Abstract
Potato virus X (PVX) contains five viral proteins as well as cis-acting elements like stem-loop 1 (SL1) RNAs at the 5′ region. SL1 RNAs are involved in PVX RNA replication, encapsidation, translation, and cell-to-cell movement. In this study, we performed two-dimensional electrophoresis Northwestern blot analysis and matrix-assisted laser desorption ionization time of flight mass spectrometry and identified 24 tobacco proteins that interact with SL1 RNAs. Interestingly, one-third of the identified host proteins have been shown to interact with many plant viral proteins. In addition, we demonstrated that PVX capsid protein can bind to both SL1(+/−) RNAs. We further selected three Nicotiana benthamiana proteins including NbMPB2Cb, NbMBF1, and NbCPIP2a, to confirm results of Northwestern blot analysis. Electrophoretic mobility shift assay showed that NbMPB2Cb and NbMBF1 bind to both SL1(+/−) RNAs in vitro. In contrast, NbCPIP2a interacts only SL1(+) RNA. Taken together, we provide a list of host proteins interacting with PVX SL1 RNAs, which would be good candidates for the study of viral RNA-host protein interaction.
Keywords: host protein, northwestern blot, PVX, RNA-protein interaction, SL1 RNAs
INTRODUCTION
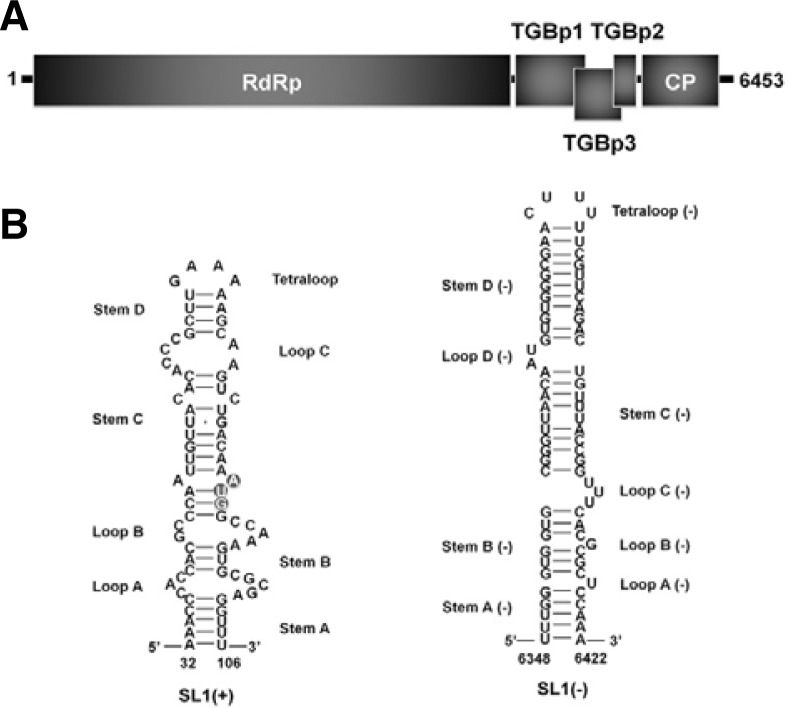
Potato virus X (PVX) is one of important plant RNA viruses in molecular plant pathology and causes very dramatic damage on crop yield. PVX is a flexuous rod-shaped virus and is a member of the genus Potexvirus in the family Alphaflexiviridae (Scholthof et al., 2011). Genome of PVX is a positive-sense single-stranded RNA and encodes a total of five viral proteins such as RNA-dependent RNA polymerase for viral RNA synthesis, triple gene block proteins for virus cell-to-cell movements, and capsid protein (CP) for viral assembly and cell-to-cell movement (Fig. 1A) (Miller et al., 1998). In addition, PVX specifically possesses cis-acting elements and structures at the 5′ end of its genome, which is defined as stem-loop 1 (SL1) RNAs (Fig. 1B). Previous studies demonstrated that the SL1 RNAs are involved in PVX translation, RNA replication, and initiation of PVX assembly (Kim and Hemenway, 1997; Kwon et al., 2005).
Fig. 1.
Genome organization and secondary structures of PVX. (A) Schematic representation of PVX RNA genome. Single-strand PVX RNA contains five open reading frames (ORFs): RNA-dependent RNA polymerase (RdRp), triple gene block proteins (TGBp1, TGBp2, and TGBp3), and CP. (B) Secondary structure of the positive strand of SL1 RNA as determined by chemical and enzymatic probing (SL1+). The initiation codon of RdRp is indicated by grey circles. Secondary structure of the negative strand of SL1 RNA (SL1−).
Until now, functional studies of plant host proteins are restricted to viral protein-host protein interaction rather than viral RNA-host protein. Numerous host proteins interacting with plant viral proteins have been identified. Little is known, however, for host proteins interacting with viral RNAs. A recent study has shown that PVX SL1 RNAs can efficiently bind to proteins extracted from tobacco protoplast as confirmed by a systematic evolution of ligands by exponential enrichment (SELEX) in vitro (Kwon and Kim, 2006).
