Abstract
AtMYB44 is a member of the R2R3 MYB subgroup 22 transcription factors and regulates diverse cellular responses in Arabidopsis thaliana. We performed quadruple 9-mer-based protein binding microarray (PBM) analysis, which revealed that full-size AtMYB44 recognized and bound to the consensus sequence AACnG, where n represents A, G, C or T. The consensus sequence was confirmed by electrophoretic mobility shift assay (EMSA) with a truncated AtMYB44 protein containing the N-terminal side R2R3 domain. This result indicates that the R2R3 domain alone is sufficient to exhibit AtMYB44 binding specificity. The sequence AACnG is the type I binding site for MYB transcription factors, including all members of the subgroup 22. EMSA showed that the R2R3 domain protein binds in vitro to promoters of randomly selected Arabidopsis genes that contain the consensus binding sequence. This implies that AtMYB44 binds to any promoter region that contains the consensus sequence, without determining their functional activity or specificity. The C-terminal side transcriptional activation domain of AtMYB44 contains an asparagine-rich fragment, NINNTTSSRHNHNN (aa 215–228), which, among the members of subgroup 22, is unique to AtMYB44. A transcriptional activation assay in yeast showed that this fragment is included in a region (aa 200–240) critical for the ability of AtMYB44 to function as a transcriptional activator. We hypothesize that the C-terminal side of the protein, but not the N-terminal side of the R2R3 domain, contributes to the functional activity and specificity of AtMYB44 through interactions with other regulators generated by each of a variety of stimuli.
Keywords: Arabidopsis, AtMYB44, protein binding microarray, protein domain, transcription factor
INTRODUCTION
We reported previously that the AtMYB44 transcription factor plays a role in the abscisic acid (ABA)-mediated signaling pathway that confers abiotic stress tolerance via the enhancement of stomatal closure in Arabidopsis thaliana (Jung et al., 2008). Transgenic Arabidopsis overexpressing the gene exhibited enhanced drought/salinity tolerance by suppressing the expression of genes encoding a group of Ser/Thr protein phosphatase 2Cs (PP2Cs) that were described as negative regulators of ABA signaling. In our recent study, transgenic soybeans transformed with the AtMYB44 gene exhibited significantly enhanced drought/salinity tolerance, without altering the amino acid and fatty acid compositions of the seeds (Seo et al., 2012). This result suggests that interaction of AtMYB44 with specific sequences in target gene promoters activates a tolerance mechanism that is conserved in Arabidopsis and soybean.
In addition to the ABA-mediated signaling, AtMYB44 mediates the suppression of jasmonate-mediated responses (Jung et al., 2010), which supports the hypothesis of mutual antagonistic actions between jasmonate-and ABA-mediated signaling pathways. Moreover, AtMYB44 regulates many other biological processes, including the expression of the ETHYLENE INSENSITIVE2 gene, to affect resistance to the green peach aphid in Arabidopsis (Liu et al., 2010; 2011; Lü et al., 2011). More recently, it was reported that AtMYB44 is phosphorylated by mitogen-activated protein (MAP) kinases and regulates seed germination (Nguyen et al., 2012). The mechanism(s) behind such functional diversity of AtMYB44 are not yet understood.
A transcription factor binds to a specific DNA sequence in promoter regions of target genes to regulate RNA polymerase activity for gene transcription. A transcription factor is composed of a DNA-binding domain and a catalytic (activation or repression) domain. MYB transcription factors contain a conserved DNA-binding domain consisting of two or three imperfect repeats of 50–53-amino acids (R1, R2 and R3) that form the helix-turn-helix motifs (Peters et al., 1987; Rosinsky and Atchley, 1998).
In plants, two-repeat (R2R3) MYB family members predominate. Among 198 genes in the MYB superfamily, a total of 126 R2R3 MYB-encoding genes have been identified in the Arabidopsis genome, making it one of the largest transcription factor groups in this plant (Yanhui et al., 2006). Extensive functional analyses using large-scale insertional mutagenesis (Meissner et al., 1999; Stracke et al., 2001) and expression profiling (Kranz et al., 1998; Yanhui et al., 2006) have been performed to characterize R2R3 MYB proteins in Arabidopsis. In parallel, the roles of individual R2R3 MYB proteins in diverse plant processes have been explored, including hormonal signaling, cell-cycle control, stress responses, secondary metabolism, cellular morphogenesis and meristem formation (Das et al., 2012; Jin and Martin, 1999; Martin and Paz-Ares, 1997).
