Abstract
We previously showed that silencing of NbBPS1 encoding an endoplasmic reticulum (ER)-localized protein results in pleiotrophic developmental defects and cell death in Nicotiana benthamiana [Kang et al. (2008)]. In this study, we investigated the mechanism of the cell death caused by NbBPS1 silencing. Affected leaf cells exhibited morphological markers of programmed cell death (PCD) and accumulated excessive amounts of reactive oxygen species. NbBPS1 silencing caused dramatic induction of the ER stress marker genes BiP-like protein (BLP) genes, HSP70, and Bax Inhibitor-1. Furthermore, NbBPS1 deficiency led to relocalization of bZIP28 transcription factor from the ER membrane to the nucleus, similar to the bZIP28 relocalization during tunicamycin-induced ER stress. Abnormal accumulation of vesicles and increased autophagy activity were also observed in the affected leaf cells. These results suggest that inactivation of NbBPS1 function in the ER leads to ER stress, autophagy, and PCD activation in N. benthamiana.
Keywords: autophagy, bZIP28 transcription factor, ER stress, programmed cell death, vesicles, virus-induced gene silencing
INTRODUCTION
Programmed cell death (PCD) is an active process associated with distinctive morphological and biochemical features. The process is essential for development and survival of multicellular organisms (Vaux and Korsmeyer, 1999; Wertz and Hanley, 1996). In plants, PCD is integral in development of xylem tracheary elements, leaf senescence, and the hypersensitive response to pathogen attack (Coll et al., 2011; Lam et al., 2001; Reape and McCabe, 2008). Environmental stresses such as nutrient deficiency, high salt, and drought also induce PCD in plants, suggesting that the process may confer resistance to stress. Recent studies have reported that endoplasmic reticulum (ER)-mediated stress can induce apoptotic pathways in animals and plants and that the ER stress-induced PCD is involved in plant responses to biotic and abiotic stresses (Costa et al., 2008; Szegezdi et al., 2006; Watanabe and Lam, 2006; 2008).
Protein folding in the ER is highly regulated under quality control surveillance. Nonetheless, inadequate nutrition, pathogen infection, hypoxia, changes in ER luminal Ca++ stores, toxic chemicals, and mutations disrupt protein folding reactions in the ER, leading to accumulation of unfolded proteins and protein aggregates. These conditions are detrimental to cell survival, and are called ER stress. To counteract ER stress, the cell activates the unfolded protein response (UPR), ER-associated degradation, and Ca++ signaling (Boyce and Yuan, 2006; Høyer-Hansen and Jäättelä, 2007; Kaufman et al., 2002; Schröder and Kaufman, 2005). These pathways eventually reduce the ER load of unfolded proteins by decreasing global translation, increasing protein folding capacity in the ER, and stimulating degradation of misfolded proteins by the 26S proteasome or autophagy.
When ER stress is excessive or sustained, these signaling pathways can activate apoptosis in mammals (Li et al., 2006; Shiraishi et al., 2006; Szegezdi et al., 2006). The ER stress also activates autophagy, which functions to mitigate ER stress by removing aggregates and suppressing cell death, although excessive autophagy may also promote cell death (Ding and Yin, 2008; Høyer-Hansen and Jäättelä, 2007; Sakaki and Kaufman, 2008). In plants, tunicamycin (an inhibitor of N-linked protein glycosylation), cyclopiazonic acid (an inhibitor of ER calcium pumps), and water stress in roots elicit the ER stress and PCD (Crosti et al., 2001; Duan et al., 2010; Watanabe and Lam, 2008; Zuppini et al., 2004), but the mechanisms of the UPR pathway and the ER stress-induced PCD and autophagy are not well understood in plants.
Arabidopsis BYPASS1 (BPS1) encodes a protein with no homology to any characterized proteins (Van Norman et al., 2004). The bypass1 (bps1) mutant was originally identified by a screen for altered leaf vein patterns, and the mutant exhibited severe defects in shoot and root growth (Van Norman et al., 2004). In a previous study, we examined the function of Nicotiana benthamiana BPS1 (NbBPS1) in plant growth and development by studying the cellular effects of NbBPS1 depletion in N. benthamiana (Kang et al., 2008b). The NbBPS1:GFP fusion protein was mainly associated with the endoplasmic reticulum. Virus-induced gene silencing (VIGS) of NbBPS1 resulted in growth retardation, abnormal leaf development, and cell death (Kang et al., 2008b). The VIGS plants exhibited hyperproliferation of the cambial cells in the stem and elevated endoreduplication in the leaves. Cell death in NbBPS1 VIGS leaves involved vacuole collapse followed by degeneration of the organelles (Kang et al., 2008b). These results suggest an essential function of NbBPS1 in plant growth and development. In this study, we investigated the mechanism of cell death induced by NbBPS1-silencing in N. benthamiana. From our results, we suggest that NbBPS1 deficiency activated ER stress and PCD at the cellular level.