In this study, we identified a total of 24 tobacco proteins which bind to PVX SL1 RNAs by Northwestern blot analysis and matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS). In addition, we further confirmed interaction of three selected proteins with SL1 RNAs by the electrophoretic mobility shift assay (EMSA).
MATERIALS AND METHODS
Preparation of SL1 RNA transcripts
The plasmid pSPVXp31 (agroinfiltration-based PVX replicon, Hemenway et al., 1990; Park and Kim, 2006) was used as a template to amplify SL1 RNAs. SL1 RNA probes containing (+) and (−) strand, respectively, were in vitro transcribed by T7 promoter. For the PCR, strand specific primers were used as described previously (Kang et al., 2010; Kim et al., 2002). Each PCR products were radioactively labeled in the presence of 50 μCi of α-32P CTP (3,000 Ci/mmol) (PerkinElmer Life Science), 5 mM CTP, and 125 mM each of ATP, GTP, and UTP. Labeled RNAs were purified by NucAway Spin Columns (Ambion). Radioactivity of acquired RNAs was measured with liquid scintillation counter (LSC) and RNA probes with 20,000 counts per minute (cpm) were used for EMSA.
Preparation of plant cell extracts and Escherichia coli expressed PVX CP
S100 protein extracts were prepared from tobacco BY-2 suspension cell as described previously (Kim and Hemenway, 1997). PVX CP gene was cloned and expressed in E. coli using pET vector system (Novagen). The purification of His-tagged CP fusion protein was purified as described previously (Kwon et al., 2005). The extracted protein contents were determined with a Bradford analysis and used as positive control for north-western analysis.
One- and two-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis
The protein pellets of S100 were solubilized in 250 μl of rehydration buffer containing 8 M urea, 2% CHAPS, 0.5% IPG 4–7 buffer (GE healthcare), 18 mM DTT, and bromophenol blue. Isoelectric focusing (IEF) was conducted at 20°C using IPG strips (pH 4–7, 13 cm, GE healthcare) and an Ettan IPGphor II system (GE healthcare). Denatured proteins (50 μg) were loaded into the IEF tray, and passive rehydration was carried out overnight under a layer of mineral oil. IEF was performed as follows: 5 h at 30 V, 1 h at 100 V, 1 h at 500 V, 3 h gradient to 8000 V, and 7 h at 8000 V at 10 μA/strip (56,000 Vh) at 20°C. Focused strips were equilibrated according to the manufacturer’s instruction and then subjected to 10% tricine acrylamide gel electrophoresis; proteins were silver stained. To detect SL1-binding proteins, proteins were transferred to nitrocellulose membranes and subjected to Northwestern blot analysis.
Northwestern blot analysis
Northwestern blot analysis was performed as previously described (Bichsel et al., 1997; Gustafson et al., 1998; Schiff et al., 1988) with slight modifications. In brief, highly purified S100 proteins and recombinant PVX CP were separated on 12% polyacrylamide (1-DE) and 10% tricine polyacrylamide (2-DE) and transferred electrophoretically to nitrocellulose membranes. To denature proteins, the membranes were incubated with 6 M guanidine hydrochloride (GuHCl) in northwestern buffer containing 10 mM Tris-HCl (pH 6.8), 25 mM NaCl, 1 mM EDTA, 0.04% BSA, 0.04% Ficoll 400, and 0.02% polyvinyl pyrrolidone-40 for 30 min. The membrane was then washed with 6, 3, 1.5, 0.75, 0.375, and 0.187 M GuHCl in northwestern buffer. The membranes were blocked with 0.5% fat-free dry milk and 10 μg/μl yeast tRNA in northwestern buffer for 30 min at room temperature. The blots were incubated with 32P-labeled transcripts at room temperature for 2 h and then washed three times for 1 min each with northwestern buffer to remove unbound or non-specifically bound RNA. After they were air-dried for 10 min, the membranes were exposed to a BAS imaging plate (IP) for autoradiography (Fuji film).
Trypsin digestion and sample preparation for peptides analysis
For in-membrane digestion, spots were excised from the membrane based on analysis of Multi Gauge V2.2 (Fuji film). Membrane slices (∼2 mm2) were incubated in digestion buffer containing 1% hydrogenated Triton X-100 (RTX), 10% acetonitrile, and 100 mM Tris-Cl (pH 8.0), for 30 min and hydrated in 5 μl of trypsin (0.5 μg/μl) (Sequencing grade, Promega) for 24 h at 37°C. Membrane pieces were sonicated for 5 min and then centrifuged. Supernatants of extracts were dried to ∼5 μl with a Speed Vac evaporator (Savant).
MALDI-TOF MS and database search based on peptide mass fingerprint spectra
The peptides were eluted with 2 μl of matrix solution containing α-cyano-4-hydroxycinnamic acid (HCCA) in acetonitrile/0.1% TFA 1:1 and transferred onto the MALDI target plate using Zip Tips (Millipore). The extracts were processed for MALDI on the ZipPlate (Millipore) according to the manufacturer’s protocol. The spectra were collected using the Voyager-DE STR Bio-spectrometry Workstation MALDI-TOF Mass Spectrometer (Applied Biosystems) at the National Instrumentation Center for Environmental Management (NICEM), Seoul National University. The mono-isotopic masses were subjected to search in the NCBI protein database with the Protein Prospector algorithm (http://prospector.ucsf.edu) MS-Fit. For MS-Fit searches, masses derived from trypsin and keratin were excluded. To increase the specificity of protein identification, we restricted searches to include only proteins derived from Nicotiana tabacum. The searches were performed with fixed default parameters: 0–100 kDa molecular mass and tryptic digest with a maximum number of one missed cleavage. The mass tolerance was set to 100 ppm.
