSD1Δrepeat construct lacking the repeat and C-terminal non-repeat regions did not result in the phenotype observed under conditions of CaSD1 overexpression (data not shown).
In addition, we found that CaSD1 is related to leaf senescence using gain-of-function study. Senescence is controlled by a genetic program, and various internal or external factors are involved (Buchanan-Wollaston et al., 2003; Lim et al., 2007). To date, genes related to senescence have been isolated using mutagenesis or transcriptomic analyses (Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006). For example, certain proteases, dehydrogenases, proteins acting on chlorophyll, etc. are known to be involved in senescence (Buchanan-Wollaston et al., 2005; Lara et al., 2004; Zapata et al., 2005; Zhou et al., 2011). In this study, CaSD1-overexpressing regions maintained greenness, while senescence and cell death were accelerated by Agrobacterium in control regions (Fig. 4). Furthermore, the fact that expression of senescence-associated genes was altered in CaSD1-overexpressing plants lends further support to a role for CaSD1 in regulating critical steps of leaf senescence. Since the greenness of CaSD1-overexpressing regions remained after the leaf was detached, CaSD1 may be involved in the maintenance or production of chlorophyll.
We also observed that CaSD1 overexpression accelerated trichome formation. Trichomes are specialized epidermal cells that respond to positional cues (Hulskamp et al., 1994). However, CaSD1-overexpressing plants produced a large number of trichomes, some even in clusters (Fig. 5). From these data, we hypothesized that CaSD1 might alter cell fate. In Arabidopsis, research on trichome formation is well known (Hulskamp, 2004; Ishida et al., 2008). A cell that is destined to become a trichome enlarges, grows, and forms branches. Although N. benthamiana trichomes have multicellular stalks but no branches, a similar mechanism is likely to operate at the initiation of trichome formation. As shown in Figs. 5 and 6, at the beginning of trichome formation, epidermal cells of CaSD1-overexpressing plants are enlarged, and this enlargement may indicate early trichome formation. This enlargement of epidermal cells caused CaSD1-overexpressing regions to bulge.
Furthermore, sim (SIAMESE) mutants and the misexpression of CYCD3;1 in Arabidopsis lead to trichome phenotypes similar to those of CaSD1-overexpressing plants, showing multicellular trichome clusters (Schnittger et al., 2002; Walker et al., 2000). However, TIS were not increased in sim mutants. SIAMESE controls endoreduplication, and CYCD3;1 is related to both DNA replication and cell division. The expression of CYCD3;1 remains stable in CaSD1-overexpressing plants as do the expression levels of other genes related to cell division. These results indicate that CaSD1 may be involved in cell division and endoreduplication. The expression level stability of these genes in CaSD1-overexpressing plants may also contribute to the delay in senescence by maintaining cell division and viability. However, in the loss-of-function approach, we observed no significant difference between GFP- and CaSD1-silenced leaves because the transcript levels of CaSD1 in leaves was low in normal condition.
Even though we could not show the mechanistic relationship between CaSD1 and senescence, we clearly show functions of CaSD1 in senescence and trichome formation. Overexpression of CaSD1, a novel secreted protein, delays senescence and induces trichome formation, possibly through changes in the expression of a subset of genes common to both senescence and cell division in N. benthamiana. We suggest that CaSD1, as a Capsicum-specific protein, has roles in growth and development by regulating cell division and differentiation.