Accumulated experimental data indicate that R2R3 MYB proteins bind to one or more of the following types of site: I, CnGTTr (= pAACnG); II, GkTwGTTr; and IIG, GkTwGGTr, where n indicates A, G, C, or T; k, R, A or G; p, T or C; w, A or T (Romero et al., 1998). Although members of a class of MYB transcription factors share a high degree of homology in the R2R3 DNA-binding domain and in corresponding DNA-binding sequences, individual MYB proteins play unique roles in diverse plant processes. Thus, it is debatable that the functional specificity of a plant MYB transcription factor is dependent on the DNA binding sequence specificity of each R2R3 domain.
The Arabidopsis R2R3-type MYB transcription factors have been categorized into subgroups on the basis of conserved amino acid sequence motifs present in the carboxy-terminal to the MYB domain (Kranz et al., 1998). For instance, members of subgroup 22, including AtMYB44, AtMYB70, AtMYB73 and AtMYB77, share two conserved motifs: TGLYMSPxSP (motif 22.1) and GxFMxVVQEMIxxEVRSYM (motif 22.2) (Stracke et al., 2001). These conserved motifs may facilitate the identification of functional domains outside of the DNA-binding domain of R2R3-type MYB factors.
To address whether the functional specificity of AtMYB44 in subgroup 22 is a direct result of the nucleotide sequence of its binding site, we determined its binding site using a protein-binding microarray (PBM) analysis. The PBM analysis eliminated any false nucleotide identified ambiguously in the binding site selection assay using a pool of synthetic oligonucleotides with degenerate sequences. The PBM analysis revealed that AtMYB44 recognizes and binds to the consensus sequence AACnG, which is the type I binding site for MYB transcription factors, including all members of subgroup 22. We hypothesized that the C-terminal side of the protein, which includes an asparagine-rich region, but not the N-terminal side R2R3 domain, contributes to the functional specificity of AtMYB44. The structural feature identified here is likely to prove useful in further studies of the functional specificity and diversity of At-MYB44.
MATERIALS AND METHODS
Protein binding microarray (PBM) analysis
Using the full-length cDNA of AtMYB44 (TAIR clone 119B8), proteins fused at the N-termini with DsRed fluorescent protein and a polyhistidine-tag were expressed in the Escherichia coli strain BL21-ColonPLus, as described previously (Kim et al., 2009). A protein-binding mixture containing 200 nM fusion protein was incubated with the quadruple 9-mer PBM (Q9-PBM) at 25°C for 1 h. Fluorescence images were obtained with a 4000B microarray scanner (Molecular Devices, USA).
The consensus binding sequence was determined based on fluorescence signal strength, as described previously (Kim et al., 2012). Two independent linear models, y = ax + b, were applied to the steep left and the extended right tail regions of the rank-ordered fluorescence signal distribution curve of the bound protein, using the R statistical language. Spots that exhibited strong fluorescence intensity and high enrichment were subject to alignment. These groups were denoted with SEQLOGO ‘Visualize information content of patterns’ [http://www.bioinf.ebc.ee/EP/EP/SEQLOGO/], yielding an intensity profile figure, sequence logos and their related statistics. P-values and position weight matrices were calculated using the Wilcoxon-Mann-Whitney test.
R2R3 domain protein
Using the full-length AtMYB44 cDNA, a DNA fragment encoding the N-terminal side of the AtMYB44 protein containing Met1 through Tyr110 was amplified by polymerase chain reaction (PCR) using a primer set containing EcoRI sites. The fragment was inserted into the pGEX-5X-1 vector (Pharmacia, USA) at an EcoRI site to fuse with the glutathione-S-transferase gene (GST). The DNA construct was transformed into E. coli BL21. Expressed GST-AtMYB44 R2R3 proteins were purified using glutathione-agarose beads (Sigma, USA).
Electrophoretic mobility shift assay
Double-stranded synthetic oligonucleotides were labeled with digoxigenin and incubated with proteins in the binding buffer at room temperature for 30 min. Protein-oligonucleotide complexes were separated from free probes on a 6% polyacrylamide gel and transferred to a nylon membrane. Horseradish peroxidase-conjugated anti-digoxigenin antibody and a chemiluminescent substrate, Supersignal West Pico (Pierce, USA), were used for detection of digoxigenin.