Isolation of full-length ORF sequences for NbMPB2Cb, NbMBF1, and NbCPIP2a
To isolate the full-length cDNAs for NbMPB2Cb, NbMBF1, and NbCPIP2a, total RNAs were extracted from N. benthamiana plants, and cDNA was synthesized using an oligo (dT) 3′ primer. Known tobacco homologous genes were used to design following primers: MPB2Cb_F (5′-ATGGCGCTTGCATTT-3′) and MPB2Cb 3′ NTR_R (5′-ACAAACTATAGTAGTCAGTTTCCTA-3′) for NbMPB2Cb, MBF1 mRNA_F (5′-TCCCGAGAAGAAG AAGAA-3′) and MBF1 mRNA_R (5′-ACTGGAACGGAGATTTGAGA-3′) for NbMBF1, and CPIP2a_F (5′-ATGGGACTTGATTACTATAA-3′) and CPIP2a_R (5′-TTAGTCAGCGCTCCTGCACA-3′) for NbCPIP2a.
EMSA
To construct a maltose binding protein (MBP)-fusion protein for EMSA, full-length fragments of individual gene were cloned into the pMAL-c2X vector (New England Biolabs Inc.) using EcoRI enzyme. EMSA was performed as described previously (Cho and Kim, 2012; Peal et al., 2011; Sriskanda et al., 1996).
RESULTS
Identification of 24 host proteins that bind to SL1 RNAs
We speculated that the 3′ end of genomic minus-strand RNA might have a similar RNA secondary structure. For that, we predicted the structure of SL1(-) (nt 6405-6299) in silico using the mfold program version 3.1 (http://mfold.rna.albany.edu/) (Fig. 1B). The predicted SL1(−) structure is quite similar to that of SL1(+). Both SL1 structures are composed of four stems, four loops, and one tetraloop. However, the stem in SL1(−), like stems C(−) and D(−), is slightly longer than that of SL1(+), and the loops of SL1(−), like loops C(−) and D(−), are relatively unstable.
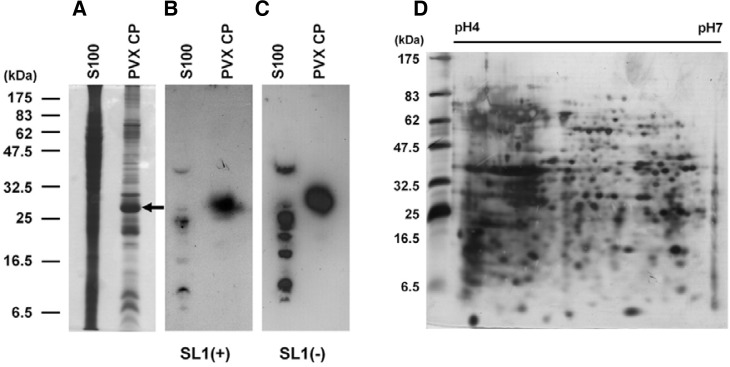
To isolate host proteins that specifically bind to SL1(+) and SL1(−) of PVX RNA, we performed 1-DE Northwestern blot analysis. As a positive control, we used PVX CP, which was previously demonstrated to bind to SL1(+) RNA in vitro (Kwon et al., 2005). Coomassie Brilliant Blue staining of the gel revealed numerous bands (Fig. 2A). Several bands were detected after hybridizing with the 32P-labeled SL1(+) and SL1(−) transcripts (Figs. 2B and 2C). Interestingly, 1-DE Northwestern blot analysis revealed that PVX CP binds not only to SL1(+) RNA transcripts but also to SL1(−) RNA transcripts. Although the same amounts of 32P-labeled RNA transcripts were used for hybridization, the total signal intensity was weaker for SL1(+) than SL1(−).
Fig. 2.
Northwestern blot analysis to confirm interaction of PVX CP with SL1 RNAs. (A) One-DE analysis of extracted S100 and recombinant PVX CP. Proteins were stained with Coomassie Brilliant Blue after electrophoresis. The arrow indicates PVX CP recombinant protein (about 30 kDa). One-DE Northwestern blot analysis using 32P-labeled SL1(+) RNA transcripts (B) and 32P-labeled SL1(−) RNA transcripts (C). (D) Two-DE analysis of extracted S100 proteins visualized by silver staining.
