Abstract
The identification of true causal loci to unravel the statistical evidence of genotype-phenotype correlations and the biological relevance of selected single-nucleotide polymorphisms (SNPs) is a challenging issue in genome-wide association studies (GWAS). Here, we introduced a novel method for the prioritization of SNPs based on p-values from GWAS. The method uses functional evidence from populations, including phenotype-associated gene expressions. Based on the concept of genetic interactions, such as perturbation of gene expression by genetic variation, phenotype and gene expression related SNPs were prioritized by adjusting the p-values of SNPs. We applied our method to GWAS data related to drug-induced cytotoxicity. Then, we prioritized loci that potentially play a role in drug-induced cytotoxicity. By generating an interaction model, our approach allowed us not only to identify causal loci, but also to find intermediate nodes that regulate the flow of information among causal loci, perturbed gene expression, and resulting phenotypic variation.
Keywords: genome-wide association study, interaction network, prioritization, SNP
INTRODUCTION
The enumeration of network components by identifying molecular-level interactions may unravel underlying mechanisms for physiological and pathological phenotypes (Taylor et al., 2009). In modern genetics, the exploration of networks of proteins and expression of quantitative trait loci (eQTL) are recognized increasingly as a promising approach to understanding various phenotypes (Jia et al., 2011; Zhong et al., 2010). Genome-wide association studies (GWAS) have been applied generally to the discovery of causal loci that influence trait variation, including disease susceptibility and drug responses (Cichon et al., 2011; Sugiyama et al., 2011). Although GWAS have revolutionized our ability to localize and identify the causal determinants of disease-related phenotypes in a statistical manner, typically, they do not inform on the broader context in which the trait-associated genes operate, thereby providing limited insights into the mechanisms driving diseases or various phenotypes. Therefore, in GWAS, there remains one open, yet very important, question pertaining to the prioritization of one or a few SNPs among hundreds or thousands of candidates, aimed at identifying functional or linked loci that would represent causative variants for phenotypes of interest.
Although the predicted genetic determinants of pathological phenotypes (Chae et al., 2010; Lee et al., 2009), including risk of disease (McPherson et al., 2007), and drug susceptibility (Wu et al., 2010), have been determined based on GWAS, the functional relevance of the loci identified remains undetermined in many cases. Current bioinformatics approaches examine the functional effects of SNPs to identify true functional loci using a priori knowledge, such as variation effects on protein coding regions (Lee and Shatkay, 2008; Ng and Henikoff, 2003) and pathway information (Saccone et al., 2008). Although these bioinformatics methods support partially the biological relevance of SNPs, quantified genetic mechanisms in given populations remain unclear, such as causal loci leading to abnormalities of gene expression that cause phenotypic changes. Namely, previous bioinformatics methods identified the functional role of candidate SNPs based on corresponding genes and genomic positions, such as mutation effects on protein translation and binding of transcription factors, without direct evidence of SNP-mediated gene expression changes in given populations. Therefore, in addition to the prioritization of causal SNPs, we also focused on developing a method that provides a detailed model for the propagation of information from genetic variation to target phenotypes: the mechanisms via which causal SNPs perturb gene/protein expression and lead to phenotypic variation in given populations. Our previous attempts (Paik et al., 2010; 2011) highlighted the fact that the unraveling of interactions between genetic variation and gene expression patterns is a promising approach to understanding the mechanisms underlying complex phenotypes in given populations. The interaction concept used in our attempts to uncover the functional role of SNPs in phenotypic variation was also a forefront issue in previous studies, via the integrative analysis of genetic variation and gene expression in disease (Li et al., 2010; Schadt et al., 2008), as well as of drug responses (Etemadmoghadam et al., 2009). These attempts have manifested that the development of an interactive model between SNPs and gene expression patterns may assist the prioritization of true functional SNPs.
To obtain functional evidence for selected loci, this study used information on interactions between SNPs and gene expression patterns in given populations (rather than a priori knowledge), such as translational modification by polymorphisms located in coding regions. Based on the concept of eQTL (Vinuela et al., 2010), we stipulated the degree of interaction between SNPs and gene expression patterns according to the p-values of genetic regulations for gene expressions per se. Our method of prioritization of SNPs, for further study of GWAS, yielded weighted p-values for true functional loci in a zero-convergence manner. We designed a weighting algorithm that displays lower values for functional SNPs according to the following principles: i) functional SNPs may have lower p-values for a phenotype or for the regulation of gene expression, and ii) perturbed gene expression may also present lower p-values for target phenotypes. Based on these principles, we constructed a priority function for the prioritization of p-values of causal SNPs, using p-values of perturbed gene expression and p-values of interactions between SNPs and gene expression patterns. In summary, the building of a priority function was performed according to the information flow in the genetic architecture model, which is a working model of the genetic variations that drive phenotypic variation via interacting transcriptional signatures (Paik et al., 2011). As this method uses functional signatures of SNPs, such as altered gene expression, we termed our method the “functional prioritization of p-value (fpp-value)” approach.
Demonstration of the fpp-value approach was achieved using the p-values of SNPs, gene expression, and their interaction in relation to the phenotype of the cytotoxicity induced by an anticancer drug (cisplatin). Cisplatin-induced cytotoxicity was determined using half-inhibition of cell growth/survival (IC50) after treatment with the drug (Gamazon et al., 2010a). For the evaluation of the fpp-value method, sensitivity and specificity were measured using hypothetical true-positive genes, given the absence of a gold-standard gene set for cisplatin-related cytotoxicity. Using identical gene sets, we also quantified the performance of p-value-based selection, to illustrate the outperformance of our method. The data dependency and robustness of the fpp-value method were validated via duplicated running of the fpp-value approach using two independent populations of HapMap: CEU and YRI (CEU, Utah residents with Northern and Western European ancestry from the CEPH collection; YRI, residents of Yoruba in Ibadan, Nigeria) (International_HapMap_Consortium, 2003). In addition, gene set enrichment analysis (GSEA) (Subramanian et al., 2005) showed that the genes and interaction network identified using our fpp-value approach confirmed the biological relevance of prioritized SNPs, compared with the typical p-value analysis of GWAS.