For the promoter binding test, ∼2 kb DNA fragments were amplified by polymerase chain reaction from the nucleotide sequences of the promoter region of the Arabidopsis genes ABI5 (AT2g36270), ERA1 (AT5g40280), ERD 1 (AT5g51070) and FCA (AT4g16280). Primer sets used were designed from the sequences of each gene promoter region; nucleotide positions-2,000∼-1,976 were used for 5′-primers and-1∼-25 for 3′-primers. The amplified DNA fragments were labeled with digoxigenin and used in the electrophoretic mobility shift assay.
Yeast transactivation assay
A DNA fragment encoding a partial AtMYB44 was amplified by PCR using proper primer sets containing BamHI (for the N-terminal side of the protein) and EcoRI sites (for the C-terminal side). The PCR products were fused in-frame to the sequence of the yeast GAL4 DNA-binding domain in the yeast expression vector pGBK-T7 (Clontech, USA). The recombinant vectors were transformed into the yeast strain Y187 using the lithium acetate-mediated method (Gietz et al., 1992). The β-galactosidase activity was quantified by liquid culture assay using o-nitrophenyl β-D-galactopyranoside (ONPG) as a substrate, according to the manufacturer’s instructions.
RESULTS
Structural features of AtMYB44
The AtMYB44 (At5g67300) gene has an open reading frame of 918 base pairs encoding a putative 305-amino acid polypeptide with a predicted molecular mass of 33.3 kDa (Fig. 1). The tryptophan residues conserved in the R2 (W9, W29 and W48) and R3 (W80 and W99) domains were found in the amino acid sequence. Two lysine residues (K42 in R2 and K96 in R3) and one asparagine residue (N97 in R3), which was identified as recognizing the specific binding sequence AACnG (Ogata et al., 1994), were also found in the AtMYB44 amino acid sequence.
Fig. 1.
Comparison of the AtMYB44 amino acid sequence and related MYB transcription factors. Amino acid sequences deduced from AtMYB44 (GenBank accession no. AY519648), AtMYB77 (AY124828), AtMYB73 (AY091267) and AtMYB70 (AY519574) were aligned using the COBALT Multiple Alignment Tool provided by the National Center for Biotechnology Information (NCBI). Gaps were represented by hyphens to maximize the alignment. Sequences under the lines represent amino acid residues conserved among the Arabidopsis subgroup 22 MYB transcription factors, R2, R3, motif 22.1 and motif 22.2. Tryptophan residues conserved in typical R2R3 MYB transcription factors are indicated by dots. Two lysine and one asparagine residues essential for specific DNA binding are indicated by asterisks. Asparagine-rich sequences found in AtMYB44 and AtMYB77 are indicated by a dotted line.
The amino acid sequence of AtMYB44 was aligned with those of other subgroup 22 transcription factors, AtMYB77 (At3-g50060), AtMYB73 (At4G37260), and AtMYB70 (At2g23290). AtMYB44 and AtMYB77 showed the highest degree of homology to each other in nucleotide (75%) and amino acid (63%) sequences. These proteins share two motifs, motifs 22.1 and 22.2 (Stracke et al., 2001), which are characteristically conserved throughout the subgroup. The motif 22.1 is included in a proline-rich region (aa 123–199). Between the two conserved motifs, the asparagine-rich fragments NINNTTSSRHNHNN (aa 215–228) and NTSSSHNNNNN were found in AtMYB44 and AtMYB77, respectively.