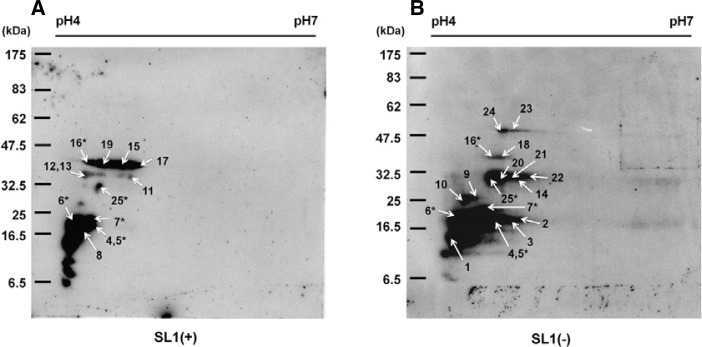
We further fractionated the 1-DE by 2-DE analysis to separate such host proteins in high resolution. The silver-stained gel revealed many well-separated protein spots (Fig. 2D). Two-DE Northwestern blot analyses were conducted three times using 32P-labeled SL1(+) and SL1(−) RNA transcripts and similar results were obtained (data not shown). The molecular mass of the separated proteins ranged from 6.5 to 55 kDa, and most of them were rather acidic (Figs. 3A and 3B). The patterns of detected signals between SL1(+) and SL1(−) were different. However, some signals were common to both SL1 RNAs. Major spots were excised and were analyzed as described in the experimental procedures.
Fig. 3.
Northwestern blot analysis to identify tobacco host proteins that interact with PVX SL1 RNAs. Two-DE Northwestern blot analysis using 32P-labeled SL1(+) RNA transcripts (A) and 32P-labeled SL1(−) RNA transcripts (B). Arrows indicate the excised spots for MALDI TOF MS analysis; the spot number adjacent to each arrow indicates each of the 24 identified proteins (see Table 1). The five locations where host proteins were commonly found by both RNA probes are indicated by asterisks.
With the criteria that at least four peptides had to match and that sequence coverage had to be greater than 15%, we identified a total of 24 proteins that interacted with SL1(+) (12 proteins) and SL1(−) (17 proteins) RNAs (Figs. 3A and 3B; Table 1); five of these 24 bind to both SL1 RNAs. Moreover, we detected PVX CP used as a positive control indicating that our 2-DE Northwestern blot analysis was reliable. The molecular mass of the 24 proteins ranged from 15 to 53 kDa, and their isoelectric points (pI) were between 5 and 10 (Table 1). To obtain information about the function of the identified proteins, we searched orthologous Arabidopsis thaliana proteins. Blast search found several orthologous proteins with low E-values, indicating high conservancy of the 24 proteins in N. tabacum and Arabidopsis. According to their known functions and conserved domains in tobacco and Arabidopsis, the identified proteins can be divided into four functional groups: transcriptional factors, heat shock proteins, signaling proteins, and others including cytoskeletal protein and rRNA binding protein. Remarkably, many isolated host proteins in our study are homologous to other host proteins interacting with various viral proteins. They included Tobacco mosaic virus (TMV)-movement protein 30 (MP30) binding protein 2C (MPB2C) (Kragler et al., 2003), putative multiprotein bridging factor 1 (MBF1) (Matsushita et al., 2002), potyviral CP interacting protein 2a and 2b (CPIP2s), Tobamovirus multiplication 3 (TOM) (Yamanaka et al., 2000), Cucumber mosaic virus (CMV) 1a interacting protein 1 (Kim et al., 2008), and DnaJ-like protein (Shimizu et al., 2009). We further investigated the interaction of three N. benthamiana proteins including MPB2C, MBF1, and CPIP2a by EMSA.
Table 1.
Host proteins that bind to PVX SL1 RNAs as determined by proteomics
| Spot # | Accession # | Protein name | MW Da/pI | SL1 strand | No. of peptide | Seq. Cov. % | MOWSE score | Accession # of Arabidopsis | E-value | Function |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 20086364 | Putative multiprotein bridging factor 1 | 15327/10.0 | (−) | 6 | 30.7 | 1856 | AT3G58680 | 7.00E-45 | TF |
| 2 | 1841462 | Elongation factor 2 | 19875/5.3 | (−) | 5 | 38 | 193 | AT1G56070 | 7.00E-97 | SP |
| 3 | 90654964 | DnaJ heat shock protein | 20774/9.3 | (−) | 6 | 31.6 | 788 | AT1G77930 | 4.00E-63 | HSP |
| 4 | 5932420 | LIM domain protein WLIM2 | 20812/5.6 | (+)(−) | 10 | 35 | 2389 | AT3G55770 | 2.00E-75 | TF |
| 5 | 5932413 | LIM domain protein WLIM1 | 21521/9.1 | (+)(−) | 6 | 30.6 | 745 | AT1G10200 | 8.00E-83 | TF |
| 6 | 3256375 | Heat shock protein 26 (Type I) | 22574/5.2 | (+)(−) | 5 | 33.3 | 381 | AT4G27670 | 2.00E-37 | HSP |
| 7 | 15778156 | 14-3-3 protein | 22981/5.6 | (+)(−) | 7 | 29.4 | 1149 | AT4G09000 | 2.00E-90 | SP |