MATERIALS AND METHODS
Prioritization of SNPs using the fpp-value approach
Using the prepared p-values of interactions between SNPs and gene expression patterns, as well as p-values of genetic loci and p-values of gene expression for target phenotypes, such as cisplatin-derived cytotoxicity, we determined the functional priority of the p-values (fpp-values) of a selected locus. These p-values were prepared using GWAS and eQTL analyses. In the population analyzed, ρ, ps(li) was defined as the p-value of a SNP for the i-th locus, and pe(gk) represented the p-value of the expression of the k-th gene for trait variation. As the goal of our method was the prioritization of SNPs using dynamic interactions between SNPs and gene expression patterns, we also prepared p-values of the interactions between the i-th locus and the expression of the k-th gene (I(li, gk)) in the same population, ρ:
li = i-th locus
gk = expression of the k-th gene
ps(li) = p-value of the i-th locus for trait variation
pe(gk) = p-value of the expression of the k-th gene for trait variation
I(li, gk) = p-value of the interactions between li and gk
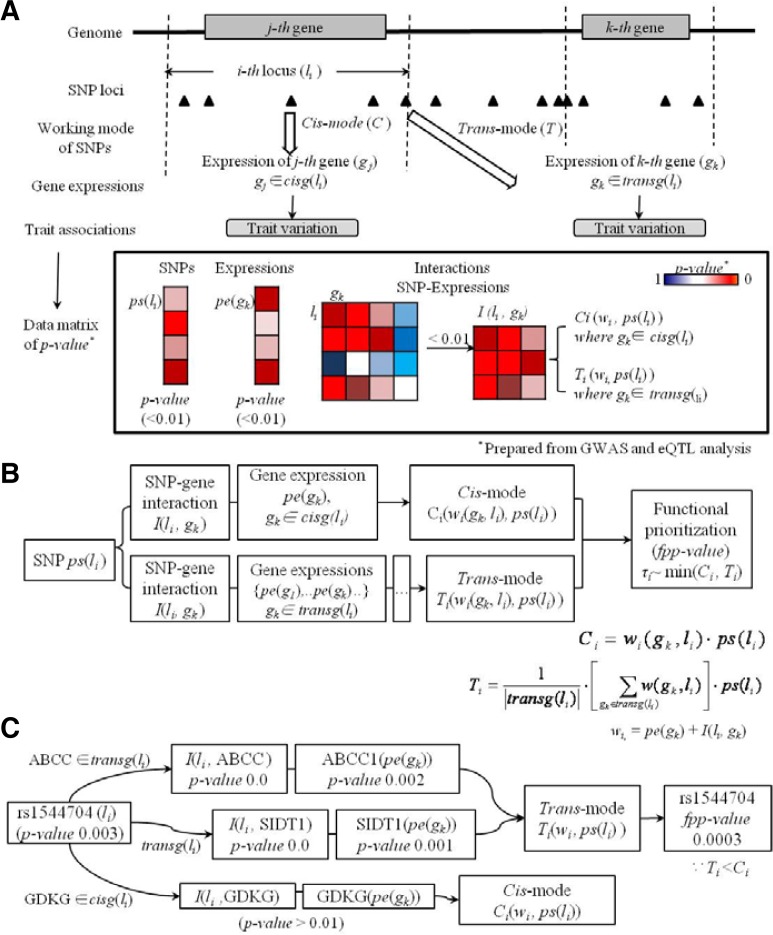
Using these prepared p-values in the data matrices (Fig. 1A), we modeled working mechanisms of the i-th locus according to the biological model of genetic regulation, such as cis- and trans-acting modes. Prepared p-values of gene expression (pe(gk)) and interactions between li and gk (I) were used for the modeling of cis and trans modes for the prioritization of SNPs based on interactions captured in the given population, ρ. According to the genetic structures used in the eQTL study, in which genetic variation perturbs gene expression and leads to phenotypic variation, these p-values of genetic regulation according to phenotype-associated gene expression and p-values of gene expression according to phenotype are functional signatures of causal SNPs. Therefore, we used these p-values as weight values (wi) to prioritize true causal SNPs:
τi = functional prioritization of p-value (fpp-value) of li according to trait variation
cisg (li) = genes associated in cis with the i-th locus, li
transg (li) = genes associated in trans with the i-th locus, li
| (1) |
Ci (w(gk,li), ps (gij)) = fpp-value of the i-th locus in cis mode
Ti (w(gk,li), ps (gij)) = fpp-value of the i-th locus in trans mode
where wi (gk, li) = weighting value for the p-value of the i-th locus, ps (li)
wi (gk, li) = pe (gk) + I (li, gk)
| (2) |
| (3) |
| (4) |
The cis and trans modes of the i-th locus were determined by the identity of the gene expressed (expression of the k-th gene) and the locus of the corresponding gene (i-th locus of the corresponding gene) (equation 1). For instance, the i-th locus of GDKG (rs1544704) showed strong interactions regarding the expression of ABCC1 and SIDT1 in the trans mode (p-value = 0.0) (Fig. 1C, equation 3, ABCC1 and SIDT1 ∈ transg (rs1544704)). Similarly, the cis mode of the i-th locus (rs1544704) regarding trait variation was determined by presenting the interaction p-value for the expression of the corresponding gene, GDKG (Fig. 1C, equation 2, GDKG ∈ cisg (rs1544704)). As the trans mode yielded more significant prioritization results (lower fpp-value) for the target phenotype, our method determined trans mode function as the fpp-value of rs1544704[0] (fpp-value = 0.0003, Fig. 1C, equation 4).
Fig. 1.
Overview of functional prioritization method. (A) Building of data matrix for the functional prioritization of SNPs after GWAS. (B) The model for the functional prioritization. The symbols beneath the boxes represent used data matrices for the functional prioritization value (fpp-value) via our modeling function, τ. (C) Example process for the functional prioritization of rs1544704.
Performance evaluations
The absence of a gold-standard set of genes and loci, particularly for complex phenotypes, represents a challenge in the evaluation of the success of SNP prioritization, as these standards are necessary for changes in phenotype. To address this challenge, we measured the sensitivity/specificity of the fpp-value approach using hypothetical true-positive genes, given the absence of a standard set of genes for drug-induced cytotoxicity. The hypothetical true genes were compiled considering their biological relevance for our target phenotypes: inhibition of cell growth/survival after treatment with cisplatin (Gamazon et al., 2010a). Existing bioinformatics resources were used for the compilation of these hypothetical true genes, which included the Database of Essential Genes (DEG 5.0) (Zhang and Lin, 2009), for human genes that are essential for cell survival, the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000), for the cytochrome P450 and membrane transport ABC pathways, and the Pharmacogenetics Knowledge Base (PharmGKB) (Hewett et al., 2002), for cisplatin- and taxane-related pathways regarding cisplatin-related metabolic features (Crul et al., 2002). In addition to the hypothetical true genes prepared, other human genes were used as nonfunctional genes for the cytotoxicity of cisplatin. True-positive (TP) results were defined as the identification of hypothetical true genes using the fpp-value approach; true-negative (TN) results were defined as the classification of genes that interacted with nonprioritized SNPs as being nonfunctional; false-positive (FP) results were defined as the incorrect classification of genes as being true genes using the fpp-value method; and false-negative (FN) results were defined as the incorrect classification of genes as being nonfunctional using our method. The performance of the fpp-value approach was determined by comparing hypothetical true genes and genes that were regulated by prioritized SNPs in the cis or trans mode functions (equations 2 and 3). Conversely, the performance of the typical p-value approach was determined using selected SNP-corresponding genes. The R package was used for this performance analysis (www.r-project.org).