Protein-binding microarray (PBM) analysis of AtMYB44
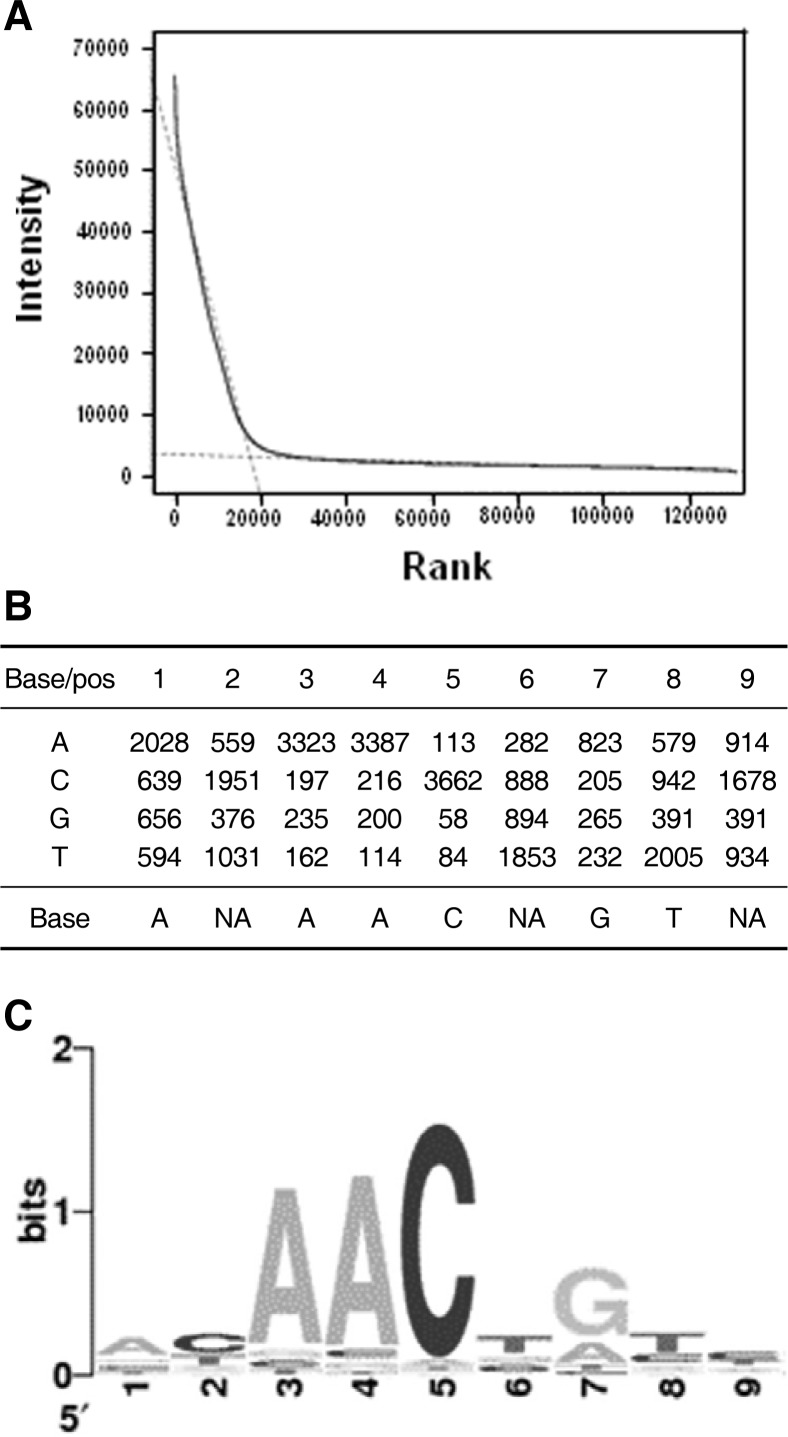
AtMYB44-DsRed fusion proteins were reacted with a Q9-PBM. The rank-ordered signal distribution curve showed a steep leftward slope and an extensive right tail region (Fig. 2A), due to a specific interaction between the protein and features on the microarray. Two independent linear models, y = ax + b, were applied to the left steep (b1 = 51384.5, slope = −2.7) and right extensive tail (b1 = 3469.8, slope = −0.021) regions. The range of the ranking was 17818. For motif extraction, 9-mers in the steep left region were clustered, based on the best alignment using the highest-ranked 9-mers among the clusters as seeds. Out of the 17818 total signals, 3917 were clustered, and the cluster’s position weight matrix (PWM) was obtained. The Wilcoxon-Mann-Whitney test was performed to yield a consensus sequence, AACnG (Fig. 2B), where n represents A, G, C or T. P-value calculated from the test was 0 (P = 0).
Fig. 2.
Schematic representation of protein binding microarray analysis. (A) Rank analysis of AtMYB44 PBM binding. From the rank-ordered signal distribution, two independent linear models, y = ax + b, were applied in the steep (b1 = 51384.5, slope = −2.7) and the heavy right (b1= 3469.8, slope = −0.021) tail regions. Rank extrapolation was 17818. (B) Total position weight matrix. Spots that exhibited strong intensity and high enrichment were subjected to alignment. These groups were denoted by SEQLOGO ‘Visualize information content of patterns’ [http://www.bioinf.ebc.ee/EP/EP/SEQLOGO/]. (C) Sequence logo of the determined consensus binding sequence of AtMYB44.