| 8 | 417063 | Floral homeotic protein GLOBOSA | 24691/8.9 | (+) | 5 | 29.7 | 231 | AT5G20240 | 5.00E-51 | TF |
| 9 | 6552359 | Myb-related transcription factor LBM1 | 31985/5.1 | (−) | 6 | 30.2 | 1141 | AT3G23250 | 6.00E-71 | TF |
| 10 | 1732247 | Transcription factor Myb1 | 32005/5.5 | (−) | 4 | 22.3 | 258 | AT3G23250 | 9.00E-68 | TF |
| 11 | 5821136 | Heat shock factor | 32106/6.0 | (+) | 4 | 15.1 | 84.9 | AT4G36990 | 2.00E-65 | HSP |
| 12 | 34811738 | Potyviral CP interacting protein 2a | 33872/9.3 | (+) | 5 | 23 | 592 | AT1G10350 | 1.00E-101 | HSP |
| 13 | 34811740 | Potyviral CP interacting protein 2b | 33930/9.3 | (+) | 5 | 16.4 | 94 | AT1G59725 | 1.00E-104 | HSP |
| 14 | 74038607 | Tobamovirus multiplication 3 | 34023/9.8 | (−) | 4 | 28.9 | 315 | AT2G02180 | 1.00E-114 | TF |
| 15 | 4760596 | DNA-binding protein NtWRKY3 | 35940/9.6 | (+) | 6 | 15.5 | 3101 | AT4G31550 | 3.00E-75 | TF |
| 16 | 19716176 | TMV-MP30 binding protein 2C | 35998/5.0 | (+)(−) | 5 | 21.2 | 102 | AT5G08120 | 7.00E-34 | CSP |
| 17 | 57014014 | Ribosomal protein L2 | 36644/10.9 | (+) | 11 | 28.1 | 756 | ATMG00560 | 1.00E-129 | RBP |
| 18 | 6179940 | DnaJ-like protein | 37117/9.1 | (−) | 4 | 27.5 | 188 | AT2G20560 | 1.00E-105 | HSP |
| 19 | 39598908 | CMV 1a interacting protein 1 | 41538/6.4 | (+) | 7 | 27.9 | 5836 | AT1G23360 | 2.00E-05 | SP |
| 20 | 78096654 | Mitogen-activated protein kinase | 42884/6.1 | (−) | 6 | 24.3 | 167 | AT4G01370 | 0 | SP |
| 21 | 18143321 | Wound induced protein kinase | 42898/5.2 | (−) | 7 | 28 | 795 | AT3G45640 | 0 | SP |
| 22 | 51458330 | Trehalose-phosphate phosphatase | 42945/7.6 | (−) | 10 | 31.4 | 2612 | AT5G51460 | 1.00E-150 | SP |
| 23 | 11133532 | Shaggy-related protein kinase NtK-1 | 46309/8.6 | (−) | 7 | 18.3 | 377 | AT5G26751 | 0 | SP |
| 24 | 1617200 NSK6; | Shaggy-like kinase6 | 53434/7.9 | (−) | 10 | 20.8 | 6019 | AT4G00720 | 0 | SP |
| 25 | 70994254 | PVX CP | 25105/7.7 | (+)(−) | 5 | 16.5 | 269 |
Table headings: Spot #, protein spot number from Fig. 3; Accession #, accession number of N. tabacum proteins from NCBI; MW Da/pI, theoretical Daltons/isoelectric point; No. of peptide, number of peptides matched to the identified protein sequences; Seq. Cov. %, the percent coverage for the protein based upon the matched peptide sequences; MOWSE score, peptide identification score; E-value, expectation value between candidate proteins of N. tabacum and Arabidopsis
TF, transcriptional factor; HSP, heat shock protein; SP, signaling protein; CSP, cytoskeletal protein; RBP, rRNA binding protein
Identification of NbMPB2Cb, NbMBF1, and NbCPIP2a homologous proteins from Nicotiana benthamiana
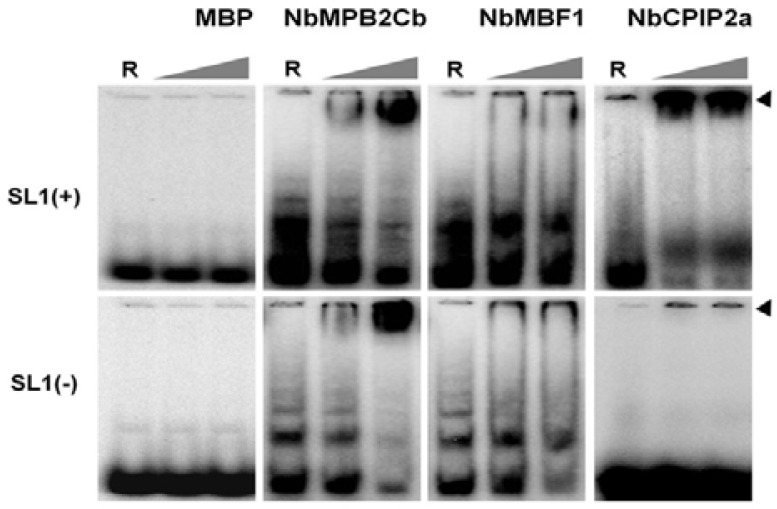
To isolate full-length cDNAs for MPB2C, MBF1, and CPIP2a homologous genes from N. benthamiana, RT-PCRs were performed using gene-specific primers based on known tobacco homologous gene sequence information. The isolated genes were referred as NbMPB2Cb, NbMBF1, and NbCPIP2a, respectively. Each host gene was cloned into a modified pMAL-c2X plasmid for expression as fusions to the C-terminus of non-secreted MBP. MBP-NbMPB2Cb, MBP-NbMBF1, and MBP-NbCPIP2a productions of the expected sizes were detected by SDS-PAGE (data not shown). To confirm the result of the 2-DE Northwestern blot analysis, EMSA was performed using 32P-labeled SL1 RNA and individual purified fusion protein. EMSA with only MBP used as a negative control showed that MBP does not interact with any SL1 RNAs. EMSA displayed reduced mobility of the NbMPB2Cb-RNA complex in both SL1(+/−) RNAs (Fig. 4 and Table 1). These data are consistent with results of Northwestern blot analysis. In case of NbMBF1, the Northwestern blot analysis showed that NbMBF1 can specifically interact with SL1(−) RNA, however, the EMSA results displayed weak interaction of NbMBF1 with both SL1 RNAs. NbCPIP2a can only bind to SL1(+) RNA, which is confirmed by both EMSA and Northwestern blot analysis.