A gene set enrichment analysis (GSEA) (Subramanian et al., 2005) was conducted in the present study to test the functional significance of genes selected using our fpp-value method. The specific aim of this enrichment analysis was the overall presentation of the biological relevance of prioritized SNPs using GO terms. Comparison of GO enrichment results with the results of the straight p-value method revealed the significance of the fpp-value method for the interpretation of prioritized SNPs using GWAS. The performance of the genes selected using the fpp-value and p-value methods was evaluated in the previous step. We used DAVID (Huang da et al., 2009) for the enrichment test of GO terms using these prepared sets (Fisher’s exact p-value < 0.1).
Interaction network between prioritized loci, gene expression, and target phenotypes
According to the suggested functions, the SNP working model using cis or trans modes (Fig. 1A) identified regulatory relationships between prioritized SNPs and gene expression, which were presented graphically at network level. Using Cytoscape (Shannon et al., 2003), we demonstrated the interactive model depicted in Fig. 1A, as well as the modeling equations 2 and 3, which derived phenotypic variations for cisplatin-induced cytotoxicity.
Dataset for the application of the fpp-value approach
Our method for the prioritization of SNPs was demonstrated using GWAS data related to drug-induced cytotoxicity. As denoted in Figs. 1A and 1B, p-values of SNPs and gene expression in target traits and drug-induced cytotoxicity should be prepared. The existing PACdb database (Gamazon et al., 2010a) fulfilled these requirements by providing the p-values of SNPs and gene expression patterns according to cisplatin-induced cytotoxicity in two different populations, CEU and YRI, of the HapMap project. In addition, p-values of interactions between SNPs and gene expression patterns were also prepared using the SCAN public database (Gamazon et al., 2010b). These p-values of interactions were retrieved using the same populations, CEU and YRI. In particular, cisplatin-induced cytotoxicity was identified using the IC50 degree of cell-growth/survival inhibition according to cisplatin concentration (Gamazon et al., 2010a). Although PACdb provides p-values for cisplatin-derived cytotoxicity using two different measurements, IC50 and area under the concentration-time curve (AUC), we used p-values determined using IC50.
RESULTS
Overview of the fpp-value approach
The principle behind the design of our fpp-value approach was based on the biological intuition that true-causal loci may regulate gene expression significantly and lead to variation of target phenotypes. We gave higher priority to selected SNPs that exhibited p-values that were more significant regarding the regulation of phenotype-associated gene expression. As the p-values of SNPs perturbed gene expression patterns and the p-values of interactions between SNPs and gene expression patterns are a functional signature of SNPs, these were used as weighting values (wi) to determine the functional priority of the p-values (fpp-value) of SNPs. Based on genetic regulation models of gene expression (Cheung et al., 2010) and eQTL studies, the fpp-values of SNPs were measured in the trans and cis modes of interaction between SNPs and gene expression patterns, respectively (Fig. 1A). Subsequently, the most significant value was selected as the fpp-value of a SNP for a target phenotype [τi ∼ min (Ci, Ti), Fig. 1B]. The interaction models were designed using the p-values of SNPs, gene expression, and their interactions, and were presented as fpp-values in cis and trans modes, to measure the functional significance of selected SNPs for target phenotypes. The p-values of a genetic locus (ps(li)) and gene expression (pe(gk)) according to phenotypic variation were prepared using previous GWAS datasets. Using a threshold of p-value (p-value < 0.01, Fig. 1A), data matrices consisted of p-values for the i-th locus (ps(li)) and p-values for the expression of the k-th gene (pe(gk)) that exhibited significance for the target phenotype. In addition, p-values of interactions between li and gk were also prepared (I(li, gk)) using an eQTL analysis of a given population with an identical p-value cut-off (0.01; Fig. 1A). Using the data matrices prepared, we used the developed functions of cis- (Ci (wi(gk,li), ps(li))) and trans-mode models (Ti(wi(gk,li), ps(li))) to determine the functional prioritization of the p-values (fpp-value) of SNPs (Fig. 1B). Figure 1C presents a graphical example of the prioritization process for a p-value of SNP rs1544704, via selection of the most significant value, i.e., the lower fpp-value in the trans mode.
Dataset preparation
The repositories of PACdb (Gamazon et al., 2010a) and SCAN (Gamazon et al., 2010b) were used to apply the method developed here to GWAS data. The PACdb consists of p-values of SNPs and gene expression for a target phenotype, cisplatin-induced cytotoxicity, and SCAN is a database that compiles the p-values of the genetic regulation of gene expression. According to the eQTL concept (causal loci perturb gene expression) (Kim et al., 2011), we used the p-values of the genetic regulation of gene expression as the p-values of interactions between SNPs and gene expression patterns. Notably, SCAN and PACdb surveyed these p-values using cisplatin-treated human cells (lymphoblastoid cell lines) of the CEU and YRI populations of the HapMap consortium (www.hapmap.org). As the CEU and YRI populations of HapMap represent two independent ethnic groups (International_HapMap_Consortium, 2003), Caucasian for CEU and African for YRI, the data dependency or robustness of the fpp-value method was validated by comparing prioritization results after duplicating the running of our algorithms in these two sets. For convenience, we prepared p-values of SNPs, gene expression, and their interactions using a threshold of 0.01. Table 1 presents the results of the data matrices (p-values) prepared using the public databases SCAN and PACdb, under our threshold (p-value < 0.01). Using these p-values of SNPs, gene expression, and their interactions, the fpp-value approach was undertaken to prioritize SNPs in the CEU and YRI populations. In the CEU population, we computed the fpp-value of SNPs using 24,719 p-values of SNPs (p-values of loci per se), 492 p-values of gene expression, and 44,501 p-values of their interactions (Table 1). In the YRI population, 34,395 p-values of SNPs, 683 p-values of gene expression, and 58,804 p-values of their interactions were analyzed to determine the fpp-value of SNPs after GWAS of cisplatin-derived cytotoxicity.
Table 1.
Prepared p-values of SNPs, gene expression, and their interactions
| Trait-associated features (Cisplatin-derived cytotoxicity, IC50 and AUC) | Population
|
Resources | |
|---|---|---|---|
| CEU^ | YRI^ | ||
| Total no. of SNPs (p-value < 0.01)# | 30,807 | 41,112 | PACdb |
| IC50-associated SNPs (loci)* | 24,719 | 34,395 | |
| Number of corresponding genes | 1,938 | 3,289 | |
| Mean of the p-value | 0.0049 | 0.0047 | |
| Standard deviation of the p-value | 0.0029 | 0.0029 | |
| Total no. of gene expression patterns (p-value < 0.01)# | 541 | 825 | PACdb |
| IC50-associated gene expression* | 492 | 683 | |
| Mean of the p-value | 0.0039 | 0.0035 | |
| Standard deviation of the p-value | 0.0029 | 0.003 | |
| Interactions between SNPs and gene expression** | 44,501 | 58,804 | SCAN |
| Mean of the p-value | 0.00006 | 0.00005 | |
| Standard deviation of the p-value | 0.00004 | 0.00004 | |
Number of features associated with cisplatin-derived cytotoxicity: IC50 and AUC (area under the concentration-time curve).