MATERIALS AND METHODS
Virus-induced gene silencing
Virus-induced gene silencing of NbBPS1 was performed as described previously (Kang et al., 2008b).
Realtime quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis
Realtime quantitative RT-PCR was performed with RNA isolated from the fourth leaf above the infiltrated leaf as described (Ahn et al., 2011). To detect BLPs, HSP70, and Bax Inhibitor-1 (BI1) transcripts, the following primers were designed, based on the published tobacco and N. benthamiana cDNA sequences: BLP1 (X60060; 5′-aacaccgtcatcccaacc-3′ and 5′-tggtctgctggtcttggtaa-3′), BLP2 (X60059; 5′-atcctgatgaggcagttgct-3′ and 5′-cctctccgctaaggatacca-3′), BLP3 (X60061; 5′-gagacagccgtaaaggaagc-3′ and 5′-ctctttctcagcgctctggt-3′), BLP4 (X60057; 5′-ttctttccacaaatggagacac-3′ and 5′-tccataatcctctggtcaaagtc-3′), BLP5 (X60058; 5′-tgtgtatcagagatctggtggag-3′ and 5′-tcatcctcgttggattcctc-3′), BI1 (AF390556; 5′-gaagaatgcatccgacaagg-3′ and 5′-ccgcttatgcattagtttctcc-3′), HSP70 (GQ354819; 5′-ggattcactgatactgagcgtct-3′ and 5′-ttgatagggttcatagctacctga-3′), and α-tubulin (AJ421411; 5′-atggcttgctgccttatgtt-3′ and 5′-cacagcagcattgacatcct-3′).
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay
TUNEL assay was performed as described (Ahn et al., 2011). Leaves from TRV control and TRV:BPS1(C) VIGS plants were fixed and treated with DNase-free RNase A (30 μg/ml; Promega) and Proteinase K (20 μg/ml) for 5 min before the TUNEL assay. The assay was carried out according the manufacturer’s manual (Promega) and counterstained with DAPI solution (0.1 μg/ml DAPI, 50 mM NaCl, 5 mM EDTA, 10 mM mercaptoethylamine, and 10 mM Tris-HCl [pH 7.4]) for 5–10 min before examination with confocal microscopy (Zeiss LSM510). For a positive control, leaves from the TRV plants were treated with RQ1 RNase-Free DNase (Promega) for 15 min at 37°C before TUNEL assay according the manual (Promega).
Measurement of in vivo H2O2 levels
H2DCFDA staining and fluorescence measurement were carried out as described previously (Kim et al., 2006).
Agrobacterium-mediated transient expression of GFP:bzip28
The GFP:bZIP28 fusion construct between GFP and Arabidopsis bZIP28 (At3g10800) under the control of the CaMV35S promoter was generated as described (Liu et al., 2007). Agroinfiltration was carried out as described (Voinnet et al., 2003). Agrobacterial cultures (GV3101) containing the GFP:bZIP28 construct fused to the CaMV35S promoter were adjusted to OD600 = 0.6 in 2-(N-morpholino)ethanesulfonic acid (MES) buffer. The suspension was incubated with acetosyringone for 2–3 h at a final concentration of 150 μM and infiltrated into leaves of TRV and TRV:BPS1(C) plants at 15 days after infiltration (DAI). Protoplasts were prepared from the infiltrated leaves collected from independent plants at 48 h post-infiltration and GFP:bZIP28 fluorescence localization was examined by fluorescence microscopy and confocal laser scanning microscopy.
Lysotracker staining and GFP:ATG8e expression
LysoTracker Red (LTR) fluorescence indicative of autophagy activity was examined by using confocal microscopy (Zeiss LSM510) as described (Liu et al., 2005). Leaf protoplasts isolated from VIGS plants were stained with 1 μM LTR (Molecular Probes) and kept for 1 min in darkness before visualization with confocal microscopy. Quantification of LTR-stained autophagosome-like structures was carried out using confocal microscopy.
The GFP:ATG8e fusion construct between GFP and Arabidopsis ATG8e (At2g45170) under the control of the CaMV35S promoter was generated as described (Ahn et al., 2011; Contento et al., 2005). Agrobacterial cultures (C58C1) containing the GFP:ATG8e construct were adjusted to OD600=0.6 in MES buffer (10 mM MES [pH 7.5], 10 mM MgSO4). The suspension was incubated with acetosyringone for 2–3 h at a final concentration of 150 μM and infiltrated into leaves of VIGS plants. Protoplasts were prepared from the infiltrated leaves at 48 h post-infiltration and localization of the GFP fluorescence was examined by using confocal microscopy.
