Abstract
Legumes have evolved symbiotic interactions with rhizobial bacteria to efficiently utilize nitrogen. Recent progress in symbiosis has revealed several key components of host plants required for nitrogen-fixing nodule organogenesis, in which complicated metabolic and signaling pathways in the host plant are reprogrammed to generate nodules in the cortex upon perception of the rhizobial Nod factor. Following the recognition of Nod factors, plant hormones are likely to be essential throughout nodule organogenesis for integration of developmental and environmental signaling cues into nodule development. Here, we review the molecular events involved in plant hormonal regulation and signaling cross-talk for nitrogen-fixing nodule development, and discuss how these signaling networks are integrated into Nod factor-mediated signaling during plant-microbe interactions.
Keywords: cross talk, hormone, legume, nitrogen fixing, symbiosis
NOD FACTOR SIGNALING DURING NODULATION
Uptake of nutrients by plants is limited by the insufficiency of biologically available forms of these compounds in the environment. As a consequence of selection pressure for survival, plants have broadly evolved beneficial symbiotic interactions with commensal microbes. The arbuscular mycorrhizal (AM) fungi have developed a symbiotic relationship with most land plants, which is highly beneficial for the uptake of minerals and water from the soil (Parniske, 2008). In contrast to general AM symbiosis, nitrogen-fixing nodule symbiosis has been strictly adopted by a few plant families, including legumes, together with their symbiotic partners, which are collectively called rhizobium bacteria (Kouchi et al., 2010; Oldroyd and Downie, 2008; Sandal et al., 2002). In general, the key signaling components for nitrogen fixing symbiosis are well conserved in the legume family (Markmann and Parniske, 2009).
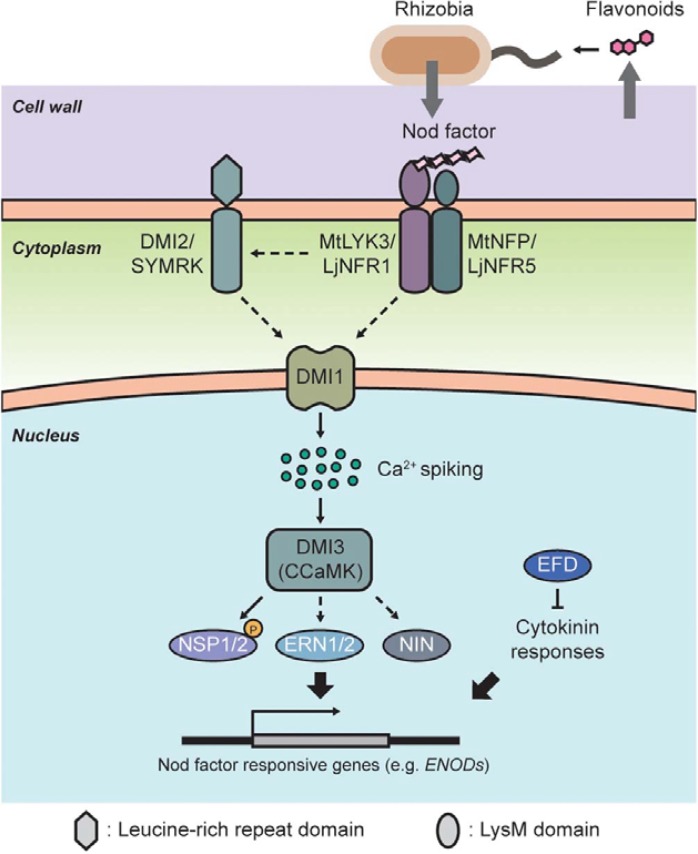
To initiate symbiotic nitrogen-fixing nodulation, the host plants provide a cocktail of flavonoids to stimulate synthesis and secretion of Nod factors from symbiotic rhizobia bacteria (Barnett and Fisher, 2006; Perret et al., 2000). The secreted Nod factors directly stimulate their putative receptor proteins (NFRs), LjNFR1/5 (Nod factor through receptor-like kinase) in Lotus japonicus and MtLYK3 (LysM-RLK3)/NFP (Nod factor perception) in Medicago trucatula, which are peptidoglycan-binding lysine domain (LysM)-containing receptor kinases (Arrighi et al., 2006; Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003). Recognition of Nod factors, and compatibility of rhizobia bacteria with host legume plants, are specifically determined by the extracellular LysM domain in the NFRs. Transgenic M. truncatula roots harboring LjNFR1/5 could make nodules upon inoculation of the L. japonicus-compatible bacterium, Mesorhizobium loti, and swapping the LysM between the NFRs of L. filicaulis and L. japonicas changed the ability of the plants to recognize their compatible symbiotic partners (Radutoiu et al., 2007). Activation of NFRs induced root hair deformation and subsequent stimulation of downstream signaling pathways through nuclear Ca2+ spiking, and induced nodule organogenesis in the inner cortex layer of roots and rhizobia infection by elongating infection threads from the root hair tips. A perinuclear-anchored cation channel, MtDMI1 (Does-not-Make-Infections1)/LjCASTOR/LjPOLLUX plays a critical role upstream of Ca2+ spiking during early rhizobia infection, and its function appears to be regulated by an upstream component, MtDMI2 (Does-not-Make-Infections2)/LjSYMRK (Symbiosis receptor kinase)/MsNORK (Nodulation receptor kinase), which is a member of the LRR-RLK family (Leucine rich repeat-receptor like kinases) (Catoira et al., 2000; Endre et al., 2002; Madsen et al., 2003; Radutoiu et al., 2003; Stracke et al., 2002). Loss-of-function mutations in these genes caused impaired Ca2+ spiking in the nucleus and nodule formation, but did not impair root hair deformation in response to rhizobia inoculation (Ane et al., 2004; Endre et al., 2002; Kistner et al., 2002; Stracke et al., 2002). However, both root hair deformation and Ca2+ spiking were disrupted in the NFR mutants, suggesting that DMI1 and DMI2 act downstream of NFRs in the Nod factor signaling pathway (Ben Amor et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003). Ca2+-related MtDMI3 was identified as a nuclear-localized Ca2+-calmodulin-dependent kinase (CCaMK) that functions downstream of Ca2+ spiking (Gleason et al., 2006; Tirichine et al., 2006). A Mtdmi3 mutant could not make root nodules, even though nuclear Ca2+ spiking was unaffected during early nodulation processes (Levy et al., 2004; Mitra et al., 2004). By contrast, constitutively activated MtDMI3 spontaneously induced pseudo-nodules in both wild type and Mtdmi1/2 mutants independently of upstream Nod factor signaling cues. The nuclear events of Ca2+ spiking and subsequent activation of CCaMK suggested that CCaMK is a regulator of transcriptional factors for early Nod factor-responsive genes such as Expression of early nodulin (ENODs), CYCLOPS, and Nodule inception (NIN) (Heckmann et al., 2006; Kalo et al., 2005; Murakami et al., 2006; Smit et al., 2005; Yano et al., 2008). Plant transcriptomes are globally changed during nodule organogenesis by nodulation-related transcriptional factors including NSP1, NSP2 (Nodulation signaling pathway), ERNs [Ethylene response factor (ERF) required for nodulation], and EFD (ERF required for nodule differentiation). Indeed, MtDMI3 directly binds to the NSP1/2 complex that regulates ENOD11 expression, which requires Nod-factor, Ca2+ spiking and CCaMK activity (Gleason et al., 2006; Hirsch et al., 2009; Imaizumi-Anraku et al., 2006; Tirichine et al., 2007). These results indicate that NFR-induced Ca2+ spiking activates nuclear localized CCaMK and then modulates transcriptional networks via NSP1/2 to initiate nodulation (Fig. 1).
Fig. 1.
Proposed model for rhizobia-legume symbiotic signaling pathways. Nod factor from rhizobia stimulates its receptor complex (NFR), MtLYK3/NFP or LjNFR1/5. Signal cascades from NFR are transduced via as-yet-unidentified pathways to induce nuclear calcium spiking and activate DMI3 (CCaMK). Activated DMI3 directly interacts with and activates nodule-related transcription factors, including NSP1/2, via phosphorylation. These transcription factors enhance the expression of Nod factor-responsive genes by directly binding to the NF-box (AATTT). It is not clear how DMI3 regulates nodule-related NIN and ERN, but their up-regulation by Nod factor signaling is essential for nodulation. EFD, an AP2/ERF family member, inhibits nodulation, probably by disrupting cytokinin signaling. The dashed arrows indicate unknown biochemical links.