Fig. 4.
EMSA to confirm the interactions of three N. benthamiana proteins and SL1 RNAs. In each image, only 32P-labeled RNAs were loaded in the first lane indicated by ‘R’ whereas 0.5 μg and 2 μg of each purified protein were loaded in lanes 2 and 3, respectively. The top arrow indicates RNA-protein complexes while the bottom arrow indicates labeled SL1 RNAs.
DISCUSSION
In this study, we tried to identify host proteins interacting with PVX SL1 RNAs by proteomic approach since only a limited number of host proteins have been shown to interact with viral RNAs (Fujisaki and Ishikawa, 2008; Nicholson et al., 2010). Although we identified 24 host proteins by selecting only the best-matched protein from individual spots on the gels, it follows that the real number of host proteins interacting with viral RNAs is probably more than 24. Blotting results showed strong hybridization signals in many 2-DE areas. This seems to be caused by many proteins or protein complexes, which have similar pI and molecular mass. Of the 24 identified proteins, 14 out of 24 proteins have basic pI values ranging from 7.6 to 10.9. In addition to different molecular masses, proteins present in multiple spots migrated at different isoelectric points. Some of these differences may occur because of changes in primary sequences as a consequence of alternative splicing or the result of post-translational modifications, such as phosphorylation, sulfation, and acetylation.
Interestingly, the functions of most host proteins identified in our study have been previously characterized. Moreover, one-third of the identified host proteins have been shown to interact with many plant viral proteins. For instance, MBF1-like protein binds to Tomato mosaic virus (ToMV) MP and this interaction could modulate host gene expression to suppress the general defense response in plants (Matsushita et al., 2002). MPB2C and NtMPIP1 are involved in TMV cell-to-cell spread by interacting with TMV MP (Curin et al., 2007; Kragler et al., 2003). NtCPIP2a and NtCPIP2b act as potyviral susceptibility factors during Potato virus Y infection and along with heat shock protein 70, they act as plant chaperones and are essential for virus movement (Hofius et al., 2007). TOM1 and TOM3 proteins in Arabidopsis and N. tabacum are associated with the multiplication of TMV and tobamoviruses (Asano et al., 2005; Yamanaka et al., 2000). Based on those results, we suspect that various host proteins can bind to viral RNAs and/or proteins of multiple plant viruses. Although our data only revealed interactions between PVX viral RNA and host proteins, the host proteins identified in our study might be good candidates for the study of viral protein-host protein interactions with other viruses and hosts.
Similarly, many host proteins interacting with the 5′ end of the plus-strand of RNA or the 3′ end of the minus-strand were identified for Rubella virus and Mouse hepatitis virus (Furuya and Lai, 1993; Nakhasi et al., 1990). Our Northwestern blot analyses revealed that the list of host proteins interacting with each SL1 RNA is different from each other, although five proteins bind to both SL1 RNAs. These data support that viral RNA-host protein interactions are highly variable and depend on sequences and/or structures. It would also be of interest to survey functional differences between SL1(+) and SL1(−) interacting proteins in near future.
To confirm results of Northwestern blot analysis, we performed EMSA confirming interaction of NbMPB2Cb and NbCPIP2a with SL1 RNAs. However, we failed to get consistent results for NbMBF1, which showed weak interaction with both SL1 RNAs by EMSA. NbMBF1 is a member of transcription factor family and its protein abundance might be relatively lower to other proteins. Therefore, it will be difficult to detect it by the proteomic analysis. In contrast, we can use high amount of MBP-NbMBF1 fusion protein produced from E. coli. Thus, we suppose that results of EMSA could be more precise.