Transmission electron microscopy
Tissue sectioning and transmission electron microscopy were carried out using the fourth or fifth leaf above the infiltrated leaf of TRV, TRV:BPS1(C), and TRV:BPS1(U) lines as described by Ahn et al. (2004).
RESULTS AND DISCUSSION
Cell death phenotypes of TRV:NbBPS1 VIGS plants
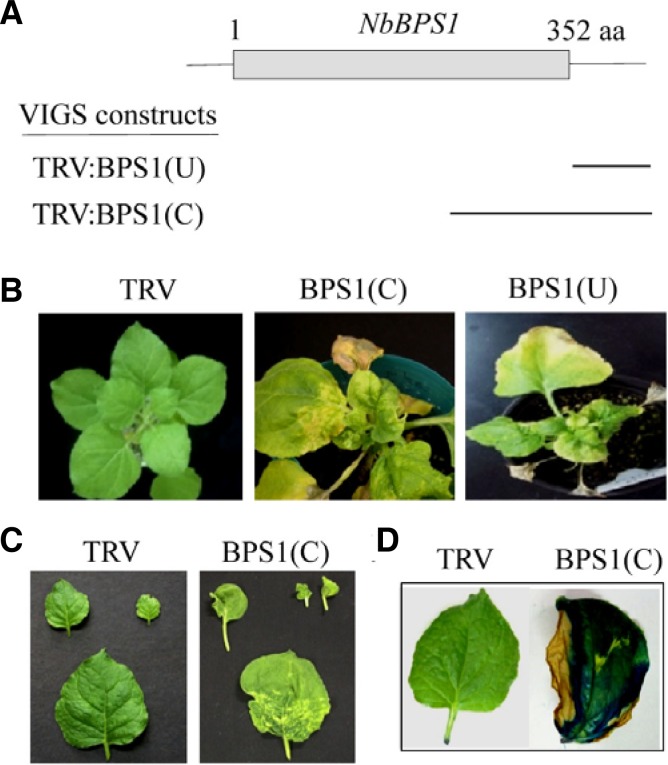
Silencing of NbBPS1 by VIGS with TRV:BPS1(U) and TRV: BPS1(C) constructs resulted in growth arrest, abnormal leaf development, and cell death (Figs. 1A–1C). Necrotic lesions developed in leaves at 15–16 days after infiltration (DAI), leading to premature plant death after 45 DAI (Fig. 1B). The intense blue pigment in Evans blue-stained leaves of TRV:BPS1(C) plants indicates localized cell death (Fig. 1D).
Fig. 1.
VIGS constructs, phenotypes, and suppression of the NbBPS1 transcripts. (A) Schematic drawing of NbBPS1 structure and two VIGS constructs, TRV:BPS1(U) and TRV:BPS1(C), containing different NbBPS1 cDNA fragments (indicated by bars). The gray box indicates the coding region of NbBPS1. (B) Cell death phenotypes of the NbBPS1 VIGS plants. Photographs of the plants were taken at 25 days after infiltration (DAI). (C) Abnormal leaf development and lesion formation in TRV:BPS1(C) plants at 15 DAI. (D) Evans blue staining visualizes leaf tissues undergoing cell death in TRV:BPS1(C) lines.
Analysis of programmed cell death phenotypes
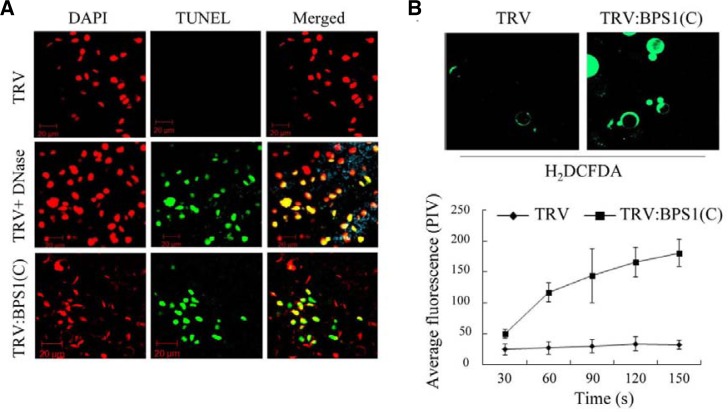
We examined nuclear DNA fragmentation as a PCD marker in TRV:BPS1(C) leaf cells using the TUNEL assay. The TUNEL assay detects single- and double-strand DNA breaks by addition of fluorescent nucleotides to free 3′ DNA termini (Gavrieli et al., 1992). TRV:BPS1(C) leaves exhibited strong fluorescent TUNEL staining in the epidermal nuclei based on confocal microscopy, suggesting single- or double-stranded DNA breaks in the nuclei (Fig. 2A). The staining was not detected in control TRV plants unless the plants had been treated with DNase (Fig. 2A). These results indicate that VIGS-mediated NbBPS1 down-regulation causes PCD in N. benthamiana. During apoptosis in animal cells, activation of the cell death pathway is initiated by modification of the mitochondrial membrane potential. We have previously shown that the mitochondrial membrane potential was significantly reduced (about 4-fold) in the leaf protoplasts of the NbBPS1 VIGS lines using TMRM (Tetramethylrhodamine methyl ester) fluorescent probes (Kang et al., 2008b).