Given the complexities of nodule development, it is not surprising that other transcription factors including ERN (ERF required for nodulation, Andriankaja et al., 2007; Middleton et al., 2007), NIN (Schauser et al., 1999), EFD (Vernie et al., 2008), and NSP1/2 are also essential for proper development of nodules. ERN1 and ERN2, which are AP2/ERF domain-containing transcriptional factors, play positive roles in nodulation. The loss-of-function mutant of ERN1, designated bit1-1 (branching infection thread 1), is impaired in early nodule formation processes, including up-regulation of nodulin gene expression, infection thread elongation, and nodule primordia initiation (Middleton et al., 2007). ERN3, ERN1 and ERN2 bind directly to the cis-element (NF-box) of the ENOD11 promoter; however, ERN3 represses ENOD11 expression (Andriankaja et al., 2007), suggesting that it functions as a negative regulator of nodule formation. EFD, another nodulation-related AP2/ERF gene, is also required for both functional bacteroid differentiation and nodule organogenesis; its loss-of-function mutant had fewer nodules defective in bacteroid differentiation. Ectopic expression of EFD in tobacco cells enhanced the promoter activity of type A-MtRR4, a negative regulator of the cytokinin signaling pathway (Vernie et al., 2008), which is involved in the regulation of nodule organogenesis. These results imply that the negative regulation of EFD during nodule formation might be linked to cytokinin signaling; however, more detailed studies of the biological functions of EFD are required. The putative transcription factor, NIN, is involved at a later stage of nodulation (Borisov et al., 2003; Roussis et al., 1999; Schauser et al., 1999). A null mutation of NIN displayed normal epidermal responses to rhizobia infection, including nodule-related gene expression, Ca2+ spiking, and root hair deformation, but nodule organogenesis and infection thread formation were completely impaired (Borisov et al., 2003; Madsen et al., 2003). NIN functions downstream of MtDMI3, as constitutively activated CCaMK could not induce spontaneous nodule formation in an Mtnin loss-of-function mutant (Marsh et al., 2007). The Nod-signaling cascade initiated from NFRs is linked to nuclear CCaMK activity, which regulates nodule-related transcription factors during initiation of nodulation at the epidermis (Fig. 1). However, the upstream signaling component(s) that are linked to phosphorylation-dependent activation of nodulation transcription factors remain to be elucidated (Fig. 1).
SYSTEMIC AUTOREGULATION OF NODULATION
Negative regulation of nodulation by systemic shoot-root communication
During nitrogen-fixing symbiosis, the number of nodules has to be strictly regulated because nodule development is an energetically expensive process for the leguminous host plants. The symbiotic relationship is formed by the legumes obtaining nitrogen from the endophytic bacteroids, while providing photosynthetic products in return. Therefore, the maintenance of a balanced number of nitrogen-fixing nodules is critical to provide optimum nutrition and prevent excessive energy drains on the host plants. To control the number of nodules, legumes have evolved negative feedback systems, such as systemic auto-regulation of nodulation (AON) and local hormonal inhibitory regulation (Figs. 2A and 2B).
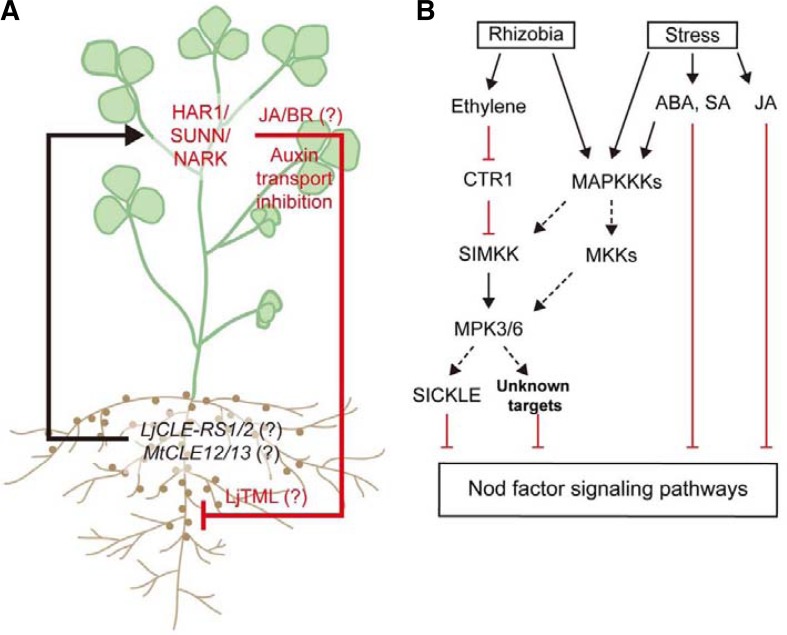
Fig. 2.
Systemic and local inhibitory regulation of nodulation. (A) Schematic model of autoregulation of nodulation (AON). Long distance inhibitory pathways for AON have been proposed, but the root-derived signals are not clearly identified although some CLE peptides including LjCLE-RS1/2 or MtCLE12/13 might function as root-to-shoot signaling molecules. Root-derived signals might directly or indirectly activate the CLV1-like receptor kinase, HAR1/SUNN/NARK. These events create shoot-derived unidentified negative signals that suppress root nodule formation by inhibiting auxin transport and by presumed TML functions in the root. JA and BR are thought to be involved in shoot-derived negative signaling pathways. (B) Local inhibition pathways mediated by the coordination of stress-related hormone actions. Rhizobia and external stress signals activate MAPK signaling cascades and the action of plant hormones including ethylene, SA, ABA, and JA. Ethylene, a major negative regulator in nodulation, probably stimulates SIMKK(AtMKK4/5)-MPK3/6 cascades by suppressing CTR1 activity. The activated MPK3/6 proteins directly or indirectly regulate their downstream signaling components, including SICKLE/EIN2 and various stress-related transcription factors (unknown targets), to inhibit Nod factor signaling pathways. ABA, SA, and JA signaling pathways directly suppress Nod factor signaling, or their negative action could be integrated into SIMKK-mediated MAPK signaling cascades. The dashed arrows indicate unknown biochemical links.
Systemic autoregulation of nodulation in legumes was proposed by Nutman in 1952 after observing a transient increase in the number of new nodules in red clover roots after excising old nodules (Nutman, 1952). The molecular mechanism of AON was actively investigated after identification of the supernodulation mutants, hyper nodulation and aberrant root 1 (har1), super numeric nodules 1 (sunn), and nodule autoregulation receptor kinase (nark) in L. japonicus, Medicago truncatula, and Glycine max, respectively (Ferguson et al., 2010; Kawaguchi and Minamisawa, 2010; Krusell et al., 2002; Nishimura et al., 2002; Oka-Kira et al., 2005; Searle et al., 2003), which encode a leucine rich repeat (LRR) CLV1 (CLAVATA)-like receptor-like kinase, and were exclusively expressed in the phloem of shoot tissues and root nodules (Nontachaiyapoom et al., 2007). Interestingly, reciprocal grafting assays between supernodulation mutants and wild-type plants showed that the supernodulation phenotype is determined by the shoot genotype (Krusell et al., 2002; Lough and Lucas, 2006; Ruiz-Medrano et al., 2001). These observations, along with the molecular mechanism of CLV1 (CLAVATA 1) triggered by a signal peptide CLV3 (CLAVATA 3), led to the hypothesis that communication between roots and shoots via a peptide-mediated mobile signal is likely involved in the systemic AON. Of the 39 potential CLE (CLV3/ESR-related) genes that have been identified in the L. japonicus genome, LjCLE-RS1/2 appeared to function as a root-derived mobile signal for AON because: 1) expression levels were rapidly up-regulated by rhizobia inoculation or KNO3 treatment; and 2) overexpression significantly suppressed nodulation in wild type plants, but not in a loss-of-function har1 mutant (Okamoto et al., 2009). It is likely that MtCLE12/13 in M. truncatula plays a similar role to that of LjCLE-RS1/2 (Mortier et al., 2010). These CLE peptides might be processed to functionally active forms in the root and transported to shoot tissues (via the phloem) where they subsequently stimulate AON-related LRR-RLKs, such as HAR1 and its orthologs (Fig. 2A). However, this hypothesis remains to be tested. The shoot-derived negative signals from the LRR-RLKs in AON are presumably delivered to roots where they are decoded to optimize nodule numbers. An as-yet-uncloned gene, LjTML (Too much love), was identified as a potential signaling decoder for shoot-derived signals in AON (Magori et al., 2009). The tml mutant exhibited a supernodulation phenotype, which was determined by the root genotype. Moreover, the nodule phenotype of tml was less sensitive to exogenous nitrogen treatment, and this decreased sensitivity was not enhanced by the har1 mutation. These results indicate that TML is one of the AON signaling components that function downstream of HAR1 in root tissues; however, further experiments are required to elucidate the detailed mechanism of its action in AON (Fig. 2A).