Number of selected features associated with the target trait (cisplatin-derived IC50).
Number of interactions between SNPs and gene expression (p-value < 0.01).
CEU, Utah residents with Northern and Western European ancestry; YRI, residents of Yoruba in Ibadan, Nigeria. Detailed information on these populations is available through the web site of the HapMap project (www.hapmap.org).
Prioritization of SNPs using the fpp-value approach
Based on the p-values of SNPs, gene expression, and interactions obtained from GWAS, our fpp-value approach reordered the p-values of over 58,000 loci in the two populations, CEU and YRI (see “Materials and Methods”). In the CEU population, 4,086 fpp-values of SNPs passed our quality-control measures (fpp-value < 0.05), whereas the previous p-value method yielded 24,719 SNPs as causal loci for cisplatin-induced cytotoxicity (p-value < 0.01) (Table 2). As we planned to perform a ROC curve analysis using various thresholds in a further step of the performance analysis, liberal p-values and fpp-values were introduced in this step. Similarly, in the YRI population, the fpp-value method prioritized 6,996 SNPs as true functional loci for the target trait, whereas the straight p-value method selected 34,395 loci under equal thresholds conditions (fpp-value < 0.05 and p-value < 0.01, Table 2). As shown in Table 2, the straight p-value method selected a number of SNPs covering 1,938 and 3,289 genes as candidates for functional analysis in the CEU and YRI populations, respectively. However, the fpp-value approach suggested 453 and 419 genes for functional analysis of prioritized loci in the two populations, respectively. Distinctively, our fpp-value approach aggregated the p-values of gene expression considering the interaction between gene expression and SNPs in the trans and cis modes of regulation, whereas the typical p-value method suggested the genes corresponding to the loci selected, without any information regarding the perturbation of gene expression by candidate loci or the contribution of gene expression patterns to drug-induced cytotoxicity. Based on the priority functions of the fpp-value approach, such as the cis and trans modes for SNPs, interactions between SNPs and gene expression patterns were identified, as presented in Table 2. Although typical p-value-based GWAS resulted in the identification of locus-corresponding genes by modeling SNP contributions in th cis mode (Table 2) (Lee and Shatkay, 2008), the trans mode of genetic regulation has been regarded as a predominant model in previous eQTL studies (Cheung et al., 2010; Noyes et al., 2010). The fpp-value method followed the trend of previous attempts that were validated experimentally and showed that most genetic interactions or regulation of gene expression happen in trans mode (Table 2).
Table 2.
Functional prioritization results of the fpp-value approach
| Results according to population | GWAS results (p-value < 0.01) | Functional prioritization (fpp-value < 0.05) |
|---|---|---|
| CEU | ||
| No. of SNPs (loci) | 24,719 | 4,086 |
| No. of gene expression patterns | 1,938* | 453** |
| Mode of SNP-gene regulation | ||
| Cis | 1,938 | - |
| Trans | - | 4,086 |
| YRI | ||
| No. of SNPs (loci) | 34,395 | 6,996 |
| No. of gene expression patterns | 3,289* | 419** |
| Mode of SNP-gene regulation | ||
| Cis | 3,289 | 8 |
| Trans | - | 6,988 |
Number of genes corresponding to loci selected in GWAS
Number of phenotype-related genes that were regulated by prioritized loci, as assessed using the fpp-value approach
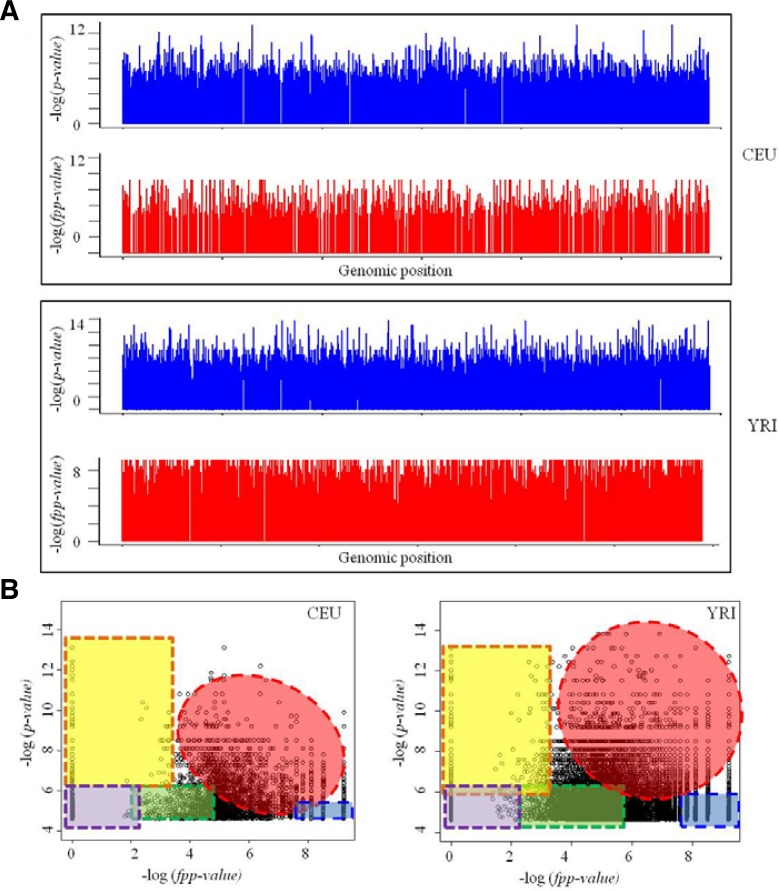
To obtain a general view of our prioritization scheme and of its application to the study of cisplatin-derived cytotoxicity, we plotted the original (nonweighted) p-values and altered the results using fpp-values against genomic position (Fig. 2A) and p-values against the overall fpp-values (Fig. 2B). As depicted in Fig. 2, the plots were analyzed using the CEU and YRI populations. The latter plots (p-value vs fpp-value) introduced a useful method to visualize the GWAS results together with the results of the prioritization of SNPs. The signals in the upper-right regions (red colored) represent p-values that were prioritized functionally (fpp-values) using lower p-values of perturbed gene expression in our interaction models for the variation of cisplatin cytotoxicity (Fig. 2B). The yellow- and blue-shaded regions in the plots denote cases of prioritization of SNPs, such as inconsistencies between the p-values of SNPs and the p-values of their interaction with gene expression for cisplatin-derived cytotoxicity (Fig. 2B). However, the incidence of these extreme cases, i.e., prioritization of SNPs using weighting values of gene expression and interactions rather than the significance of loci, was negligible.
Fig. 2.