It appears that T or C is preferred over for the nucleotide located in front of the consensus sequence. When considering the rank of signal strength (affinity), the best 13 oligomers contained TAACnG (Table 1). It appears that AtMYB44 binds preferentially to pAACnG, which is the type I site for R2R3 MYB transcription factors.
Table 1.
The top 50 nine-mer oligonucleotides that exhibited strong affinity to AtMYB441
| Rank | Oligomer | P-value (Median) | Median intensity | Max intensity |
|---|---|---|---|---|
| 1 | AATTAACCG | 2.245e−21 | 65,535.0 | 65,535.0 |
| 2 | TCGTAACTG | 2.659e−21 | 65,095.0 | 65,535.0 |
| 3 | TAACTGTCA | 2.861e−21 | 64,905.0 | 65,535.0 |
| 4 | ATAACGGTA | 3.385e−21 | 64,470.0 | 65,535.0 |
| 5 | CATAACCGT | 4.140e−21 | 63,952.0 | 65,535.0 |
| 6 | TCATACCTG | 4.660e−21 | 63,649.0 | 65,535.0 |
| 7 | TAACTGTAA | 6.056e−21 | 62,982.0 | 65,535.0 |
| 8 | TAACGGTCG | 1.116e−20 | 61,445.0 | 65,535.0 |
| 9 | CATAACGGT | 1.224e−20 | 61,216.0 | 65,535.0 |
| 10 | TAACTGCCA | 1.770e−20 | 60,305.0 | 65,535.0 |
| 11 | CCGTAACCG | 2.547e−20 | 59,415.0 | 65,535.0 |
| 12 | GAATTAACT | 3.011e−20 | 59,009.0 | 65,535.0 |
| 13 | TACCGGTAA | 3.111e−20 | 58,930.0 | 64,979.0 |
| 14 | TCATACCGG | 3.791e−20 | 58,453.0 | 65,535.0 |
| 15 | TAACTGACA | 3.837e−20 | 58,424.0 | 65,535.0 |
| 16 | TAACTACCA | 3.845e−20 | 58,419.0 | 65,535.0 |
| 17 | GTAACCGTC | 4.049e−20 | 58,295.0 | 65,535.0 |
| 18 | TACCTGTAA | 4.058e−20 | 58,290.0 | 65,535.0 |
| 19 | CCGTAACTG | 4.119e−20 | 58,254.0 | 65,535.0 |
| 20 | CCTAACCGT | 4.362e−20 | 58,117.0 | 65,535.0 |
| 21 | GAAATAACG | 4.600e−20 | 57,990.0 | 65,535.0 |
| 22 | CCATAACCG | 4.671e−20 | 57,953.0 | 65,535.0 |
| 23 | TAACCGTAA | 4.695e−20 | 57,941.0 | 64,826.0 |
| 24 | AATTACCTG | 4.902e−20 | 57,838.0 | 65,535.0 |
| 25 | GTAACGGTA | 4.987e−20 | 57,797.0 | 65,535.0 |
| 26 | AATACCGTT | 5.968e−20 | 57,370.0 | 65,535.0 |
| 27 | GAAATAACT | 6.047e−20 | 57,339.0 | 63,696.0 |
| 28 | TATAACGGT | 6.899e−20 | 57,027.0 | 65,535.0 |
| 29 | TTACCGGAA | 7.035e−20 | 56,981.0 | 64,488.0 |
| 30 | AAACGGTCA | 8.746e−20 | 56,469.0 | 63,900.0 |
| 31 | TTACCAGAA | 9.065e−20 | 56,385.0 | 63,916.0 |
| 32 | CTATAACGG | 9.706e−20 | 56,225.0 | 65,202.0 |
| 33 | TCCTAACTG | 1.060e−19 | 56,019.0 | 65,535.0 |
| 34 | AATTACCCG | 1.077e−19 | 55,982.0 | 64,589.0 |
| 35 | TAACCGCTA | 1.260e−19 | 55,618.0 | 65,194.0 |
| 36 | CTAACGGTT | 1.312e−19 | 55,523.0 | 65,535.0 |
| 37 | AAACGGTAT | 1.335e−19 | 55,484.0 | 65,492.0 |
| 38 | GAATTAACG | 1.345e−19 | 55,467.0 | 65,535.0 |
| 39 | CACAACGGT | 1.439e−19 | 55,310.0 | 64,793.0 |
| 40 | AGTTCGGTA | 1.448e−19 | 55,296.0 | 57,868.0 |
| 41 | AATAACTGG | 1.457e−19 | 55,281.0 | 56,572.0 |
| 42 | CAACGGTCT | 1.561e−19 | 55,122.0 | 65,386.0 |
| 43 | TAACCGACA | 1.648e−19 | 54,998.0 | 65,535.0 |
| 44 | CACAACTGT | 1.826e−19 | 54,762.0 | 65,535.0 |
| 45 | AACTAACCG | 1.921e−19 | 54,645.0 | 57,296.0 |
| 46 | CTAACCGCT | 1.986e−19 | 54,569.0 | 65,161.0 |
| 47 | AACTAACTT | 2.220e−19 | 54,315.0 | 63,956.0 |
| 48 | AACGAACTT | 2.229e−19 | 54,306.0 | 65,155.0 |
| 49 | TACCGGAAA | 2.281e−19 | 54,253.0 | 61,092.0 |
| 50 | CAACTGCCA | 2.383e−19 | 54,154.0 | 61,702.0 |
The consensus sequence AACnG in the oligomers is indicated by bold characters. Nucleotides not fitting the consensus sequence are underlined.