Plant hormones as candidates for shoot-derived signals in AON
The plant hormones auxin, jasmonic acid (JA), and brassinosteroids (BR) appear to act as signaling mediators in the AON signaling pathway (Nakagawa and Kawaguchi, 2006; Oka-Kira and Kawaguchi, 2006; Terakado et al., 2006). Foliar application of JA decreased the nodule number in AON-defective hypernodulation mutants of G. max and L. japonicus (Kinkema and Gresshoff, 2008; Nakagawa and Kawaguchi, 2006). JA biosynthesis in the foliar region of soybean was enhanced by inoculation of symbiotic rhizobia, but this increase was impaired in AON-deficient Gmnark mutants (Kinkema and Gresshoff, 2008), suggesting that rhizobia bacteria-mediated activation of JA biosynthesis is controlled by a GmNARK-mediated AON signaling pathway.
Exogenous BR treatment effectively reduced the number of nodules in a soybean hypernodulation mutant, but not in wild type plants. Exogenous BR also enhanced the biosynthesis of spermidine (Terakado et al., 2006), which caused a decrease in the number of nodules. Thus, negative regulation of nodulation by BR might be mediated through the action of spermidine in shoots. In spite of the immobility of BR in the pea (Pisum sativum) (Symons and Reid, 2004), defective nodulation in a BR-deficient pea mutant was recovered in wild-type shoot grafting experiments, implying that BRs might regulate shoot-derived AON signaling via unknown pathways (Ferguson et al., 2005). It is clear that BR play a critical role in AON; however, the discrepancy between the negative effects of exogenous BR on nodulation and impaired nodule formation in BR-biosynthetic or BR-insensitive pea mutants suggests that BR might differentially control nodulation with respect to local and systemic regulation. Indeed, BR facilitates polar auxin transport by enhancing PIN expression in Arabidopsis (Bao et al., 2004; Nakamura et al., 2004), and auxin transport and its subsequent local accumulation are important in nodule organogenesis (Pacios-Bras et al., 2003; van Noorden et al., 2006; 2007). These results lead to a possibility that lower auxin levels in the roots of BR-deficient legumes decrease the initiation of nodule primordia, although BR-mediated AON signaling from the shoot might be defective.
Polar auxin transport also appears to be important in AON signaling pathways (van Noorden et al., 2006; Wasson et al., 2006). The inoculation of rhizobia normally decreased auxin loading from shoots to roots by increasing the level of flavonoids, inhibitors of auxin transport, in the shoot. However, the hypernodulation Mtsunn, a loss-of-function mutant of AON-related LRR-RLK in M. truncatula, showed much higher auxin transport than the wild type, and rhizobia inoculation could not reduce the polar auxin transport to root tissues in this mutant (Penmetsa et al., 2008). These results support a hypothesis that AON signaling pathways could inhibit acropetal polar auxin transport; thus, decreased auxin levels in root tissues could inhibit the development of newly initiated nodule primordia (Fig. 2A).
LOCAL REGULATION OF NODULATION VIA HORMONES
Ethylene, a major local inhibitory signal in symbiosis
Legumes have evolved local inhibitory regulation systems of nodulation via hormones such as Abscisic acid (ABA), Jasmonic acid (JA), ethylene, and SA (Salicylic acid), in addition to systemic AON signaling (Biswas et al., 2009; Ding et al., 2008; Oldroyd et al., 2001; Penmetsa and Cook, 1997; Sun et al., 2006; Suzuki et al., 2004) (Fig. 2B). The hypernodulating sickle (sik) mutant of M. truncatula was caused by a loss-of-function mutation (Q893stop) in the ortholog of Arabidopsis Ethylene insensitive 2 (EIN2), a key component of the ethylene signal transduction pathway (Penmetsa and Cook, 1997). The sickle mutant was defective in ethylene responses and showed increases in nodule numbers by generating pairs of nodules at opposite regions of both the xylem and phloem (Penmetsa and Cook, 1997; Penmetsa et al., 2008). Furthermore, numerous uncontrolled infection threads and root hair hypersensitivity to Nod factors were observed in the Mtsickle mutant. This genetic evidence of the negative role of ethylene was supported by the effects of an ethylene precursor, Aminocyclopropane-1-carboxylic acid (ACC), and an ethylene biosynthetic inhibitor, AVG (Aminoethoxyvinylglycine), on nodulation (Engstrom et al., 2001; Oldroyd et al., 2001). ACC or ethylene treatments strongly suppressed the nodulation and Nod factor signaling pathways, but inhibition of ethylene signaling or biosynthesis significantly increased the nodule numbers. Moreover, ethylene treatment effectively inhibited Nod factor-inducible Ca2+ spiking and the expression of nodule-related ENODs (Engstrom et al., 2001; Oldroyd et al., 2001; Penmetsa and Cook, 1997), supporting the evidence for a negative effect of ethylene on nodulation by disrupting Nod factor signal transduction pathways. Ethylene inhibits nodulation by altering the nuclear Ca2+ spiking frequency and suppressing nodulin gene expression, suggesting that EIN2-mediated ethylene signaling and Nod factor signaling pathways are probably integrated downstream of CCaMK signaling. The elucidation of how ethylene signaling pathways regulate calcium spiking during early nodulation will provide critical clues to understand ethylene-mediated local inhibition of nodulation.
Stress-related JA, SA, and ABA signaling inhibits nodulation
Stress-related hormones such as JA and ABA greatly reduce nodulation by disrupting Nod factor-induced Ca2+ spiking and nodulation-related gene expression (Akune et al., 2004; Cardoza et al., 2006; Ding et al., 2008; Stacey et al., 2006; Sun et al., 2006; Suzuki et al., 2001). JA-treated Medicago roots rarely induced symbiotic nodules and consistently showed reduced expression of ENOD11 and RIP1 (Rhizobium meliloti-induced peroxidase gene). However, these JA effects were suppressed by an ethylene biosynthetic inhibitor, AVG, and the sickle mutant was less sensitive to exogenous JA in nodulation (Sun et al., 2006). These results imply that JA-mediated inhibition of nodulation is linked to ethylene signaling pathways. This hypothesis is supported by the previous results that JA and ethylene share downstream target genes, including ERF1 and PDFs, which function to stimulate plant defenses and repel insects, respectively (Lorenzo et al., 2003). Similar to JA, ABA decreased nodule numbers and disrupted Nod factor-induced calcium spiking (Ding et al., 2008). ABA-insensitive Medicago roots were generated by overexpressing Arabidopsis abi1-1, an ABA-insensitive dominant allele of ABI1 (ABA insensitive 1), and these roots formed vigorous nitrogen-fixing nodules. However, the sickle mutation could not suppress the negative effect of ABA on nodule formation. Furthermore, ABA repressed the expression of a different set of nodulation-related genes, including cytokinin-responsive ENOD40 and NIN. It is clear that ABA negatively regulates nodulation via a different regulatory mechanism from JA.
Nodulation and rhizobia infection in legumes was promoted by reduction of the endogenous SA level by ectopic expression of a bacterial nahG gene, which encodes an SA hydroxylase (Stacey et al., 2006). Exogenous SA represses the expression of nodule-related genes and nodule development, but it also suppresses the growth of rhizobia (Stacey et al., 2006). SA is a key signaling molecule for defense responses to diverse plant invaders. Recent studies show that defense and stress responses are elicited during rhizobia infection, and these responses are important factors for determining host range or nodule numbers (Mellor and Collinge, 1995; Santos et al., 2001). However, the molecular mechanisms that mediate SA-induced defense signaling during nodulation are largely unknown. In the future, the precise roles of SA and SA-mediated defense signaling during nodulation should be investigated.