Results of functional prioritization approach after GWAS. (A) The original p-values plotted against genomic position (blue colored peaks). Results of functional prioritization method (red colored plot). CEU and YRI means analyzed populations. (B) The original p-values plotted against results of functional prioritization values (fpp-values). Signals in the redcolored regions correspond to the prioritized SNPs by both of the significant p-value and weighting effect via fpp-value analysis. The yellow and blue shaded regions show the differences between these two methods. The purple shaded region displays non-significant SNPs by both of methods. The green colored region denotes SNPs which have p-values and fpp-values at intermediated levels.
Validation of the performance of the fpp-value approach via comparison with the straight p-value method
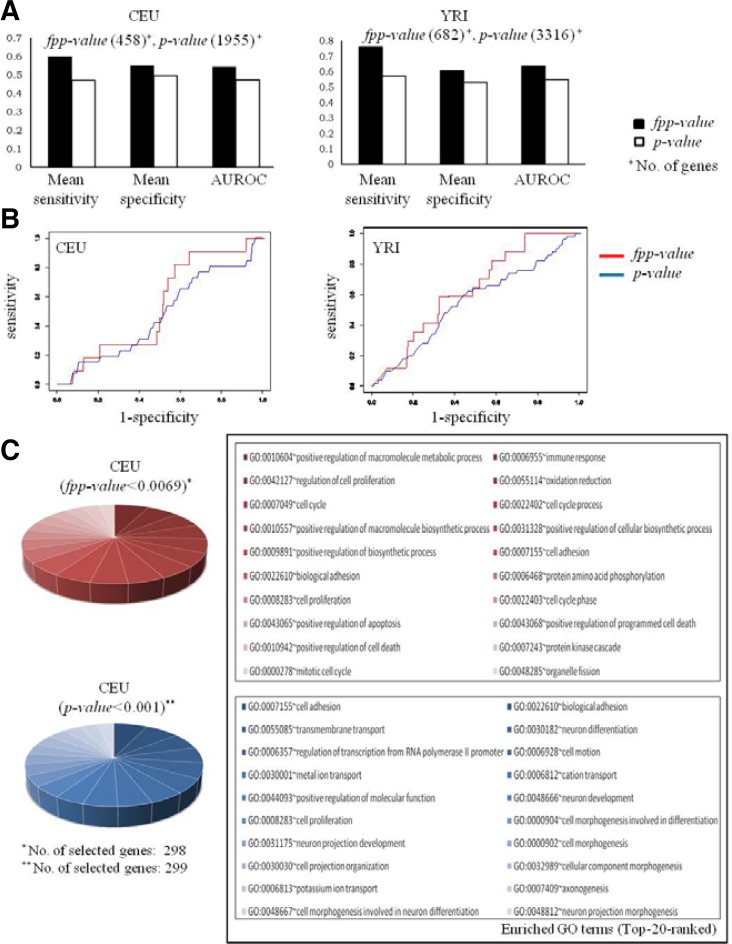
A key element in assessing the role of a given SNP is the determination of whether the variation is likely to result in trait variation, such as cisplatin-induced cytotoxicity. Using the scheme presented here, the p-values of selected loci were prioritized using captured dynamic interactions between genetic variation and phenotype-associated gene expression (Table 2). For the evaluation of our fpp-value approach, we assessed whether the fpp-value approach suggested “true” genes as key players in phenotype differences. Because of the absence of standard gene sets for the cytotoxicity of cisplatin, part of the database was based on a set of determined hypothetical true genes. The hypothetical true genes were prepared considering their biological relevance for the phenotype and given condition: inhibition of cell-growth/survival after treatment with the anticancer drug cisplatin. Existing bioinformatics resources were used for the preparation of these hypothetical true genes, which included DEG 5.0 (Zhang and Lin, 2009), KEGG (Kanehisa and Goto, 2000) (for the cytochrome P450 and membrane transport ABC pathways), and PharmGKB (Hewett et al., 2002) (for the cisplatin- and taxane-related pathways), as described in the “Materials and Methods”. The hypothetical true genes used in the present study included 252 human genes: 118 genes involved in cell survival functions, 55 genes involved in the cisplatin-related pathway, 39 genes involved in the cytochrome P450 pathway (for drug-response metabolism) (Wrighton and Stevens, 1992), 27 genes involved in the membrane transport ABC pathway (for drug resistance) (Brinkmann and Eichelbaum, 2001), and 16 genes involved in the taxane-related pathway (for cisplatin-related metabolic features) (Crul et al., 2002). In addition to the hypothetical true genes prepared, other human genes were defined as nonfunctional (negative) for cisplatin cytotoxicity. Figures 3A and 3B depict the results of the sensitivity/ specificity analysis using various cut-off thresholds for our fpp-value and for the general p-value methods in the CEU and YRI populations. As the fpp-value method exhibited a favorable performance, as it yielded improved AUROC values for both populations compared with the p-value approach (AUROC in CEU, 0.54 vs 0.47; in YRI, 0.63 vs 0.54, respectively), our method exhibited enhanced biological relevance regarding SNPs prioritized using an interaction model for the regulation of gene expression (cis and trans modes).
Fig. 3.
Performance evaluation and enrichment test for the functional annotation of selected genes. (A) Performances of functional prioritization approach and previous straight p-value approach. Both of panels are measured performance in CEU (left) and YRI (right) populations. (B) Panels for ROC analysis. (C) Results of enrichment test using GO terms (category of Biological Process).
A gene set enrichment analysis (GSEA) was performed to identify overrepresented and conserved biological functions among the candidate genes obtained using the fpp-value approach. As depicted in Fig. 3C, the genes identified using our method were enriched for cell-growth- and cytotoxicity-related functions, such as cell proliferation, immune response (Zagozdzon and Golab, 2001), and apoptosis. In contrast, the results of the straight p-value analysis, in GO terms, yielded insufficient biological connections regarding the cytotoxicity of cisplatin (Fig. 3C, bottom). Our method selected prioritized SNPs and perturbed gene expressions that had significant biological relevance for the target traits. Therefore, the fpp-value approach fulfilled our aim of identifying functional loci. We confirmed this outperformance of the fpp-value method in CEU (Fig. 3C) and YRI populations, consistently (Fig. 3C; Supplementary Fig. 1).