Previous studies have demonstrated that the sequences of upper region and C:C mismatch in SL structures of PVX and Bamboo mosaic virus (BaMV) play important roles in accumulation of genomic RNA (Chen et al., 2010; Miller et al., 1998). It has been shown that SL1(+) RNA binds to PVX CP and several tobacco proteins including the N. tabacum WRKY1 transcription factor (NtWRKY1) (Kim et al., 2002; Part and Kim, 2011). Silencing of NtWRKY1 in N. benthamiana caused plants to exhibit lethal apical necrosis by increasing PVX accumulation, suggesting that NtWRKY1 functions to regulate multiple defense response genes (Park and Kim, 2012). Recently, we identified NbDnaJ which interacts with SL1(−) RNA and showed that it plays a role in the early stages of viral infection by acting as a negative regulator of PVX replication and movement (Cho et al., 2011). In this regard it is worth noting that the 5′ or 3′ end of RNA behaves as cis-acting elements and can affect cell-to-cell movement of PVX (Lough et al., 2006). These results suggest that these interactions are necessary for PVX replication and assembly. Altogether, it is quite sure that the identified host proteins will play roles in PVX replication, assembly, and cell-to-cell movement. However, it will be our next studies to elucidate the identity and function of the host proteins associated with PVX infection cycles.
Acknowledgments
This research was supported in part by grants from the National Research Foundation grant (No. 20110012328) funded by the Ministry of Education, Science, and Technology; the Vegetable Breeding Research Center (No. 710001-03) through Agriculture Research Center program from the Ministry for Food, Agriculture, Forestry and Fisheries; and the Next-Generation Bio-Green 21 Program of Rural Development Administration (No. PJ00819801). WKC was supported by a post-graduate research fellowship from the Brain Korea 21 Project.
REFERENCES
- Asano M., Satoh R., Mochizuki A., Tsuda S., Yamanaka T., Nishiguchi M., Hirai K., Meshi T., Naito S., Ishikawa M. Tobamovirus-resistant tobacco generated by RNA interference directed against host genes. FEBS Lett. 2005;579:4479–4484. doi: 10.1016/j.febslet.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Bichsel V.E., Walz A., Bickel M. Identification of proteins binding specifically to the 3′-untranslated region of granulocyte/macrophage-colony stimulating factor mRNA. Nucleic Acids Res. 1997;25:2417–2423. doi: 10.1093/nar/25.12.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.C., Desprez A., Olsthoorn R.C.L. Structural homology between Bamboo mosaic virus and its satellite RNAs in the 5′ untranslated region. J. Gen. Virol. 2010;91:782–787. doi: 10.1099/vir.0.015941-0. [DOI] [PubMed] [Google Scholar]
- Cho S.-Y., Kim K.-H. Identification of the capsid protein-binding region of the SL1(+) RNA located at the 5′ region of the Potato virus X genome. Plant Pathol. J. 2012;28:75–80. [Google Scholar]
- Cho S.-Y., Cho W.-K., Sohn S.-H., Kim K.-H. Interaction of the host protein NbDnaJ with Potato virus X minus-strand stem-loop 1 RNA and capsid protein affects viral replication and movement. Biochem. Biophys. Res. Commun. 2012;417:451–456. doi: 10.1016/j.bbrc.2011.11.137. [DOI] [PubMed] [Google Scholar]
- Curin M., Ojangu E.L., Trutnyeva K., Ilau B., Truve E., Waigmann E. MPB2C, a microtubule-associated plant factor, is required for microtubular accumulation of Tobacco mosaic virus movement protein in plants. Plant Physiol. 2007;143:801–811. doi: 10.1104/pp.106.091488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaki K., Ishikawa M. Identification of an Arabidopsis thaliana protein that binds to Tobacco mosaic virus genomic RNA and inhibits its multiplication. Virology. 2008;380:402–411. doi: 10.1016/j.virol.2008.07.033. [DOI] [PubMed] [Google Scholar]
- Furuya T., Lai M.M. Three different cellular proteins bind to complementary sites on the 5′-end-positive and 3′-end-negative strands of mouse hepatitis virus RNA. J. Virol. 1993;67:7215–7222. doi: 10.1128/jvi.67.12.7215-7222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson W.C., Taylor C.W., Valdez B.C., Henning D., Phippard A., Ren Y., Busch H., Durban E. Nucleolar protein p120 contains an arginine-rich domain that binds to ribosomal RNA. Biochem. J. 1998;331:387–393. doi: 10.1042/bj3310387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemenway C., Weiss J., O’Connell K., Tumer N.E. Characterization of infectious transcripts from a potato virus X cDNA clone. Virology. 1990;175:365–371. doi: 10.1016/0042-6822(90)90421-m. [DOI] [PubMed] [Google Scholar]
- Hofius D., Maier A.T., Dietrich C., Jungkunz I., Bornke F., Maiss E., Sonnewald U. Capsid protein-mediated recruitment of host DnaJ-like proteins is required for Potato virus Y infection in tobacco plants. J. Virol. 2007;81:11870–11880. doi: 10.1128/JVI.01525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Oh S.-K., Kim J.-J., Choi D., Baek K.-H. NMMP1, a matrix metalloprotease in Nicotiana benthamiana has a role in protection against bacterial infection. Plant Pathol. J. 2010;26:402–408. [Google Scholar]