Fig. 2.
Phenotypes of programmed cell death. (A) Confocal images of the TUNEL assay in leaves of TRV control and TRV:BPS1(C) plants at 15 DAI. Leaves were counterstained with DAPI. As a positive control, fixed leaf material of TRV samples was subjected to DNase treatment. Scale bars = 20 μm. (B) ROS production. Leaf protoplasts from TRV control and TRV:BPS1(C) plants were incubated with the ROS indicator H2DCFDA (2 μM) (top). H2DCFDA is a ROS indicator that becomes fluorescent when oxidized by ROS within the cell. Fluorescence of protoplasts from the VIGS lines was quantified by pixel intensity (bottom). Data points represent means ± SD of 30 individual protoplasts. PIV, pixel intensity values.
To test whether reactive oxygen species (ROS) were produced in the TRV:BPS1(C) leaf cells undergoing PCD, we prepared leaf protoplasts from TRV or TRV:BPS1(C) plants and incubated the protoplasts with H2DCFDA, a membrane-permeable indicator of ROS. Activation of the indicator depends on the presence of H2O2 and produces a green fluorescent signal (Fig. 2B). The rate of accumulation of fluorescent H2DCFDA in TRV:BPS1 protoplasts was approximately six times greater than that of the TRV control protoplasts, suggesting involvement of ROS in the cell death program (Fig. 2B).
Expression of ER stress-related genes
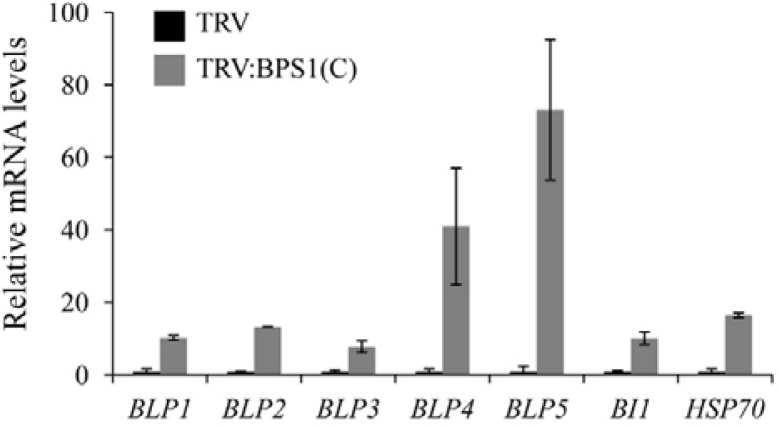
NbBPS1 is localized in the ER in N. benthamiana (Kang et al., 2008b). The cell death phenotype of the NbBPS1 VIGS plants observed may indicate that NbBPS1-deficient cells were subjected to ER stress. Previously, transcriptome analyses revealed that transcription of BiPs, BI1 (Bax-Inhibitor-1), and HSP70 was up-regulated in response to tunicamycin-induced ER stress in Arabidopsis (Urade, 2007; 2009; Watanabe and Lam, 2008). Functions of these genes during ER stress have been reported: overexpression of BLPs alleviated ER stress in tobacco (Leborgne-Castel et al., 1999) and BI-1 modulated ER stress-mediated PCD in Arabidopsis (Watanabe and Lam, 2008). Thus, we examined transcript levels of those ER stress-related genes in TRV and TRV:BPS1(C) lines (Fig. 3). Realtime quantitative RT-PCR revealed that the transcript levels of N. benthamiana BLP1 (BiP-like protein 1), BLP2, BLP3, BLP4, BLP5, BI1, and Hsp70 genes were all induced to a much higher level in TRV:BPS1(C) leaves at 15 DAI than in TRV control leaves (Fig. 3). Since BLPs encoding ER-localized chaperones and BI1 have been used as marker genes for ER stress and UPR activation in plants (Martinez and Chrispeels, 2003; Watanabe and Lam, 2008; Zuppini et al., 2004), our results suggest that NbBPS1 deficiency induces ER stress in N. bethamiana.
Fig. 3.
Induction of ER stress-related genes in TRV:BPS1 plants. Real-time quantitative RT-PCR analyses were performed to determine transcript levels of ER stress-related genes in TRV and TRV:BPS1(C) leaves at 15 DAI. Each value represents the mean ± SD of three replicates per experiment. The α-tubulin mRNA level was used as a control.