Information flows identified between prioritized SNPs and phenotypes
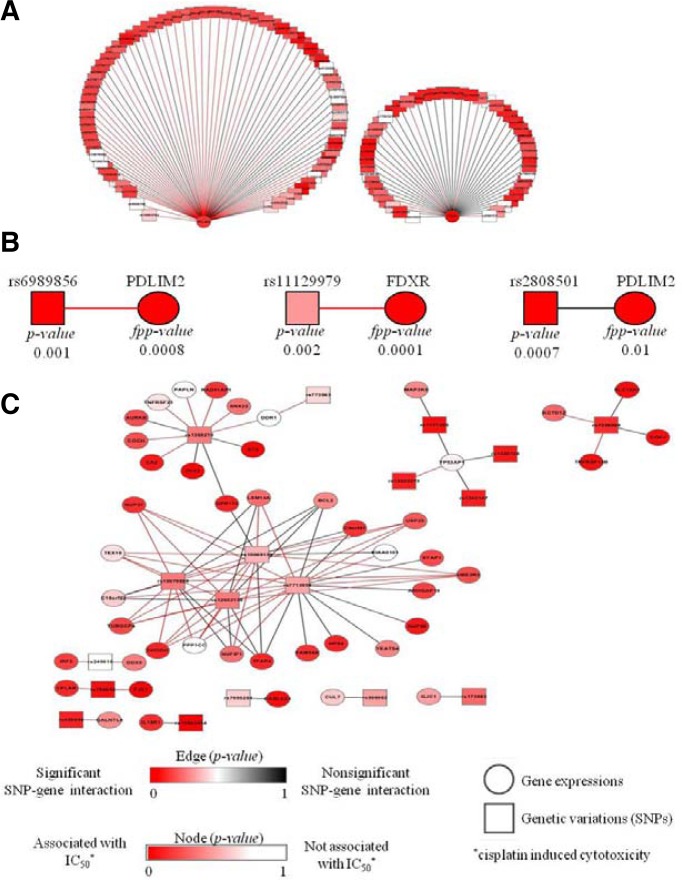
Even if a gene that is potentially causal can be identified, such p-value analysis in GWAS usually does not provide immediate insight into the molecular mechanisms that link the gene to the complex phenotype (McCarthy et al., 2008). In our model of interaction between gene expression and genetic variation, the fpp-value method selected true causal loci that are likely to explain the association of the expression of target genes with trait variations based on p-values obtained for genetic regulation (Fig. 1A). Therefore, our approach may bridge the gap between the statistical evidence stemming from genetic association studies and mechanistic factors, such as gene expression, that lead to phenotypic variation. Figure 4 shows graphically our modeled propagation of information from prioritized loci to perturbed gene expression and to the affected target phenotype, i.e., the cytotoxicity of cisplatin.
Fig. 4.
Underlying network of prioritized loci for cisplatin induced cytotoxicity (IC50). (A) Identified interaction examples between prioritized SNPs and gene expressions, which derives phenotypic variations, cytotoxicity (IC50). (B) Detailed examples of functional prioritization of SNPs. (Left and Middle) Prioritized case due to the significant p-value of SNPs, associated expressions and lower p-value for the interactions between SNPs and expressions. (Right) Non-prioritized case for the significant p-value of SNP due to the higher p-value for interactions between SNPs and gene expressions. (C) Conserved interactions network of causal loci for trait variation, IC50, in CEU and YRI populations.
Figure 4A depicts interaction networks between prioritized SNPs and perturbed gene expression patterns, which mediate phenotype variation, as an example of the information flow identified here. These interaction networks showed that the expression of FDXR and PDLIM2 was regulated by causal loci and yielded individual variation of the cytotoxicity of cisplatin. Previously, the expression of FDXR and PDLIM2 was considered as a promising marker of resistance to anticancer drugs and arrest of cell growth (Qu et al., 2010a; 2010b; Trapasso et al., 2008), albeit with limited understanding of the underlying regulatory mechanisms. Using our fpp-value approach, we identified an information flow from genetic variation to phenotypic variation via intermediate nodes of gene expression, such as FDXR and PDLIM2.
Because the major feature of our prioritization scheme was the use of dynamic interactions between SNPs and gene expression patterns, the strength of the information flow between SNPs and perturbed gene expression adjusted the p-value of loci. Regarding the concept of information flow, Fig. 4B profiles examples of the prioritization procedures used in the fpp-value approach. The p-values of rs6989856 and rs11129979 were prioritized using the fpp-value approach, based on the significance of interactions for gene expression (FDXR and PDLIM2) and on the significance of perturbed gene expression for the target trait, i.e., the cytotoxicity of cisplatin. Conversely, the significance of rs2808501 for the target trait was downplayed because of the lack of perturbation effects of this SNP on gene expression (Fig. 4B, p-value = 0.0007 and fpp-value = 0.01).
The comparison of these networks in CEU and YRI populations allowed us to assess in detail the conserved interaction patterns that lead to variation of cisplatin-derived cytotoxicity. Figure 4C presents the interactions that were conserved among different ethnic groups and regulated gene expression, which indicates the presence of individual variations of cytotoxicity under cisplatin treatment. For instance, the prioritized SNPs rs10079889 and rs12022273 were identified as conserved causal loci that perturbed the expression of CA2 and p53-activated protein (TP53AP1) genes, which are related to cell viability and are known targets of cisplatin resistance for cancer prognosis (Ju et al., 2009; Wang and Bourguignon, 2006). In addition to these conserved interactions, population-specific interactions were also identified in this analysis (Supplementary Fig. 2). These results highlight the superiority of the fpp-value approach for the identification of the contributions of causal loci to gene expression and phenotypic variation according to specific groups of individuals, which goes beyond simple genetic association with target phenotypes.
DISCUSSION
We introduced a novel approach for the prioritization of causal loci using GWAS data of cisplatin-derived cytotoxicity, by integrating phenotypic, genomic, and transcriptomic data. Simultaneously, our fpp-value method identified underlying mechanisms between true causal loci, perturbed gene expression, and phenotypes, which formed an interaction network. This perturbation of gene expression may include potential drivers of the cytotoxicity of cisplatin, as well as potential genes related to cisplatin resistance (Supplementary Table 1). To prioritize loci that were associated with cisplatin-derived cytotoxicity in the CEU and YRI populations, we used interactions between genetic variation and altered expression of target genes using eQTL analysis. The combination of statistical evidence (p-values) from genetic association studies and p-values of gene expression derived from eQTL analyses, considering interaction networks between causal loci, gene expression, and target traits in a stepwise manner, is the key concept of our fpp-value approach. The systematic evaluation of our method, the fpp-value approach, also demonstrated the power of our interaction-focused method to unravel a map of the information flow between causal loci and gene expression, which mediated individual phenotypic variations.