- Kim K.-H., Hemenway C. Mutations that alter a conserved element upstream of the potato virus X triple block and coat protein genes affect subgenomic RNA accumulation. Virology. 1997;232:187–197. doi: 10.1006/viro.1997.8565. [DOI] [PubMed] [Google Scholar]
- Kim K.-H., Kwon S.-J., Hemenway C. Cellular protein binds to sequences near the 5′ terminus of Potato virus X RNA that are important for virus replication. Virology. 2002;301:305–312. doi: 10.1006/viro.2002.1559. [DOI] [PubMed] [Google Scholar]
- Kim M.J., Huh S.U., Ham B.K., Paek K.H. A novel methyltransferase methylates Cucumber mosaic virus 1a protein and promotes systemic spread. J. Virol. 2008;82:4823–4833. doi: 10.1128/JVI.02518-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragler F., Curin M., Trutnyeva K., Gansch A., Waigmann E. MPB2C, a microtubule-associated plant protein binds to and interferes with cell-to-cell transport of Tobacco mosaic virus movement protein. Plant Physiol. 2003;132:1870–1883. doi: 10.1104/pp.103.022269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S.-J., Kim K.-H. The SL1 stem-loop structure at the 5′-end of Potato virus X RNA is required for efficient binding to host proteins and for viral infectivity. Mol. Cells. 2006;21:63–75. [PubMed] [Google Scholar]
- Kwon S.-J., Park M.-R., Kim K.-W., Plante C.A., Hemenway C.L., Kim K.-H. cis-Acting sequences required for coat protein binding and in vitro assembly of Potato virus X. Virology. 2005;334:83–97. doi: 10.1016/j.virol.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Lough T.J., Lee R.H., Emerson S.J., Forster R.L., Lucas W.J. Functional analysis of the 5′ untranslated region of potexvirus RNA reveals a role in viral replication and cell-to-cell movement. Virology. 2006;351:455–465. doi: 10.1016/j.virol.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Matsushita Y., Miyakawa O., Deguchi M., Nishiguchi M., Nyunoya H. Cloning of a tobacco cDNA coding for a putative transcriptional coactivator MBF1 that interacts with the tomato mosaic virus movement protein. J. Exp. Bot. 2002;53:1531–1532. [PubMed] [Google Scholar]
- Miller E.D., Plante C.A., Kim K.-H., Brown J.W., Hemenway C. Stem-loop structure in the 5′ region of potato virus X genome required for plus-strand RNA accumulation. J. Mol. Biol. 1998;284:591–608. doi: 10.1006/jmbi.1998.2174. [DOI] [PubMed] [Google Scholar]
- Nakhasi H.L., Rouault T.A., Haile D.J., Liu T.Y., Klausner R.D. Specific high-affinity binding of host cell proteins to the 3′ region of rubella virus RNA. New Biol. 1990;2:255–264. [PubMed] [Google Scholar]
- Nicholson B.L., Wu B.D., Chevtchenko I., White K.A. Tombusvirus recruitment of host translational machinery via the 3′ UTR. RNA. 2010;16:1402–1419. doi: 10.1261/rna.2135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-H., Kim K.-H. Agroinfiltration-based Potato virus X replicons to dissect the requirements of viral infection. Plant Pathol. J. 2006;22:386–390. [Google Scholar]
- Park S.-H., Kim K.-H. Virus-induced silencing of the WRKY1 transcription factor that interacts with the SL1 structure of Potato virus X leads to higher viral RNA accumulation and severe necrotic symptoms. Plant Pathol. J. 2012;28:40–48. [Google Scholar]
- Peal L., Jambunathan N., Mahalingam R. Phylogenetic and expression analysis of RNA-binding proteins with triple RNA recognition motifs in plants. Mol. Cells. 2011;31:55–64. doi: 10.1007/s10059-011-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff L.A., Nibert M.L., Co M.S., Brown E.G., Fields B.N. Distinct binding sites for zinc and double-stranded RNA in the reovirus outer capsid protein sigma 3. Mol. Cell. Biol. 1988;8:273–283. doi: 10.1128/mcb.8.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof K.B., Adkins S., Czosnek H., Palukaitis P., Jacquot E., Hohn T., Hohn B., Saunders K., Candresse T., Ahlquist P., et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011;12:938–954. doi: 10.1111/j.1364-3703.2011.00752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Yoshii A., Sakurai K., Hamada K., Yamaji Y., Suzuki M., Namba S., Hibi T. Identification of a novel tobacco DnaJ-like protein that interacts with the movement protein of Tobacco mosaic virus. Arch. Virol. 2009;154:959–967. doi: 10.1007/s00705-009-0397-6. [DOI] [PubMed] [Google Scholar]
- Sriskanda V.S., Pruss G., Ge X., Vance V.B. An eight-nucleotide sequence in the potato virus X 3′ untranslated region is required for both host protein binding and viral multiplication. J. Virol. 1996;70:5266–5271. doi: 10.1128/jvi.70.8.5266-5271.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T., Ohta T., Takahashi M., Meshi T., Schmidt R., Dean C., Naito S., Ishikawa M. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl. Acad. Sci. USA. 2000;97:10107–10112. doi: 10.1073/pnas.170295097. [DOI] [PMC free article] [PubMed] [Google Scholar]