Binding of R2R3 domain protein
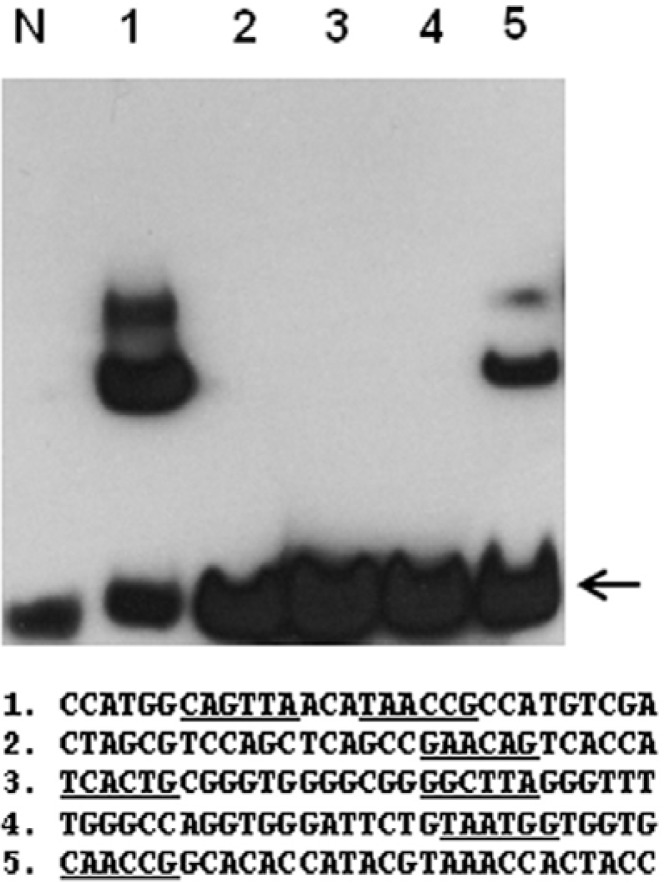
Based on the PBM analysis data, a group of digoxigenin-labeled oligonucleotides containing the AACnG sequences were synthesized and analyzed using an electrophoretic gel mobility shift assay (EMSA). In this experiment, a truncated form of the AtMYB44 protein containing the R2R3 binding domain (AtMYB44 R2R3 domain protein) was used.
The EMSA with individual sequences of oligonucleotides showed that GST-AtMYB44 R2R3 proteins exhibited relatively high binding activity on Oligo-1 (Fig. 3). Oligo-1 contains two sequences, TAACcG and TAACtG, thus showing a stronger binding affinity to the protein. The first nucleotide could be T or C because Oligo-5 containing CAACcG was also bound to the protein. Oligonucleotides containing GAACaG (Oligo-2), TAAG-cC and TCACtG (Oligo-3) or TAATgG (Oligo-4) were not bound to the protein. This result is consistent with the binding consensus sequence for the whole AtMYB44 protein determined by PBM.
Fig. 3.
Electrophoretic mobility shift assay (EMSA) with the AtMYB44 R2R3 domain protein. Synthetic DNA fragments containing an oligonucleotide, oligo-1 through oligo-5, were labeled with digoxigenin and incubated with purified GST-AtMYB44 R2R3 fusion proteins. Sequences of interest are underlined. Arrows indicate unbound free probes.