Most studies link disease-associated loci to eQTL data by identifying target genes whose expression is associated with phenotypes of interest, such as disease and drug responses. Although previous attempts have been discussed about the functional signatures of causal loci, such as gene expression, protein modification and metabolic individuality, our prioritization model utilized perturbed gene expression as a signature of functional SNPs. Therefore, designed mode of action for true causal loci is: i) genetic variation alters the expression of genes, and, in turn, ii) this change affects individual phenotypes. Regarding these stepwise genetic mechanisms, our fpp-value method used interaction networks to measure the phenotypic influence of prioritized loci according to the general model of true causal SNPs. To obtain a quantified information flow between prioritized causal SNPs and the target phenotype, we interrogated prioritized and nonprioritized SNPs within genetic pathways (Table 3). As denoted in Table 3, rs17308365 and rs4757718 were prioritized over rs4510585 because the information flow of the former SNPs had a more significant phenotypic effect. It remains unclear the detailed mechanisms for the increase and decrease of gene expressions under the variation of genetic loci, such as direct or indirect regulation of transcription factor bindings. However, a pathway enrichment analysis was conducted to assess the biological relevance of information flows with SNP-perturbed gene expressions (Table 3). The oxidative stress response pathway was an enriched metabolic process for the target trait (cisplatin-derived cell death), and rs17308365 perturbed the expression of MAPKAPK3, a key player in the response to cisplatin treatment, as reported previously (Preta et al., 2010). Conversely, the nonprioritization of rs4510585 was also reached based on the decreased level of interactions between this SNP and the expression of BCL2 and the decreased significance of the p-value for BCL2 (Table 3). Our method also showed buffered information flows between SNPs and phenotypes, based on the decreased strength of the interactions between SNPs and gene expression. Therefore, our method identified intermediate routes between SNPs and phenotypes, such as propagation and decrease of genetic contributions, via a stepwise process of genetic mechanisms (Table 3, information flow step).
Table 3.
Information flow between SNPs and the target phenotype (cytotoxicity of cisplatin)
| Population (ρ) | Step of the information flow | Example of related features | Enriched pathways | |||
|---|---|---|---|---|---|---|
| Top rank | Medium | Low rank | Bottom | |||
| CEU | Step 1: Causal loci: li
|
rs17308365 (p-value = 0.0016) | ||||
| rs4757718 (p-value = 0.006) | ||||||
| rs4510585 (p-value = 0.0017) | ||||||
|
| ||||||
Step 2: SNP–gene interactions: I(li, gk) 
|
I(rs17308365, MAPKAPK3) = 0.0 | |||||
| I(rs4757718, MAPKAPK3) = 0.0001 | ||||||
| I(rs4510585, BCL2) = 0.0001 | ||||||
|
| ||||||
Step 3: Perturbed gene expression: {g1, gk} 
|
MAPKAPK3 (p-value = 0.0007) | Oxidative stress response (PMID: 20643100) | ||||
| BCL2 (p-value = 0.0048) | ||||||
|
| ||||||
Phenotype variation: τi = min (Ci,Ti) 
|
rs17308365 (fpp-value = 0.000801) | |||||
| rs4757718 (fpp-value = 0.000805) | ||||||
| rs4510585 (fpp-value = 0.010718) | ||||||
The fpp-value approach has no limitations regarding data types, such as target phenotypes, input data platforms, species, and population size. Nevertheless, gene expression and genetic variation data should be produced to identify true causal loci for a target phenotype. The typical p-value method of GWAS simply uses the SNPs of individuals to uncover true causal loci for phenotypes, whereas the eQTL approach uses SNP data to describe changes of gene expression. Utilization of expression and interaction p-values as weighting factors for the prioritizing p-values of SNPs remained as methodological issue to determine; whether it is best-way for the prioritization of functional SNPs or heuristic approaches with given dataset, such as eQTL and expression p-values. However, the interaction model of the fpp-value approach suggested for the first time the mechanisms that underlie true-causal SNPs, which were prioritized via biological relevance for gene expressions and statistical evidences of GWAS results. Regarding the propagation of information between SNPs and phenotypes, the details of the trans model of action of causal loci remain an open question. To address these challenges, further research should be performed to develop: 1) eQTL approaches to identify true positive interactions between SNPs and gene expression patterns, and 2) a systematic method for the identification of regulatory routes between causal SNPs and gene expression patterns in the trans mode.
As the network-based integrative analysis of GWAS using high-throughput data promises successful results (Jia et al., 2011), there is great interest in the development of new frameworks including data integration and interaction modeling. Independent from the populations analyzed, several bioinformatics approaches were established to identify the biological relevance of causal loci, by combining knowledge-based concepts [including potential genes related to the phenotype and mutations in splice sites (Saccone et al., 2010)]. To the best of our knowledge, the fpp-value method is the first computational framework that prioritized true functional SNPs with statistical evidence from GWAS and provided an authentic and quantitative genetic network between gene expression and phenotype in a given population. Therefore, because of the successful suggestion of genetic networks for causal SNPs achieved here, our method represents a breakthrough compared with the typical GWAS and bioinformatics attempts to find functional loci.
Supplementary Material
Acknowledgments
This work was supported by World Class University (WCU) program (R32-2008-000-10218-0) of the Ministry of Education, Science and Technology (MEST) through the National Research Foundation of Korea. HP, HSH and CGH were supported by the Korea Research Institute of Bioscience and Biotechnology (KRIBB) research program: A omics-based integration database for cancer interpretation (KGM0661113). HP was also supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (No. 2-2010-114-0 and 0634-20100002).
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Brinkmann U., Eichelbaum M. Polymorphisms in the ABC drug transporter gene MDR1. Pharmacogenomics J. 2001;1:59–64. doi: 10.1038/sj.tpj.6500001. [DOI] [PubMed] [Google Scholar]
- Chae S.C., Yu J.I., Oh G.J., Choi C.S., Choi S.C., Yang Y.S., Yun K.J. Identification of single nucleotide polymorphisms in the TNFRSF17 gene and their association with gastrointestinal disorders. Mol. Cells. 2010;29:21–28. doi: 10.1007/s10059-010-0002-6. [DOI] [PubMed] [Google Scholar]
- Cheung V.G., Nayak R.R., Wang I.X., Elwyn S., Cousins S.M., Morley M., Spielman R.S. Polymorphic cis- and trans-regulation of human gene expression. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichon S., Muhleisen T.W., Degenhardt F.A., Mattheisen M., Miro X., Strohmaier J., Steffens M., Meesters C., Herms S., Weingarten M., et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am. J. Hum. Genet. 2011;88:372–381. doi: 10.1016/j.ajhg.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crul M., van Waardenburg R.C., Beijnen J.H., Schellens J.H. DNA-based drug interactions of cisplatin. Cancer Treat. Rev. 2002;28:291–303. doi: 10.1016/s0305-7372(02)00093-2. [DOI] [PubMed] [Google Scholar]