Are local inhibitory hormone signals integrated into MAPK cascades?
Constitutive triple response 1 (CTR1) MAPKKK-mediated MAPK (Mitogen activated protein kinase) signaling modules act as major signaling components in ethylene signal transduction (Kieber et al., 1993). A gain-of-function mutant in Arabidopsis, ctr1, exhibited a constitutively activated ethylene-related triple response, and ethylene or ACC treatment activated two specific MAPK proteins in plants (Cardinale et al., 2002; Yoo et al., 2008). In Medicago sativa, specific MAPKs, such as MMK3 (AtMPK13), SAMK (Stress activated MAPK, AtMPK3), and SIMK (Salt stress induced MAPK, AtMPK6), were rapidly activated by ACC treatment via SIMKK (Salt stress induced MAPKK, AtMKK4/5) (Cardinale et al., 2002; Kiegerl et al., 2000). Overexpression of SIMKK phenocopied the ctr1-like constitutive ethylene response phenotype and enhanced the expression of ethylene-responsive genes. Furthermore, the SIMKK (AtMKK4/5)-SIMK (AtMPK6) module also plays an essential role in ethylene biosynthesis and stress responses in other plants (Kim et al., 2003; Liu and Zhang, 2004). It is therefore likely that CTR1-mediated MAPK signaling cascades mediate the local inhibitory regulation of nodulation during ethylene signaling in legumes (Fig. 2B). In addition to ethylene signaling, MAPK signaling cascades are implicated in MAMP (Microbial associated molecular pattern)-triggered immunity (MTI) and diverse stress responses in plants (Asai et al., 2002). Inoculation of symbiotic rhizobia could effectively activate MPK3/6 and subsequently enhance the expression of defense/stress-related genes in legumes (Duzan et al., 2004; Lopez-Gomez et al., 2011). Ethylene biosynthesis was also increased in response to rhizobia inoculation in L. japonicus (Lopez-Gomezet al., 2011; Sandal et al., 2002), but these defense/stress-related responses rapidly disappeared during nodulation (Lohar et al., 2006). However, inoculation of non-compatible rhizobia strains, or a rhizobia nodC mutant that is impaired in Nod factor biosynthesis, induces SA accumulation and diverse stress responses in legume roots (Stacey et al., 2006). It is possible that the symbiotic rhizobia could induce plant defense responses through MAPK cascades or ethylene signaling at the initial stage of interaction, but unidentified signals from the symbiotic bacteria or plant-microbe interactions might suppress the plant defense response to allow invasion of the rhizobia. Consistent with this idea, MAPK signaling stimulators, such as abiotic stresses, and a bacterial MAMP, flg22, greatly reduced nodule formation (Duzan et al., 2004; Lopez-Gomez et al., 2011; Sandal et al., 2002). A recent report shows that LjSYMRK directly interacts with SymRK-interacting protein 2 (SIP2), a SIMKK ortholog, and inhibits the kinase activity of SIP2 for MPK6 activation in L. japonicus (Chen et al., 2012). SIP2 and its homologous MKKs are central signaling components for defense, ethylene responses, and abiotic stress responses (Ronald and Beutler, 2010; Sinha et al., 2011; Zhang and Klessig, 2001). Together, SYMRK-mediated Nod factor signaling might suppress diverse defense and stress responses induced by the SIP2-mediated MAPK signaling cascade upon rhizobia infection; however, SIP2 knockdown transgenic hairy roots were defective in nodulation and infection thread formation, which indicates that SIP2 acts as a positive regulator of nodulation. A gain-of-function study of MAPK cascades during nodulation could elucidate the role of MAPK signaling and cross-talk with Nod factor signaling.
Ethylene, JA, SA, and ABA are involved in plant responses to pathogens, wounding, and various abiotic stresses, and are known to stimulate MAPK signaling cascades. In turn, MAPK signaling pathways are probably coordinated with Nod factor signaling in legumes to optimize nodulation during the symbiotic interaction (Fig. 2B). More detailed studies on signaling interplays of stress-related hormones, MAPKs and Nod factors will be necessary to understand the negative regulation of nodulation.
Positive roles for hormonal regulation of nodulation: the essential roles of cytokinin in nodule organogenesis
As we expected, given the complexity of signlaing networks in nodule organogenesis, not only negative action in nitorgen-fixing nodulation, but some phytohormones including cytokinins, auxin, GAs, BRs and strigolactone are also positively involved in the nodule organogenesis and development.
Cytokinin is a key signaling molecule for nodule organogenesis in symbiotic interactions between leguminous plants and rhizobia. The earliest evidence for cytokinin action in nodulation was the morphogenic rescue of nodule formation in Medicago sativa with nonsymbiotic bacteria carrying the isopentenyl transferase (IPT) gene of Agrobacterium tumefaciens (Cooper and Long, 1994). Moreover, a cytokinin primary responsive type A-RR (response regulator) is activated in nodule primordial cells in the cortex layer during nodulation, and overexpression of cytokinin-degrading cytokinin oxidase (CKX) decreases nodule organogenesis in L. japonicus (Lohar et al., 2004). Silencing 3-hydroxy-3-methylglutaryl CoA reductase 1 (HMGR1), a gene encoding an enzyme involved in the synthesis of the cytokinin precursor isoprenoid, reduces the number of nodules in Medicago truncatula roots (Kevei et al., 2007).
Suppression of MtCRE1, a highly conserved gene for the Arabidopsis cytokinin receptor Cytokinin resistant 1 (CRE1)/Arabidopsis histidine kinase 4 (AHK4), resulted in severely defective nodulation in Medicago truncatula (Gonzalez-Rizzo et al., 2006). Loss-of-function Lotus histidine kinase 1 (LHK1), a cytokinin receptor of L. japonicus, designated as hit1, completely abolished nodule primodial development, but did not affect infection thread formation enabling rhizobia invasion (Murray et al., 2007). By contrast, Spontaneous nodule formation 2 (snf2), a gain-of-function mutant of LHK1, spontaneously developed root nodules in spite of the absence of rhizobia inoculation, and it exhibited constitutively activated cytokinin responses (Tirichine et al., 2007). The activation of cytokinin signaling is deemed as necessary to initiate nodule organogenesis.
Cytokinin signaling is integrated into Nod factor signaling pathways
The manner in which cytokinin and Nod factor signaling pathways are integrated for nodule development has been the subject of considerable research. The expression of a critical nodulation gene, NIN, was induced by both cytokinin and Nod factor perception (Gonzalez-Rizzo et al., 2006; Murray et al., 2007), but its expression was completely blocked in the loss-of-function mutants Mtcre1 and Ljhit1 (Plet et al., 2011), whereas expression was up-regulated in the constitutively activated snf2 mutant (Murray et al., 2007; Plet et al., 2011; Tirichine et al., 2007; Wasson et al., 2011). Furthermore, the snf2 mutation rescued the aborted nodulation phenotype of loss-of-function mutants of Nod factor receptors, nfr1 and nfr5, and a symbiosis related LRR-RLK mutant, symrk. However, snf2 could not rescue the impaired nodulation phenotype of the nin and nsp2 loss-of-function mutants. Therefore, cytokinin signaling is likely to be regulated by NFRs and integrated into transcriptional regulation of NIN and NSP2 during nodule organogenesis (Sandal et al., 2007; Tirichine et al., 2007). The long-distance crosstalk networks between the infection signaling pathways in the epidermis and the cytokinin-mediated organogenic pathways in the cortex still remain to be elucidated.