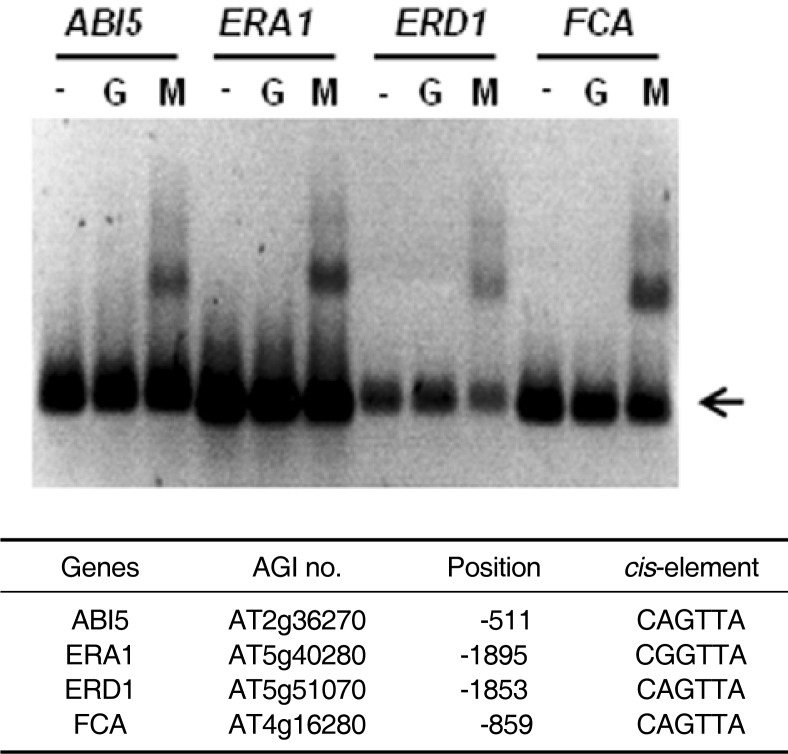
Using the Patmatch program (TAIR), we found that numerous Arabidopsis genes, including ABI5, ERA1, ERD1 and FCA, contain the consensus binding sequence in their promoter regions. An EMSA revealed that AtMYB44 proteins bind to the promoters (∼2 kb) of these genes (Fig. 4).
Fig. 4.
Binding of Arabidopsis gene promoters and the AtMYB44 R2R3 domain protein. DNA fragments amplified from promoter regions (∼2 kb) of the Arabidopsis genes ABI5, ERA1, ERD1 and FCA were labeled with digoxigenin and incubated without protein (−), with GST protein (G) or with GST-AtMYB44 R2R3 protein (M). Arrows indicate unbound free probes.
Transcriptional activation domain
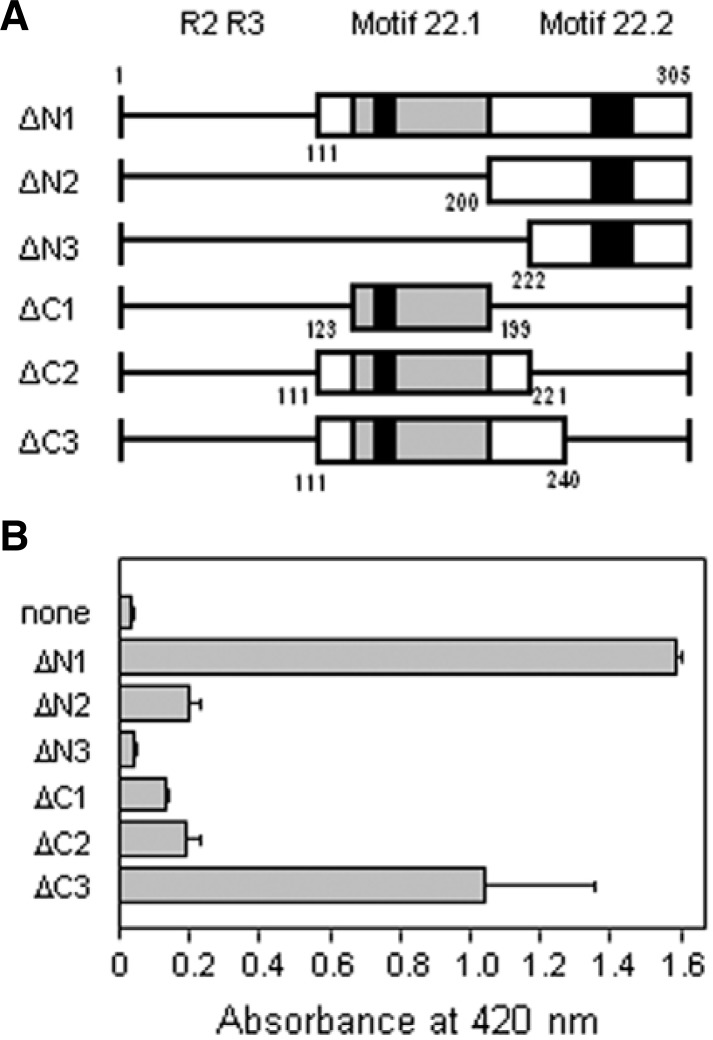
The ability of the C-terminal domain of the AtMYB44 protein to activate transcription in yeast was determined. We fused various deletion mutants (Fig. 5A) to the GAL4 DNA-binding domain (GAL4 BD) expression vector and examined the expression of the lacZ reporter gene using a liquid culture assay. When compared with the protein fragment containing the intact C-terminal domain (ΔN1), the mutants from which the proline-rich domain including motif 22.1 (ΔN2, ΔN3) was deleted showed lower β-galactosidase activity (Fig. 5B). An N-terminal deletion that did not contain the aa 200–221 region (ΔN3) showed little, if any, activity when compared to ΔN2. Deletion of the motif 22.2 (ΔC1, ΔC2, ΔC3) region also resulted in loss of transcriptional activation activity. However, the deletion mutant, ΔC3, containing 20 amino acid residues (aa 221–240) in addition to ΔC2 at the flanking C-terminal side displayed two-thirds of the activity observed with the entire C-terminal domain (ΔN1). These results suggest that the aa 200–240 region is critical for transcriptional activation by AtMYB44.
Fig. 5.
Yeast trans-activation assay. (A) Schematic representation of the truncated C-terminal catalytic domain of AtMYB44. Various deletion constructs of the AtMYB44 protein were fused to the GAL4 DNA-binding domain in the yeast expression vector pGBK-T7. The proline-rich region in the activation domain is indicated by grey boxes. The two conserved motifs within the subgroup 22 MYB genes 22.1 and 22.2 are indicated by black boxes. Small letters under the bars represent