Processing of bZIP28 transcription factor indicating ER stress
In Arabidopsis, tunicamycin-induced ER stress causes proteolytic processing of bZIP28 and bZIP60, ER membrane-associated bZIP transcription factors, and translocation of their released N-terminal domains to the nucleus (Iwata and Koizumi, 2005; Liu and Howell, 2010; Liu et al., 2007a). In the nucleus, the N-terminal domains of the bZIP factors control transcriptional induction of ER stress response genes (Liu and Howell, 2010). A similar processing and nuclear translocation of Arabidopsis bZIP17 occurs in response to salt stress (Liu et al., 2007b). To confirm that NbBPS1 deficiency causes ER stress, we examined localization of bZIP28 in control TRV and TRV: BPS1(C) lines (Fig. 4). A GFP fusion protein of bZIP28, GFP: bZIP28, was transiently expressed in TRV and TRV:BPS1(C) leaves at 15 DAI by agro-infiltration. Protoplasts (mostly from mesophyll cells) were isolated from the infiltrated leaves and examined by confocal laser scanning microscopy (Fig. 4A). In the TRV protoplasts, GFP:bZIP28 displayed a network pattern of green fluorescence throughout the cytoplasm, indicating localization of bZIP28 to the ER. This pattern of bZIP28 localization is consistent with the results by Liu et al. (2007a). Most of the protoplasts from TRV:BPS1(C) leaves also exhibited the network-like pattern, but 9% of the protoplasts (n = 200) showed strong GFP fluorescence only in the nucleus, suggesting relocation of the bZIP28 factor to the nucleus (Figs. 4A and 4C). Confocal microscopy of the leaf epidermal cells revealed green fluorescent signals of GFP:bZIP28 in the vicinity of the plasma membrane and the nucleus in both TRV and TRV:BPS1(C) leaves (Fig. 4B). However, some epidermal cells in TRV:BPS1(C) lines clearly exhibited strong fluorescent signal only in the nucleus, similar to the pattern in tunicamycin-treated TRV leaf cells (Fig. 4B). Since nuclear accumulation of GFP:bZIP28 was not observed in TRV cells, these results further support that NbBPS1 silencing induces the ER stress response.
Fig. 4.
Subcellular localization of GFP:bZIP28. (A) Protoplasts were isolated from TRV and TRV:BPS1(C) plants (15 DAI) infiltrated with Agrobacterium containing GFP:bZIP28, and localization of fluorescent signals was examined by fluorescence microscopy. Representative images of ER and nuclear localization of GFP:bZIP28 in the protoplasts are shown. (B) TRV control, tunicamycin (TM)-treated TRV, and TRV:BPS1(C) plants (15 DAI) were infiltrated with Agrobacterium containing GFP:bZIP28 construct. After 24 h, localization of GFP fluorescent signals in leaf epidermal cells was observed by confocal microscopy (top). Merged images of GFP:bZIP28 fluorescence and chlorophyll autofluorescence (pseudo-colored blue; mostly from underlying mesophyll cells) are also shown (bottom). Representative images are shown. (C) The frequency of ER and nuclear patterns of GFP:bZIP28 localization in the protoplasts (n = 200) from TRV and TRV:BPS1(C) lines shown in (A).
Induction of autophagy
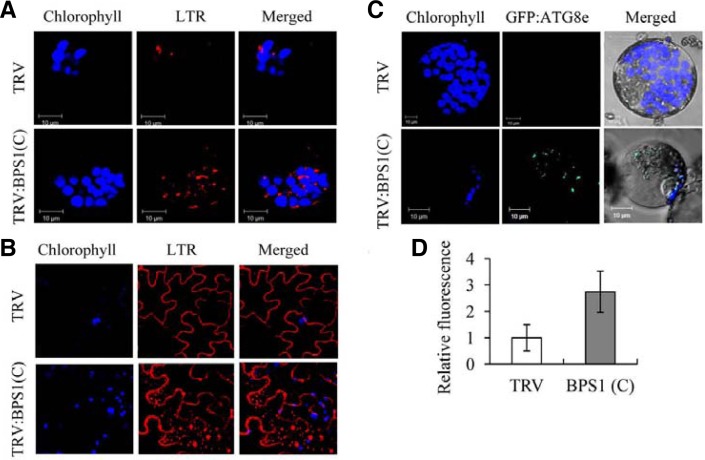
Autophagy is a conserved catabolic pathway in eukaryotic cells that controls recycling of bulk cytoplasmic materials from proteins to whole organelles (Bassham, 2007; Lum et al., 2005). During autophagy, cytoplasm is enclosed in small vesicles (autophagosomes) and delivered to the lysosome or lytic vacuole for degradation. Because ER stress can activate autophagy (Ding and Yin, 2008; Høyer-Hansen and Jäättelä, 2007; Sakaki and Kaufman, 2008), we examined the autophagic activity of TRV:BPS1(C) and TRV control plants at 15 DAI (Fig. 5). Epidermal cells and leaf protoplasts from the VIGS plants were stained with Lysotracker Red (LTR) dye as a probe to detect autolysosome structures (Figs. 5A, 5B, and 5D). The LTR dye labels acidic organelles such as autolysosomes, but it also stains the cell wall in epidermal cells (Hofius et al., 2009; Liu et al., 2005). Many LTR-stained particles were observed in the cytosol of the epidermal cells and the protoplasts from NbBPS1-silenced plants, but were rarely observed in the cytoplasm of TRV control (Figs. 5A and 5B). The LTR fluorescence in the protoplasts was quantified: TRV:BPS1(C) protoplasts exhibited almost three times the level of fluorescence of TRV (Fig. 5D).