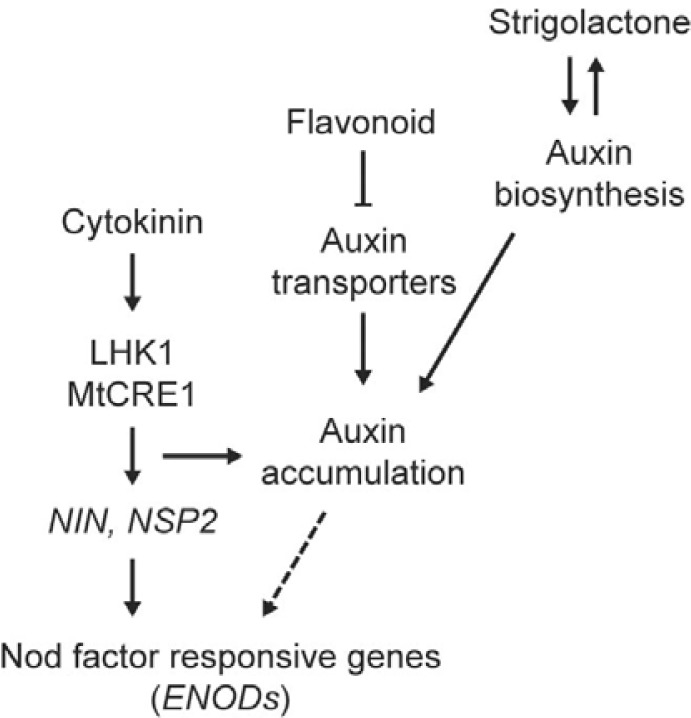
Cytokinin signaling interacts with other hormones such as ethylene and auxin during nodule organogensis. The skl/cre1 double mutant reduced root nodule numbers compared with the wild type, but had more nodules than cre1 (Plet et al., 2011; Wasson et al., 2011). The cre1 mutation restored the skl irregular nodulation pattern to the wild-type nodulation pattern. Cytokinin signaling functions in parallel with ethylene-mediated local inhibitory signaling pathways to control nodule numbers, but integrates with ethylene signaling to determine the pattern of nodule formation. Interestingly, MtCRE1-mediated cytokinin signaling induced the expression of a subset of MtPINs, leading to local auxin accumulation during initiation of nodule primodia (Plet et al., 2011; Wasson et al., 2011). Nod factors also induced auxin accumulation by inhibiting polar auxin transport in dividing pericycle and cortex cells, which is critical for nodule initiation (Hirsch et al., 1989; Pellerone et al., 2006; Plet et al., 2011; Wasson et al., 2011). Although it is still unclear whether Nod factors or flavonoids induced by the bacteria control cytokinin signaling for auxin accumulation, local auxin accumulation induced by cytokinin in primordial dividing cells is a critical event for nodule organogenesis (Fig. 3).
Fig. 3.
Positive regulation of nodulation through the action of plant hormones. The interplay of auxin and cytokinin plays a major role in nodule organogenesis. Cytokinins positively regulate nodule-related NSP2 and NIN expression, and expression of PINs that lead to local auxin accumulation. The local accumulation of auxin during the initiation of nodule primordia is also promoted by cooperation between flavonoids and strigolactone. The dashed arrows indicate unknown biochemical links.
Local Auxin accumulation is important for nodule organogenesis
During plant organogenesis, the balance of auxin and cytokinin signaling is critical for generating secondary organs and maintaining the meristematic activity of plant stem cells (Stahl and Simon, 2010). The tight regulation of auxin accumulation is necessary and sufficient to initiate organ formation (Benkova et al., 2003). The first evidence of auxin-dependent nodule organogenesis in legumes was the observation that auxin transport inhibitors could induce pseudo-nodule formation in Medicago sativa without rhizobia or Nod factor inoculation, and auxin transport inhibitors also activated the expression of early nodule-related ENOD2 (Bhuvaneswari et al., 1989; Hirsch et al., 1989). Activation of the auxin-responsive reporter, GH3, was observed in nodule primordial cells during symbiotic interaction with rhizobia (Mathesius et al., 1998). Furthermore, inoculation of rhizobia harboring an auxin biosynthetic enzyme increased the nodule number compared to inoculation of wild-type rhizobia (Pii et al., 2007). Therefore, it seems that local auxin accumulation is a prerequisite for nodule primordia initiation, and auxin signaling pathways function in parallel, or downstream of, the Nod factor-mediated signaling pathway.
Symbiotic rhizobia dependent flavonoid production in legumes is a primary regulatory step for local auxin accumulation during early nodulation processes. A molecular mechanism underlying flavonoid-mediated auxin accumulation and nodule formation has been proposed in a genetic study of Arabidopsis (Peer et al., 2004; Taylor and Grotewold, 2005). Flavonoid biosynthetic mutants exhibited an increased acropetal transport of auxin in roots compared with the wild type, and auxin transport was completely blocked by exogenous application of flavonoids in Arabidopsis. Flavonoids could bind to the auxin transporters AtMDR (Multi-drug resistance), AtAPM (Aminopeptidase M1), and AtPINs (Pin-formed) (Peer et al., 2004), and prevent their intracellular trafficking (Taylor and Grotewold, 2005), suggesting that rhizobia-induced flavonoids might regulate local auxin accumulation in legume roots in early nodulation. Silencing MtCHS (Chalcone synthase) and GmIFS (Isoflavone synthase) that encode key enzymes for flavonoid production dramatically inhibited nodulation in host legume plants (Pellerone et al., 2006; Subramanian et al., 2006; Wasson et al., 2006). The inhibition of polar auxin transport by rhizobia-mediated flavonoid biosynthesis and the subsequent accumulation of auxin in nodule initiation sites are early physiological events that induce cell differentiation in nodule primordia (Fig. 3). Despite the critical roles of auxin in plant organogenesis and differentiation, the way in which auxin signaling and local accumulation are integrated into Nod factor-dependent signaling modules is still largely unknown. The characterization of functional auxin transporters and signaling elements that induce local auxin accumulation during nodule initiation would help to decipher how auxin signaling regulates nitrogen fixing nodule development with diverse signal cues.
Gibberellin and strigolactone regulate nodule development
Gibberellins (GAs) are a class of growth-promoting hormones in plants. Early studies suggested that GAs play a positive role in nodulation. GAs, along with the biosynthetic oxidase gene, GA20, accumulate in developing nodules, bacterial invasion tracks, and pre-infection zones in the lima bean (Phaseolus lunatus) and cowpea (Vigna unguiculata) (Dobert et al., 1992; Lievens et al., 2005). Application of GA biosynthesis inhibitors causes severe nodulation defects in Sesbania (Sesbania rostrata) (Lievens et al., 2005). A GA-deficient pea mutant also showed decreased nodule numbers (Ferguson et al., 2005; Ross et al., 2005). However, recent genetic and pharmacological studies of GA during nodulation have revealed a negative effect of GA on nodule development by L. japonicus. Both overexpression and gain-of-function mutations of LjSLEEPY1, a positive regulator of GA signaling, significantly reduced nodule numbers (Maekawa et al., 2009). The root hair curling triggered by rhizobia was clearly inhibited by exogenous GA treatment. Furthermore, GA treatment decreased the expression of NSP2 and NIN during early nodulation processes. These results demonstrate that GA negatively regulates nodule formation by interfering with nodulation signaling pathways (Maekawa et al., 2009). These contradictory observations regarding GA effects indicate that there are some evolutionary differences (or alternative GA signaling networks) in nodulation in different legume species.
Strigolactone was initially identified as a host recognition signal for mycorrhizal fungi in AM symbiotic interactions (Yoneyama et al., 2008), and its biological role has been recently defined as that of a novel plant hormone involved in the inhibition of shoot branching (Gomez-Roldan et al., 2008; Umehara et al., 2008). Strigolactone also affects nodulation (Foo and Davies, 2011; Soto et al., 2010). Exogenous application of GR24, a synthetic strigolactone, increased the number of nodules in Medicago sativa, but did not affect symbiotic bacterial growth and flavonoid biosynthesis (Soto et al., 2010). The strigolactone-deficient pea rms1 mutant showed a significantly decreased number of nodules, and exogenous treatment with GR24 rescued the defects of nodule development in rms1 (Foo and Davies, 2011). Therefore, it is now evident that strigolactone acts as a positive regulator of nodule development; however, the molecular mechanisms underlying strigolactone action in nodulation are still obscure. A recent report shows that low nitrogen or phosphate conditions in soil stimulate strigolactone biosynthesis in plants (Xie et al., 2010). Strigolactone also promotes auxin biosynthesis and modulates auxin and cytokinin signaling pathways (Ferguson and Beveridge, 2009; Xie et al., 2010; Yoneyama et al., 2010). The availability of nitrogen in the rhizosphere is one of the main factors determining nodule numbers, and auxin and cytokinin are positively involved in nodule development. These observations lead us to speculate that the strigolactone biosynthesis and signaling pathways are closely connected to nutritional conditions, whereas other plant hormones control nodule numbers (Fig. 3).