Supplementary Material
Acknowledgments
We appreciate Dr. S.P. Dinesh-Kumar for providing the pTRV vector and Dr. Jin-Kyung Kwon for her assistance in confocal microscopy. This work was supported by grants from the Agricultural Research Center Program and the Screening Center for Disease Resistant Vegetable Crops of Technology Development Program for the Ministry for Food, Agriculture, Forestry, and Fisheries of Korean Government.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Agrawal G.K., Jwa N.S., Lebrun M.H., Job D., Rakwal R. Plant secretome: unlocking secrets of the secreted proteins. Proteomics. 2010;10:799–827. doi: 10.1002/pmic.200900514. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J.H., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Ringli C., Keller B. The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Gen. Dev. 2001;15:1128–1139. doi: 10.1101/gad.200201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Earl S., Harrison E., Mathas E., Navabpour S., Page T., Pink D. The molecular analysis of leaf senescence--a genomics approach. Plant Biotechnol. J. 2003;1:3–22. doi: 10.1046/j.1467-7652.2003.00004.x. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P.O., Nam H.G., Lin J.F., Wu S.H., Swidzinski J., Ishizaki K., et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- Cho H.T., Cosgrove D.J. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2000;97:9783–9788. doi: 10.1073/pnas.160276997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005;6:850–861. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- Davis S.J., Vierstra R.D. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 1998;36:521–528. doi: 10.1023/a:1005991617182. [DOI] [PubMed] [Google Scholar]
- Dewitte W., Scofield S., Alcasabas A.A., Maughan S.C., Menges M., Braun N., Collins C., Nieuwland J., Prinsen E., Sundaresan V., et al. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA. 2007;104:14537–14542. doi: 10.1073/pnas.0704166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A.J., McQueenMason S., Mandel T., Kuhlemeier C. Induction of leaf primordia by the cell wall protein expansion. Science. 1997;276:1415–1418. [Google Scholar]
- Hematy K., Cherk C., Somerville S. Host-pathogen warfare at the plant cell wall. Curr. Opin. Plant Biol. 2009;12:406–413. doi: 10.1016/j.pbi.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Huckelhoven R. Cell wall-associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 2007;45:101–127. doi: 10.1146/annurev.phyto.45.062806.094325. [DOI] [PubMed] [Google Scholar]
- Hulskamp M. Plant trichomes: a model for cell differentiation. Nat. Rev. Mol. Cell Biol. 2004;5:471–480. doi: 10.1038/nrm1404. [DOI] [PubMed] [Google Scholar]
- Hulskamp M., Misra S., Jurgens G. Genetic dissection of trichome cell development in Arabidopsis. Cell. 1994;76:555–566. doi: 10.1016/0092-8674(94)90118-x. [DOI] [PubMed] [Google Scholar]
- Humphrey T.V., Bonetta D.T., Goring D.R. Sentinels at the wall: cell wall receptors and sensors. New Phytol. 2007;176:7–21. doi: 10.1111/j.1469-8137.2007.02192.x. [DOI] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T. A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 2008;59:365–386. doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- Lara M.E.B., Garcia M.C.G., Fatima T., Ehness R., Lee T.K., Proels R., Tanner W., Roitsch T. Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell. 2004;16:1276–1287. doi: 10.1105/tpc.018929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.J., Saravanan R.S., Damasceno C.M., Yamane H., Kim B.D., Rose J.K. Digging deeper into the plant cell wall proteome. Plant Physiol. Biochem. 2004;42:979–988. doi: 10.1016/j.plaphy.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Lerouxel O., Cavalier D.M., Liepman A.H., Keegstra K. Biosynthesis of plant cell wall polysaccharides - a complex process. Curr. Opin. Plant Biol. 2006;9:621–630. doi: 10.1016/j.pbi.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Lim P.O., Kim H.J., Nam H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- Lohman K.N., Gan S.S., John M.C., Amasino R.M. Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol. Plantarum. 1994;92:322–328. [Google Scholar]
- Matsuzaki Y., Ogawa-Ohnishi M., Mori A., Matsubayashi Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science. 2010;329:1065–1067. doi: 10.1126/science.1191132. [DOI] [PubMed] [Google Scholar]
- Oh I.S., Park A.R., Bae M.S., Kwon S.J., Kim Y.S., Lee J.E., Kang N.Y., Lee S., Cheong H., Park O.K. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell. 2005;17:2832–2847. doi: 10.1105/tpc.105.034819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.K., Kim S.B., Yeom S.I., Lee H.A., Choi D. Positive-selection and ligation-independent cloning vectors for large scale in planta expression for plant functional genomics. Mol. Cells. 2010;30:557–562. doi: 10.1007/s10059-010-0156-2. [DOI] [PubMed] [Google Scholar]