Fig. 5.
Activation of autophagy. (A) LysoTracker Red (LTR) staining of leaf protoplasts isolated from TRV and TRV:BPS1(C) lines at 15 DAI. LTR-stained punctuate autophagosome-like structures, chloro-phyll autofluorescence (pseudo-colored blue), and merged images were observed by confocal laser scanning microscopy. (B) LTR staining of leaf epidermal cells of TRV and TRV:BPS1(C) lines at 15 DAI. Note that LTR also stains the epidermal cell wall. (C) Confocal microscopy of transiently expressed GFP:ATG8e in leaf protoplasts isolated from TRV and TRV:BPS1(C) lines at 15 DAI. (D) LTR-derived red fluorescence of the protoplasts shown in (A) was quantified by confocal microscopy. Data points represent means ± SD of 30 individual protoplasts.
Autophagy activity was further monitored by transient expression of a GFP:ATG8e fusion protein, which has been used to visualize autophagy in plant cells (Contento et al., 2005). GFP:ATG8e was transiently expressed in the VIGS plants (15 DAI) by agroinfiltration. After 2 days, protoplasts were generated from the leaves and examined by confocal microscopy (Fig. 5C). GFP fluorescence was evident as multiple punctuate structures in TRV:BPS1(C) protoplasts, but was not detectable in TRV protoplasts (Fig. 5C; Supplementary Fig. 1A). Transient expression of the GFP:ATG8e transcript in the infiltrated leaves of TRV control and TRV:BPS1(C) lines was determined by using RT-PCR (Supplementary Fig. 1B). These confocal microscopy observations indicate that NbBPS1 silencing results in activation of autophagy.
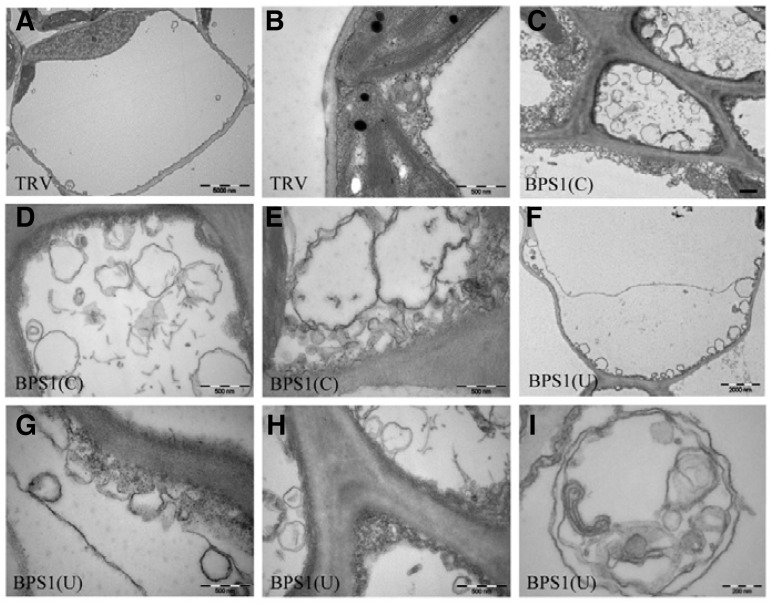
Ultrastructural analyses using transmission electron microscopy
Using transmission electron microscopy, cell ultrastructure of TRV:BPS1 lines was examined compared with TRV control plants at 15 DAI. Leaf cells of TRV:BPS1(C) and TRV:BPS1(U) lines accumulated a large number of small vesicles or vacuoles near the plasma membrane (Figs. 6C–6H) compared to TRV control plants (Figs. 6A and 6B). Dramatic degeneration of cellular ultrastructure was evident in TRV:BPS1 lines. High magnification showed that most membrane vesicles had double or multiple membrane boundaries, a morphological feature of autophagosomes (Fig. 6I).