CONCLUSIONS
Nodulation is strictly controlled by the host plant to ensure appropriate levels of nitrogen fixation without excessive depletion of photosynthetic products. The host leguminous plants negotiate with symbiotic bacterial partners to optimize the levels of nodulation during nitrogen fixing symbiotic interactions. The coordination of plant hormones is broadly and tightly associated with the control of nodule formation, which likely co-evolved with symbiotic partners to adjust the availability of nitrogen and to reduce the carbon drain to a minimum. Because sessile land plants strictly regulate their resources in response to environmental changes, it is reasonable to postulate that legumes utilize stress- and growth-related hormones as developmental cues to regulate nodulation.
It is clear that the coordination of hormonal signaling path-ways is essential for nodule organogenesis. However, the mechanisms of spatio-temporal cooperative regulation between Nod factor and hormone signaling pathways during symbiotic interactions are not clear. Although all legumes can produce nitrogen-fixing nodules with their symbiotic partners, the efficiency of nitrogen fixation can differ by more than 10-fold between legume species (Den Herder and Parniske, 2009). Regulation of nodule number is one of the critical factors causing this large difference in nitrogen fixing efficiency. Recently, the overuse of nitrogen fertilizers has caused many serious global problems such as soil acidification and dead zones in the ocean caused by overgrowth of algae. Understanding the mechanics and regulation of nodulation, and translating this knowledge to optimize nodule development through agricultural biotechnology, will enable significant progress toward resolving these global issues.
Acknowledgments
This work was supported by grants to I.H. from the Advanced Biomass R&D Center (2010-0029720) and National Research Foundation (20110000212) funded by the Korean Ministry of Education, Science and Technology. H.R. was supported by a grant from Next-Generation BioGreen 21 Program (PJ008039). H.C. was a recipient of a Brain Korea 21 fellowship.
REFERENCES
- Andriankaja A., Boisson-Dernier A., Frances L., Sauviac L., Jauneau A., Barker D.G., de Carvalho-Niebel F. AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell. 2007;19:2866–2885. doi: 10.1105/tpc.107.052944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ane J.M., Kiss G.B., Riely B.K., Penmetsa R.V., Oldroyd G.E., Ayax C., Levy J., Debelle F., Baek J.M., Kalo P., et al. Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science. 2004;303:1364–1367. doi: 10.1126/science.1092986. [DOI] [PubMed] [Google Scholar]
- Arrighi J.F., Barre A., Ben Amor B., Bersoult A., Soriano L.C., Mirabella R., de Carvalho-Niebel F., Journet E.P., Gherardi M., Huguet T., et al. The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- Bao F., Shen J., Brady S.R., Muday G.K., Asami T., Yang Z. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 2004;134:1624–1631. doi: 10.1104/pp.103.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett M.J., Fisher R.F. Global gene expression in the rhizobial-legume symbiosis. Symbiosis. 2006;42:1–24. [Google Scholar]
- Ben Amor B., Shaw S.L., Oldroyd G.E.D., Maillet F., Penmetsa R.V., Cook D., Long S.R., Denarie J., Gough C. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 2003;34:495–506. doi: 10.1046/j.1365-313x.2003.01743.x. [DOI] [PubMed] [Google Scholar]
- Benkova E., Michniewicz M., Sauer M., Teichmann T., Seifertova D., Jurgens G., Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Biswas B., Chan P.K., Gresshoff P.M. A novel ABA insensitive mutant of Lotus japonicus with a wilty phenotype displays unaltered nodulation regulation. Mol Plant. 2009;2:487–499. doi: 10.1093/mp/ssp009. [DOI] [PubMed] [Google Scholar]
- Borisov A.Y., Madsen L.H., Tsyganov V.E., Umehara Y., Voroshilova V.A., Batagov A.O., Sandal N., Mortensen A., Schauser L., Ellis N., et al. The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol. 2003;131:1009–1017. doi: 10.1104/pp.102.016071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale F., Meskiene I., Ouaked F., Hirt H. Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell. 2002;14:703–711. [PMC free article] [PubMed] [Google Scholar]
- Catoira R., Galera C., de Billy F., Penmetsa R.V., Journet E.P., Maillet F., Rosenberg C., Cook D., Gough C., Denarie J. Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell. 2000;12:1647–1666. doi: 10.1105/tpc.12.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Zhu H., Ke D., Cai K., Wang C., Gou H., Hong Z., Zhang Z. A MAP kinase kinase interacts with SymRK and regulates nodule organogenesis in Lotus japonicus. Plant Cell. 2012;24:823–838. doi: 10.1105/tpc.112.095984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.B., Long S.R. Morphogenetic rescue of Rhizobium meliloti nodulation mutants by trans-zeatin secretion. Plant Cell. 1994;6:215–225. doi: 10.1105/tpc.6.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Herder G., Parniske M. The unbearable naivety of legumes in symbiosis. Curr. Opin. Plant Biol. 2009;12:491–499. doi: 10.1016/j.pbi.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Ding Y., Kalo P., Yendrek C., Sun J., Liang Y., Marsh J.F., Harris J.M., Oldroyd G.E. Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell. 2008;20:2681–2695. doi: 10.1105/tpc.108.061739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duzan H.M., Zhou X., Souleimanov A., Smith D.L. Perception of Bradyrhizobium japonicum Nod factor by soybean [Glycine max (L.) Merr.] root hairs under abiotic stress conditions. J. Exp. Bot. 2004;55:2641–2646. doi: 10.1093/jxb/erh265. [DOI] [PubMed] [Google Scholar]
- Endre G., Kereszt A., Kevei Z., Mihacea S., Kalo P., Kiss G.B. A receptor kinase gene regulating symbiotic nodule development. Nature. 2002;417:962–966. doi: 10.1038/nature00842. [DOI] [PubMed] [Google Scholar]
- Ferguson B.J., Beveridge C.A. Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiol. 2009;149:1929–1944. doi: 10.1104/pp.109.135475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B.J., Ross J.J., Reid J.B. Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol. 2005;138:2396–2405. doi: 10.1104/pp.105.062414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B.J., Indrasumunar A., Hayashi S., Lin M.-H., Lin Y.-H., Reid D.E. Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 2010;52:61–76. doi: 10.1111/j.1744-7909.2010.00899.x. [DOI] [PubMed] [Google Scholar]
- Foo E., Davies N.W. Strigolactones promote nodulation in pea. Planta. 2011;234:1073–1081. doi: 10.1007/s00425-011-1516-7. [DOI] [PubMed] [Google Scholar]
- Gleason C., Chaudhuri S., Yang T., Munoz A., Poovaiah B.W., Oldroyd G.E. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature. 2006;441:1149–1152. doi: 10.1038/nature04812. [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., Fermas S., Brewer P.B., Puech-Pages V., Dun E.A., Pillot J.P., Letisse F., Matusova R., Danoun S., Portais J.C., et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S., Crespi M., Frugier F. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell. 2006;18:2680–2693. doi: 10.1105/tpc.106.043778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann A.B., Lombardo F., Miwa H., Perry J.A., Bunnewell S., Parniske M., Wang T.L., Downie J.A. Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 2006;142:1739–1750. doi: 10.1104/pp.106.089508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch A., Bhuvaneswari T., Torrey J., Bisseling T. Early nodulin genes are induced in alfalfa root outgrowths elicited by auxin transport inhibitors. Proc. Natl. Acad. Sci USA. 1989;86:1244–1248. doi: 10.1073/pnas.86.4.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Munoz A., Heckmann A.B., Downie J.A., Oldroyd G.E. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell. 2009;21:545–557. doi: 10.1105/tpc.108.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalo P., Gleason C., Edwards A., Marsh J., Mitra R.M., Hirsch S., Jakab J., Sims S., Long S.R., Rogers J., et al. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science. 2005;308:1786–1789. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]
- Kawaguchi M., Minamisawa K. Plant-microbe communications for symbiosis. Plant Cell Physiol. 2010;51:1377–1380. doi: 10.1093/pcp/pcq125. [DOI] [PubMed] [Google Scholar]