- Etemadmoghadam D., deFazio A., Beroukhim R., Mermel C., George J., Getz G., Tothill R., Okamoto A., Raeder M.B., Harnett P., et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin. Cancer Res. 2009;15:1417–1427. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon E.R., Duan S., Zhang W., Huang R.S., Kistner E.O., Dolan M.E., Cox N.J. PACdb: a database for cell-based pharmacogenomics. Pharmacogenet. Genomics. 2010a;20:269–273. doi: 10.1097/FPC.0b013e328337b8d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon E.R., Zhang W., Konkashbaev A., Duan S., Kistner E.O., Nicolae D.L., Dolan M.E., Cox N.J. SCAN: SNP and copy number annotation. Bioinformatics. 2010b;26:259–262. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewett M., Oliver D.E., Rubin D.L., Easton K.L., Stuart J.M., Altman R.B., Klein T.E. PharmGKB: the pharmacogenetics knowledge base. Nucleic Acids Res. 2002;30:163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- International_HapMap_Consortium The international Hap map project. Nature. 2003;426:789–796. [Google Scholar]
- Jia P., Zheng S., Long J., Zheng W., Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;27:95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju W., Yoo B.C., Kim I.J., Kim J.W., Kim S.C., Lee H.P. Identification of genes with differential expression in chemoresistant epithelial ovarian cancer using high-density oligonucleotide microarrays. Oncol. Res. 2009;18:47–56. doi: 10.3727/096504009789954672. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.A., Wuchty S., Przytycka T.M. Identifying causal genes and dysregulated pathways in complex diseases. PLoS Comput. Biol. 2011;7:e1001095. doi: 10.1371/journal.pcbi.1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.H., Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008;36:D820–824. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.O., Cheong H.S., Park B.L., Bae J.S., Sim W.C., Chun J.Y., Isbat M., Uh S.T., Kim Y.H., Jang A.S., et al. MYLK polymorphism associated with blood eosinophil level among asthmatic patients in a Korean population. Mol. Cells. 2009;27:175–181. doi: 10.1007/s10059-009-0022-2. [DOI] [PubMed] [Google Scholar]
- Li Y., Sheu C.C., Ye Y., de Andrade M., Wang L., Chang S.C., Aubry M.C., Aakre J.A., Allen M.S., Chen F., et al. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol. 2010;11:321–330. doi: 10.1016/S1470-2045(10)70042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P., Hirschhorn J.N. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- McPherson R., Pertsemlidis A., Kavaslar N., Stewart A., Roberts R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., et al. A common allele on chromo-some 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes H.A., Agaba M., Anderson S., Archibald A.L., Brass A., Gibson J., Hall L., Hulme H., Oh S.J., Kemp S. Genotype and expression analysis of two inbred mouse strains and two derived congenic strains suggest that most gene expression is trans regulated and sensitive to genetic background. BMC Genomics. 2010;11:361. doi: 10.1186/1471-2164-11-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik H., Lee E., Lee D. Relationships between genetic polymorphisms and transcriptional profiles for outcome prediction in anticancer agent treatment. BMB Rep. 2010;43:836–841. doi: 10.5483/BMBRep.2010.43.12.836. [DOI] [PubMed] [Google Scholar]
- Paik H., Lee E., Park I., Kim J., Lee D. Prediction of cancer prognosis with the genetic basis of transcriptional variations. Genomics. 2011;97:350–357. doi: 10.1016/j.ygeno.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Preta G., de Klark R., Chakraborti S., Glas R. MAP kinase-signaling controls nuclear translocation of tripeptidyl-peptidase II in response to DNA damage and oxidative stress. Biochem. Biophys. Res. Commun. 2010;399:324–330. doi: 10.1016/j.bbrc.2010.06.133. [DOI] [PubMed] [Google Scholar]
- Qu Z., Fu J., Yan P., Hu J., Cheng S.Y., Xiao G. Epigenetic repression of PDZ-LIM domain-containing protein 2: implications for the biology and treatment of breast cancer. J. Biol. Chem. 2010a;285:11786–11792. doi: 10.1074/jbc.M109.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z., Yan P., Fu J., Jiang J., Grusby M.J., Smithgall T.E., Xiao G. DNA methylation-dependent repression of PDZ-LIM domain-containing protein 2 in colon cancer and its role as a potential therapeutic target. Cancer Res. 2010b;70:1766–1772. doi: 10.1158/0008-5472.CAN-09-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone S.F., Saccone N.L., Swan G.E., Madden P.A., Goate A.M., Rice J.P., Bierut L.J. Systematic biological prioritization after a genome-wide association study: an application to nicotine dependence. Bioinformatics. 2008;24:1805–1811. doi: 10.1093/bioinformatics/btn315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone S.F., Bolze R., Thomas P., Quan J., Mehta G., Deelman E., Tischfield J.A., Rice J.P. SPOT: a web-based tool for using biological databases to prioritize SNPs after a genome-wide association study. Nucleic Acids Res. 2010;38:W201–209. doi: 10.1093/nar/gkq513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M., Tanaka Y., Nakanishi M., Mizokami M. Novel findings for the development of drug therapy for various liver diseases: Genetic variation in IL-28 B is associated with response to the therapy for chronic hepatitis C. J. Pharmacol. Sci. 2011;115:263–269. doi: 10.1254/jphs.10r15fm. [DOI] [PubMed] [Google Scholar]
- Taylor I.W., Linding R., Warde-Farley D., Liu Y., Pesquita C., Faria D., Bull S., Pawson T., Morris Q., Wrana J.L. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat. Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- Trapasso F., Pichiorri F., Gaspari M., Palumbo T., Aqeilan R.I., Gaudio E., Okumura H., Iuliano R., Di Leva G., Fabbri M., et al. Fhit interaction with ferredoxin reductase triggers generation of reactive oxygen species and apoptosis of cancer cells. J. Biol. Chem. 2008;283:13736–13744. doi: 10.1074/jbc.M709062200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Vinuela A., Snoek L.B., Riksen J.A., Kammenga J.E. Genome–wide gene expression regulation as a function of genotype and age in C. elegans. Genome Res. 2010;20:929–937. doi: 10.1101/gr.102160.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.J., Bourguignon L.Y. Hyaluronan-CD44 promotes phospholipase C-mediated Ca2+ signaling and cisplatin resistance in head and neck cancer. Arch. Otolaryngol. Head Neck Surg. 2006;132:19–24. doi: 10.1001/archotol.132.1.19. [DOI] [PubMed] [Google Scholar]
- Wrighton S.A., Stevens J.C. The human hepatic cytochromes P450 involved in drug metabolism. Crit. Rev. Toxicol. 1992;22:1–21. doi: 10.3109/10408449209145319. [DOI] [PubMed] [Google Scholar]
- Wu C., Xu B., Yuan P., Miao X., Liu Y., Guan Y., Yu D., Xu J., Zhang T., Shen H., et al. Genome-wide interrogation identifies YAP1 variants associated with survival of small-cell lung cancer patients. Cancer Res. 2010;70:9721–9729. doi: 10.1158/0008-5472.CAN-10-1493. [DOI] [PubMed] [Google Scholar]
- Zagozdzon R., Golab J. Immunomodulation by anticancer chemotherapy: more is not always better (review) Int. J. Oncol. 2001;18:417–424. doi: 10.3892/ijo.18.2.417. [DOI] [PubMed] [Google Scholar]
- Zhang R., Lin Y. DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res. 2009;37:D455–458. doi: 10.1093/nar/gkn858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H., Yang X., Kaplan L.M., Molony C., Schadt E.E. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am. J. Hum. Genet. 2010;86:581–591. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.