Fig. 6.
Accumulation of vesicles in TRV:BPS1 leaf cells. Transmission electron micrographs of leaf transverse sections from TRV (A, B), TRV: BPS1(C) (C-E), and TRV: BPS1(U) (F-I) lines at 15 DAI. Autophagosome-like structures were observed near plasma membranes in TRV:BPS1 lines (C-I). Scale bars = 5 μm (A); 0.5 μm (B-H); 0.2 μm (I).
Previously, NbBPS1 VIGS led to severe developmental abnormalities and cell death in N. benthamiana (Kang et al., 2008b). NbBPS1 shows no homology to any known proteins. NbBPS1 localization in the ER has been shown (Kang et al., 2008b), but its biochemical function in the ER remains unclear. Our results in this study suggest that severe deficiency of NbBPS1 in N. benthamiana induces ER stress, autophagy, and PCD, leading to premature death of the plants. NbBPS1 dysfunction may result in accumulation of misfolded proteins in the ER, either directly or indirectly, leading to the ER stress. Induction of autophagy in the NbBPS1 VIGS plants can be explained by the findings that ER stress is a potent trigger of autophagy, and that the phenomenon is conserved from yeast to mammals (Ding and Yin, 2008; Høyer-Hansen and Jäättelä, 2007; Sakaki and Kaufman, 2008). Further study should illustrate the mole-cular function of NbBPS1 and its connection to the regulatory mechanism of plant growth and development.
Supplementary Material
Acknowledgments
This research was supported by the Next-Generation BioGreen 21 Program (PMBC, No. PJ009079/SSAC, No. PJ008214) from Rural Development Administration (RDA) of Korea.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Ahn J.-W., Kim M., Lim J.H., Kim G.-T., Pai H.-S. Phytocalpain controls the proliferation and differentiation fates of cells in plant organ development. Plant J. 2004;38:969–981. doi: 10.1111/j.1365-313X.2004.02102.x. [DOI] [PubMed] [Google Scholar]
- Ahn C.S., Han J.-A, Lee H.-S., Lee S., Pai H.-S. The PP2A regulatory subunit Tap46, a component of TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell. 2011;23:185–209. doi: 10.1105/tpc.110.074005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D.C. Plant autophagy--more than a starvation response. Curr. Opin. Plant Biol. 2007;10:587–593. doi: 10.1016/j.pbi.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Boyce M., Yuan J. Cellular response to endoplasmic reticulum stress: a matter of life or death. Cell Death Differ. 2006;13:363–373. doi: 10.1038/sj.cdd.4401817. [DOI] [PubMed] [Google Scholar]
- Coll N.S., Epple P., Dangl J.L. Programmed cell death in the plant immune system. Cell Death Differ. 2011;18:1247–1256. doi: 10.1038/cdd.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento A.L., Xiong Y., Bassham D.C. Visualization of autophagy in Arabidopsis using the fluorescent dye mono-dansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 2005;42:598–608. doi: 10.1111/j.1365-313X.2005.02396.x. [DOI] [PubMed] [Google Scholar]
- Costa M.D., Reis P.A., Valente M.A., Irsigler A.S., Carvalho C.M., Loureiro M.E., Aragão F.J., Boston R.S., Fietto L.G., Fontes E.P. A new branch of endoplasmic reticulum stress signaling and the osmotic signal converge on plant-specific asparagine-rich proteins to promote cell death. J. Biol. Chem. 2008;283:20209–20219. doi: 10.1074/jbc.M802654200. [DOI] [PubMed] [Google Scholar]
- Crosti P., Malerba M., Bianchetti R. Tunicamycin and Brefeldin A induce in plant cells a programmed cell death showing apoptotic features. Protoplasma. 2001;216:31–38. doi: 10.1007/BF02680128. [DOI] [PubMed] [Google Scholar]
- Ding W.X., Yin X.M. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- Duan Y., Zhang W., Li B., Wang Y., Li K., Sodmergen, Han C., Zhang Y., Li X. An endoplasmic reticulum response pathway mediates programmed cell death of root tip induced by water stress in Arabidopsis. New Phytol. 2010;186:681–695. doi: 10.1111/j.1469-8137.2010.03207.x. [DOI] [PubMed] [Google Scholar]
- Gavrieli Y., Sherman Y., Ben-Sasson S.A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høyer-Hansen M., Jäättelä M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H., Mattsson O., Jørgensen L.B., Jones J.D., Mundy J., Petersen M. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137:773–783. doi: 10.1016/j.cell.2009.02.036. [DOI] [PubMed] [Google Scholar]
- Iwata Y., Koizumi N. An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. USA. 2005;102:5280–5285. doi: 10.1073/pnas.0408941102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.W., Lee J.Y., Jeon Y., Cheong G.W., Kim M., Pai H.S. In vivo effects of NbSiR silencing on chloroplast development in Nicotiana benthamiana. Plant Mol. biol. 2008a;72:569–583. doi: 10.1007/s11103-009-9593-8. [DOI] [PubMed] [Google Scholar]