- Park J.A., Ahn J.W., Kim Y.K., Kim S.J., Kim J.K., Kim W.T., Pai H.S. Retinoblastoma protein regulates cell proliferation, differentiation, and endoreduplication in plants. Plant J. 2005;42:153–163. doi: 10.1111/j.1365-313X.2005.02361.x. [DOI] [PubMed] [Google Scholar]
- Park S.Y., Yu J.W., Park J.S., Li J., Yoo S.C., Lee N.Y., Lee S.K., Jeong S.W., Seo H.S., Koh H.J., et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell. 2007;19:1649–1664. doi: 10.1105/tpc.106.044891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S., Wyrzykowska J., McQueen-Mason S., Smart C., Fleming A. Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc. Natl. Acad. Sci. USA. 2001;98:11812–11817. doi: 10.1073/pnas.191380498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D., Gan S., Amasino R.M., Roby D., Lam E. Markers for hypersensitive response and senescence show distinct patterns of expression. Plant Mol. Biol. 1999;39:1243–1255. doi: 10.1023/a:1006133311402. [DOI] [PubMed] [Google Scholar]
- Quirino B.F., Noh Y.S., Himelblau E., Amasino R.M. Molecular aspects of leaf senescence. Trends Plant Sci. 2000;5:278–282. doi: 10.1016/s1360-1385(00)01655-1. [DOI] [PubMed] [Google Scholar]
- Schnittger A., Schobinger U., Bouyer D., Weinl C., Stierhof Y.D., Hulskamp M. Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc. Natl. Acad. Sci. USA. 2002;99:6410–6415. doi: 10.1073/pnas.092657299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter A.M. Structure and function of plant cell wall proteins. Plant Cell. 1993;5:9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K., Roberts K. “Big it up”: endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 2003;6:544–553. doi: 10.1016/j.pbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Suh M.C., Choi D., Liu J.R. Cadmium resistance in transgenic tobacco plants expressing the Nicotiana glutinosa L. metallothionein-like gene. Mol. Cells. 1998;8:678–684. [PubMed] [Google Scholar]
- van der Graaff E., Schwacke R., Schneider A., Desimone M., Flugge U.I., Kunze R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 2006;141:776–792. doi: 10.1104/pp.106.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.D., Oppenheimer D.G., Concienne J., Larkin J.C. SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development. 2000;127:3931–3940. doi: 10.1242/dev.127.18.3931. [DOI] [PubMed] [Google Scholar]
- Weaver L.M., Gan S.S., Quirino B., Amasino R.M. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 1998;37:455–469. doi: 10.1023/a:1005934428906. [DOI] [PubMed] [Google Scholar]
- Woo H.R., Chung K.M., Park J.H., Oh S.A., Ahn T., Hong S.H., Jang S.K., Nam H.G. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell. 2001;13:1779–1790. doi: 10.1105/TPC.010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Eannetta N.T., Xu Y., Durrett R., Mazourek M., Jahn M.M., Tanksley S.D. A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum. Theor. Appl. Genet. 2009;118:1279–1293. doi: 10.1007/s00122-009-0980-y. [DOI] [PubMed] [Google Scholar]
- Yeom S.I., Baek H.K., Oh S.K., Kang W.H., Lee S.J., Lee J.M., Seo E., Rose J.K., Kim B.D., Choi D. Use of a secretion trap screen in pepper following Phytophthora capsici infection reveals novel functions of secreted plant proteins in modulating cell death. Mol. Plant Microbe Int. 2011;24:671–684. doi: 10.1094/MPMI-08-10-0183. [DOI] [PubMed] [Google Scholar]
- Yeom S.I., Seo E., Oh S.K., Kim K.W., Choi D. A common plant cell-wall protein HyPRP1 has dual roles as a positive regulator of cell death and a negative regulator of basal defense against pathogens. Plant J. 2012;69:655–768. doi: 10.1111/j.1365-313X.2011.04828.x. [DOI] [PubMed] [Google Scholar]
- Yoo E.Y., Kim S., Kim Y.H., Lee C.J., Kim B.D. Construction of a deep coverage BAC library from Capsicum annuum, ‘CM334’. Theor. Appl. Genet. 2003;107:540–543. doi: 10.1007/s00122-003-1279-z. [DOI] [PubMed] [Google Scholar]
- Yoon J., Chung W.I., Choi D. NbHB1, Nicotiana benthamiana homeobox 1, is a jasmonic acid-dependent positive regulator of pathogen-induced plant cell death. New Phytol. 2009;184:71–84. doi: 10.1111/j.1469-8137.2009.02967.x. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Ito M., Callis J., Nishida I., Watanabe A. A delayed leaf senescence mutant is defective in arginyl-tRNA: protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis. Plant J. 2002;32:129–137. doi: 10.1046/j.1365-313x.2002.01407.x. [DOI] [PubMed] [Google Scholar]
- Zapata J.M., Guera A., Esteban-Carrasco A., Martin M., Sabater B. Chloroplasts regulate leaf senescence: delayed senescence in transgenic ndhF-defective tobacco. Cell Death Differ. 2005;12:1277–1284. doi: 10.1038/sj.cdd.4401657. [DOI] [PubMed] [Google Scholar]
- Zhou X., Jiang Y., Yu D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells. 2011;31:303–313. doi: 10.1007/s10059-011-0047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.