- Kevei Z., Lougnon G., Mergaert P., Horvath G.V., Kereszt A., Jayaraman D., Zaman N., Marcel F., Regulski K., Kiss G.B., et al. 3-hydroxy-3-methylglutaryl coenzyme A reductase1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell. 2007;19:3974–3989. doi: 10.1105/tpc.107.053975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kiegerl S., Cardinale F., Siligan C., Gross A., Baudouin E., Liwosz A., Eklof S., Till S., Bogre L., Hirt H., et al. SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell. 2000;12:2247–2258. doi: 10.1105/tpc.12.11.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.Y., Liu Y., Thorne E.T., Yang H., Fukushige H., Gassmann W., Hildebrand D., Sharp R.E., Zhang S. Activation of a stress-responsive mitogen-activated protein kinase cascade induces the biosynthesis of ethylene in plants. Plant Cell. 2003;15:2707–2718. doi: 10.1105/tpc.011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M., Gresshoff P.M. Investigation of downstream signals of the soybean autoregulation of nodulation receptor kinase GmNARK. Mol. Plant Microbe Interact. 2008;21:1337–1348. doi: 10.1094/MPMI-21-10-1337. [DOI] [PubMed] [Google Scholar]
- Kouchi H., Imaizumi-Anraku H., Hayashi M., Hakoyama T., Nakagawa T., Umehara Y., Suganuma N., Kawaguchi M. How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 2010;51:1381–1397. doi: 10.1093/pcp/pcq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L., Madsen L.H., Sato S., Aubert G., Genua A., Szczyglowski K., Duc G., Kaneko T., Tabata S., de Bruijn F., et al. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature. 2002;420:422–426. doi: 10.1038/nature01207. [DOI] [PubMed] [Google Scholar]
- Levy J., Bres C., Geurts R., Chalhoub B., Kulikova O., Duc G., Journet E.P., Ane J.M., Lauber E., Bisseling T., et al. A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science. 2004;303:1361–1364. doi: 10.1126/science.1093038. [DOI] [PubMed] [Google Scholar]
- Lievens S., Goormachtig S., Den Herder J., Capoen W., Mathis R., Hedden P., Holsters M. Gibberellins are involved in nodulation of Sesbania rostrata. Plant Physiol. 2005;139:1366–1379. doi: 10.1104/pp.105.066944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E., Franken C., Smit P., Willemse J., Bisseling T., Geurts R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell. 2004;16:3386–3399. doi: 10.1105/tpc.104.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohar D., Schaff J., Laskey J., Kieber J., Bilyeu K., Bird D. Cytokinins play opposite roles in lateral root formation, and nematode and rhizobial symbioses. Plant J. 2004;38:203–214. doi: 10.1111/j.1365-313X.2004.02038.x. [DOI] [PubMed] [Google Scholar]
- Lohar D.P., Sharopova N., Endre G., Penuela S., Samac D., Town C., Silverstein K.A., VandenBosch K.A. Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol. 2006;140:221–234. doi: 10.1104/pp.105.070326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gomez M., Sandal N., Stougaard J., Boller T. Interplay of flg22-induced defence responses and nodulation in Lotus japonicus. J. Exp. Bot. 2011;63:393–401. doi: 10.1093/jxb/err291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sanchez-Serrano J.J., Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15:165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lough T.J., Lucas W.J. Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 2006;57:203–232. doi: 10.1146/annurev.arplant.56.032604.144145. [DOI] [PubMed] [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- Maekawa T., Maekawa-Yoshikawa M., Takeda N., Imaizumi-Anraku H., Murooka Y., Hayashi M. Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 2009;58:183–194. doi: 10.1111/j.1365-313X.2008.03774.x. [DOI] [PubMed] [Google Scholar]
- Magori S., Oka-Kira E., Shibata S., Umehara Y., Kouchi H., Hase Y., Tanaka A., Sato S., Tabata S., Kawaguchi M. Too much love, a root regulator associated with the long-distance control of nodulation in Lotus japonicus. Mol. Plant Microbe Int. 2009;22:259–268. doi: 10.1094/MPMI-22-3-0259. [DOI] [PubMed] [Google Scholar]
- Markmann K., Parniske M. Evolution of root endosymbiosis with bacteria: How novel are nodules? Trends Plant Sci. 2009;14:77–86. doi: 10.1016/j.tplants.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Marsh J.F., Rakocevic A., Mitra R.M., Brocard L., Sun J., Eschstruth A., Long S.R., Schultze M., Ratet P., Oldroyd G.E. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007;144:324–335. doi: 10.1104/pp.106.093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U., Schlaman H.R., Spaink H.P., Of Sautter C., Rolfe B.G., Djordjevic M.A. Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 1998;14:23–34. doi: 10.1046/j.1365-313X.1998.00090.x. [DOI] [PubMed] [Google Scholar]
- Mellor R.B., Collinge D.B. A simple-model based on known plant defense reactions is sufficient to explain most aspects of nodulation. J. Exp. Bot. 1995;46:1–18. [Google Scholar]
- Middleton P.H., Jakab J., Penmetsa R.V., Starker C.G., Doll J., Kalo P., Prabhu R., Marsh J.F., Mitra R.M., Kereszt A., et al. An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell. 2007;19:1221–1234. doi: 10.1105/tpc.106.048264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R.M., Gleason C.A., Edwards A., Hadfield J., Downie J.A., Oldroyd G.E., Long S.R. A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci USA. 2004;101:4701–4705. doi: 10.1073/pnas.0400595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V., Den Herder G., Whitford R., Van de Velde W., Rombauts S., D’Haeseleer K., Holsters M., Goormachtig S. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 2010;153:222–237. doi: 10.1104/pp.110.153718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Miwa H., Imaizumi-Anraku H., Kouchi H., Downie J.A., Kawaguchi M., Kawasaki S. Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res. 2006;13:255–265. doi: 10.1093/dnares/dsl017. [DOI] [PubMed] [Google Scholar]
- Murray J.D., Karas B.J., Sato S., Tabata S., Amyot L., Szczyglowski K. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science. 2007;315:101–104. doi: 10.1126/science.1132514. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kawaguchi M. Shoot-applied MeJA suppresses root nodulation in Lotus japonicus. Plant Cell Physiol. 2006;47:176–180. doi: 10.1093/pcp/pci222. [DOI] [PubMed] [Google Scholar]
- Nakamura A., Goda H., Shimada Y., Yoshida S. Brassinosteroid selectively regulates PIN gene expression in Arabidopsis. Biosci. Biotechnol. Biochem. 2004;68:952–954. doi: 10.1271/bbb.68.952. [DOI] [PubMed] [Google Scholar]
- Nishimura R., Hayashi M., Wu G.J., Kouchi H., Imaizumi-Anraku H., Murakami Y., Kawasaki S., Akao S., Ohmori M., Nagasawa M., et al. HAR1 mediates systemic regulation of symbiotic organ development. Nature. 2002;420:426–429. doi: 10.1038/nature01231. [DOI] [PubMed] [Google Scholar]
- Nontachaiyapoom S., Scott P.T., Men A.E., Kinkema M., Schenk P.M., Gresshoff P.M. Promoters of orthologous Glycine max and Lotus japonicus nodulation autoregulation genes interchangeably drive phloem-specific expression in transgenic plants. Mol. Plant Microbe Int. 2007;20:769–780. doi: 10.1094/MPMI-20-7-0769. [DOI] [PubMed] [Google Scholar]
- Nutman P.S. Host-factors influencing infection and nodule development in leguminous plants. Proc. R Soc. Lond. B Biol. Sci. 1952;139:176–185. doi: 10.1098/rspb.1952.0003. discussion 202–177. [DOI] [PubMed] [Google Scholar]
- Oka-Kira E., Kawaguchi M. Long-distance signaling to control root nodule number. Curr. Opin. Plant Biol. 2006;9:496–502. doi: 10.1016/j.pbi.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Oka-Kira E., Tateno K., Miura K., Haga T., Hayashi M., Harada K., Sato S., Tabata S., Shikazono N., Tanaka A., et al. klavier (klv), a novel hypernodulation mutant of Lotus japonicus affected in vascular tissue organization and floral induction. Plant J. 2005;44:505–515. doi: 10.1111/j.1365-313X.2005.02543.x. [DOI] [PubMed] [Google Scholar]
- Okamoto S., Ohnishi E., Sato S., Takahashi H., Nakazono M., Tabata S., Kawaguchi M. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 2009;50:67–77. doi: 10.1093/pcp/pcn194. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Downie J.A. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Engstrom E.M., Long S.R. Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell. 2001;13:1835–1849. doi: 10.1105/TPC.010193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacios-Bras C., Schlaman H.R., Boot K., Admiraal P., Langerak J.M., Stougaard J., Spaink H.P. Auxin distribution in Lotus japonicus during root nodule development. Plant Mol. Biol. 2003;52:1169–1180. doi: 10.1023/b:plan.0000004308.78057.f5. [DOI] [PubMed] [Google Scholar]
- Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- Peer W.A., Bandyopadhyay A., Blakeslee J.J., Makam S.N., Chen R.J., Masson P.H., Murphy A.S. Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell. 2004;16:1898–1911. doi: 10.1105/tpc.021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa R.V., Cook D.R. A legume ethylene-Insensitive mutant hyperinfected by its rhizobial symbiont. Science. 1997;275:527–530. doi: 10.1126/science.275.5299.527. [DOI] [PubMed] [Google Scholar]
- Penmetsa R.V., Uribe P., Anderson J., Lichtenzveig J., Gish J.C., Nam Y.W., Engstrom E., Xu K., Sckisel G., Pereira M., et al. The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J. 2008;55:580–595. doi: 10.1111/j.1365-313X.2008.03531.x. [DOI] [PubMed] [Google Scholar]
- Perret X., Staehelin C., Broughton W.J. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 2000;64:180–201. doi: 10.1128/mmbr.64.1.180-201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pii Y., Crimi M., Cremonese G., Spena A., Pandolfini T. Auxin and nitric oxide control indeterminate nodule formation. BMC Plant Biol. 2007;7:21–31. doi: 10.1186/1471-2229-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plet J., Wasson A., Ariel F., Le Signor C., Baker D., Mathesius U., Crespi M., Frugier F. MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 2011;65:622–633. doi: 10.1111/j.1365-313X.2010.04447.x. [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Gronlund M., Sato S., Nakamura Y., Tabata S., Sandal N., et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Jurkiewicz A., Fukai E., Quistgaard E.M., Albrektsen A.S., James E.K., Thirup S., Stougaard J. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007;26:3923–3935. doi: 10.1038/sj.emboj.7601826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald P.C., Beutler B. Plant and animal sensors of conserved microbial signatures. Science. 2010;330:1061–1064. doi: 10.1126/science.1189468. [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano R., Xoconostle-Cazares B., Lucas W.J. The phloem as a conduit for inter-organ communication. Curr. Opin. Plant Biol. 2001;4:202–209. doi: 10.1016/s1369-5266(00)00162-x. [DOI] [PubMed] [Google Scholar]
- Sandal N., Krusell L., Radutoiu S., Olbryt M., Pedrosa A., Stracke S., Sato S., Kato T., Tabata S., Parniske M., et al. A genetic linkage map of the model legume Lotus japonicus and strategies for fast mapping of new loci. Genetics. 2002;161:1673–1683. doi: 10.1093/genetics/161.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R., Herouart D., Sigaud S., Touati D., Puppo A. Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol. Plant Microbe Int. 2001;14:86–89. doi: 10.1094/MPMI.2001.14.1.86. [DOI] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- Searle I.R., Men A.E., Laniya T.S., Buzas D.M., Iturbe-Ormaetxe I., Carroll B.J., Gresshoff P.M. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science. 2003;299:109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- Sinha A.K., Jaggi M., Raghuram B., Tuteja N. Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 2011;6:196–203. doi: 10.4161/psb.6.2.14701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debelle F., Gough C., Bisseling T., Geurts R. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308:1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- Soto M.J., Fernandez-Aparicio M., Castellanos-Morales V., Garcia-Garrido J.M., Ocampo J.A., Delgado M.J., Vierheilig H. First indications for the involvement of strigolactones on nodule formation in alfalfa (Medicago sativa) Soil Biol. Biochem. 2010;42:383–385. [Google Scholar]
- Stacey G., McAlvin C.B., Kim S.Y., Olivares J., Soto M.J. Effects of endogenous salicylic acid on nodulation in the model legumes Lotus japonicus and Medicago truncatula. Plant Physiol. 2006;141:1473–1481. doi: 10.1104/pp.106.080986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y., Simon R. Plant primary meristems: shared functions and regulatory mechanisms. Curr. Opin. Plant Biol. 2010;13:53–58. doi: 10.1016/j.pbi.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Szczyglowski K., et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417:959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- Subramanian S., Stacey G., Yu O. Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J. 2006;48:261–273. doi: 10.1111/j.1365-313X.2006.02874.x. [DOI] [PubMed] [Google Scholar]
- Sun J., Cardoza V., Mitchell D.M., Bright L., Oldroyd G., Harris J.M. Crosstalk between jasmonic acid, ethylene and Nod factor signaling allows integration of diverse inputs for regulation of nodulation. Plant J. 2006;46:961–970. doi: 10.1111/j.1365-313X.2006.02751.x. [DOI] [PubMed] [Google Scholar]
- Suzuki A., Akune M., Kogiso M., Imagama Y., Osuki K., Uchiumi T., Higashi S., Han S.Y., Yoshida S., Asami T., et al. Control of nodule number by the phytohormone abscisic acid in the roots of two leguminous species. Plant Cell Physiol. 2004;45:914–922. doi: 10.1093/pcp/pch107. [DOI] [PubMed] [Google Scholar]
- Symons G.M., Reid J.B. Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol. 2004;135:2196–2206. doi: 10.1104/pp.104.043034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L.P., Grotewold E. Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 2005;8:317–323. doi: 10.1016/j.pbi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Terakado J., Yoneyama T., Fujihara S. Shoot-applied polyamines suppress nodule formation in soybean (Glycine max) J. Plant Physiol. 2006;163:497–505. doi: 10.1016/j.jplph.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Tirichine L., Imaizumi-Anraku H., Yoshida S., Murakami Y., Madsen L.H., Miwa H., Nakagawa T., Sandal N., Albrektsen A.S., Kawaguchi M., et al. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature. 2006;441:1153–1156. doi: 10.1038/nature04862. [DOI] [PubMed] [Google Scholar]
- Tirichine L., Sandal N., Madsen L.H., Radutoiu S., Albrektsen A.S., Sato S., Asamizu E., Tabata S., Stougaard J. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science. 2007;315:104–107. doi: 10.1126/science.1132397. [DOI] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455:195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- van Noorden G.E., Ross J.J., Reid J.B., Rolfe B.G., Mathesius U. Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol. 2006;140:1494–1506. doi: 10.1104/pp.105.075879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noorden G.E., Kerim T., Goffard N., Wiblin R., Pellerone F.I., Rolfe B.G., Mathesius U. Overlap of proteome changes in Medicago truncatula in response to auxin and Sinorhizobium meliloti. Plant Physiol. 2007;144:1115–1131. doi: 10.1104/pp.107.099978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernie T., Moreau S., de Billy F., Plet J., Combier J.P., Rogers C., Oldroyd G., Frugier F., Niebel A., Gamas P. EFD Is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. Plant Cell. 2008;20:2696–2713. doi: 10.1105/tpc.108.059857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson A.P., Pellerone F.I., Mathesius U. Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell. 2006;18:1617–1629. doi: 10.1105/tpc.105.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X.N., Yoneyama K., Yoneyama K. The strigolactone story. Ann. Rev. Phytopathol. 2010;48:93–117. doi: 10.1146/annurev-phyto-073009-114453. [DOI] [PubMed] [Google Scholar]
- Yano K., Yoshida S., Muller J., Singh S., Banba M., Vickers K., Markmann K., White C., Schuller B., Sato S., et al. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci USA. 2008;105:20540–20545. doi: 10.1073/pnas.0806858105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K., Xie X., Sekimoto H., Takeuchi Y., Ogasawara S., Akiyama K., Hayashi H., Yoneyama K. Strigolactones, host recognition signals for root parasitic plants and arbuscular mycorrhizal fungi, from Fabaceae plants. New Phytol. 2008;179:484–494. doi: 10.1111/j.1469-8137.2008.02462.x. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Tena G., Xiong Y., Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature. 2008;451:789–795. doi: 10.1038/nature06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. MAPK cascades in plant defense signaling. Trends Plant Sci. 2001;6:520–527. doi: 10.1016/s1360-1385(01)02103-3. [DOI] [PubMed] [Google Scholar]