- Kang Y.W., Kim R.N., Cho H.S., Kim W.T., Choi D., Pai H.S. Silencing of a BYPASS1 homolog results in root-independent pleiotrophic developmental defects in Nicotiana benthamiana. Plant Mol. Biol. 2008b;68:423–437. doi: 10.1007/s11103-008-9384-7. [DOI] [PubMed] [Google Scholar]
- Kaufman R.J., Scheuner D., Schröder M., Shen X., Lee K., Liu C.Y., Arnold S.M. The unfolded protein response in nutrient sensing and differentiation. Nat. Rev. Mol. Cell Biol. 2002;3:411–421. doi: 10.1038/nrm829. [DOI] [PubMed] [Google Scholar]
- Kim M., Lim J.-H., Ahn C.S., Park K., Kim G.T., Kim W.T., Pai H.-S. Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell. 2006;18:2341–2355. doi: 10.1105/tpc.106.041509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E., Kato N., Lawton M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature. 2001;411:848–853. doi: 10.1038/35081184. [DOI] [PubMed] [Google Scholar]
- Leborgne-Castel N., Jelitto-Van Dooren E.P., Crofts A.J., Denecke J. Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell. 1999;11:459–470. doi: 10.1105/tpc.11.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lee B., Lee A.S. Endoplasmic reticulum stress-induced apoptosis: multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. J. Biol. Chem. 2006;281:7260–7270. doi: 10.1074/jbc.M509868200. [DOI] [PubMed] [Google Scholar]
- Liu J.-X., Howell S.H. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22:782–796. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Tallóczy Z., Levine B., Dinesh-Kumar S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Liu J.X., Srivastava R., Che P., Howell S.H. An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell. 2007a;19:4111–4119. doi: 10.1105/tpc.106.050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Srivastava R., Che P., Howell S.H. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 2007b;51:897–909. doi: 10.1111/j.1365-313X.2007.03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum J.J., DeBerardinis R.J., Thompson C.B. Autophagy in metazoans: cell survival in the land of plenty. Nat. Rev. Mol. Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- Martínez I.M., Chrispeels M.J. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell. 2003;15:561–576. doi: 10.1105/tpc.007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reape T.J., McCabe P.F. Apoptotic-like programmed cell death in plants. New Phytol. 2008;180:13–26. doi: 10.1111/j.1469-8137.2008.02549.x. [DOI] [PubMed] [Google Scholar]
- Sakaki K., Kaufman R.J. Regulation of ER stress-induced macroautophagy by protein kinase C. Autophagy. 2008;4:841–843. doi: 10.4161/auto.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder M., Kaufman R.J. ER stress and the unfolded protein response. Mutat. Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- Shiraishi H., Okamoto H., Yoshimura A., Yoshida H. ER stress-induced apoptosis and caspase-12 activation occurs downstream of mitochondrial apoptosis involving Apaf-1. J. Cell Sci. 2006;119:3958–3966. doi: 10.1242/jcs.03160. [DOI] [PubMed] [Google Scholar]
- Szegezdi E., Logue S.E., Gorman A.M., Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urade R. Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS J. 2007;274:1152–1171. doi: 10.1111/j.1742-4658.2007.05664.x. [DOI] [PubMed] [Google Scholar]
- Urade R. The endoplasmic reticulum stress signaling pathways in plants. BioFactors. 2009;35:326–331. doi: 10.1002/biof.45. [DOI] [PubMed] [Google Scholar]
- Van Norman J.M., Sieburth L.E. Dissecting the biosynthetic pathway for the bypass1 root-derived signal. Plant J. 2007;49:619–628. doi: 10.1111/j.1365-313X.2006.02982.x. [DOI] [PubMed] [Google Scholar]
- Vaux D.L., Korsmeyer S.J. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Lam E. Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J. 2006;45:884–894. doi: 10.1111/j.1365-313X.2006.02654.x. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Lam E. BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J. Biol. Chem. 2008;283:3200–3210. doi: 10.1074/jbc.M706659200. [DOI] [PubMed] [Google Scholar]
- Wertz I.E., Hanley M.R. Diverse molecular provocation of programmed cell death. Trends Biochem. Sci. 1996;21:359–364. [PubMed] [Google Scholar]
- Zuppini A., Navazio L., Mariani P. Endoplasmic reticulum stress-induced programmed cell death in soybean cells. J. Cell Sci. 2004;117:2591–2598. doi: 10.1242/jcs.01126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.