amino acid residue numbers in the AtMYB44 protein. (B) Trans-activation activities of the truncated AtMYB44 protein. Quantitative analysis of β-galactosidase activity was conducted with o-nitrophenyl β-D-galactopyranoside as a substrate. Data are means and standard deviations (error bars) of four independent experiments with triplicate samples.
In this experiment, the full-length AtMYB44 fused to GAL4 BD did not activate the transcription of lacZ due to an unknown reason (data not shown). The whole AtMYB44 protein expressed in yeast might be inactive due to, for example, misfolding. Or GAL4 BD may be too far from the catalytic domain near the R2R3 domain to activate the yeast RNA polymerase. Alternatively, the R2R3 domain of the whole protein might be bound to AACnG sequences in the yeast expression vector, hindering the binding of GAL4 BD to the lacZ promoter region.
DISCUSSION
In general, animal R2R3-MYB proteins recognize type I sequences (pAACnG) (Howe and Watson, 1991; Stober-Glässer et al., 1992). Heteronuclear multidimensional NMR has been used to demonstrate the solution structure of a specific DNA complex of the R2R3 domain. Both R2 and R3 contain three helices, and the third helix in each constitutes a recognition helix. R2 and R3 are closely packed in the major groove, and thus the two recognition helices contact each other directly to cooperatively bind to the specific base sequence AACnG (Ogata et al., 1994). The three key base pairs (the first A, third C, and fifth G) in this sequence are specifically recognized by Asn-183 (R3), Lys-182 (R3), and Lys-128 (R2). These amino acid residues are well conserved in the Arabidopsis MYB subgroup 22 transcription factors (Fig. 1).
We determined the DNA-binding sequence specificity of AtMYB44 by a comprehensive genome-wide method, using a universal PBM (Kim et al., 2009). The PBM was designed to contain quadruples of all possible 9-mer combinations, permitting unequivocal interpretation of the cis-acting elements. Proteins were labeled by N-terminal fusion with DsRed fluorescent protein, which circumvents the need for a multi-step incubation with antibodies of the protein of interest. The PBM analysis revealed that AtMYB44 binds specifically to the consensus sequence AACnG (Fig. 2).
The EMSA also revealed that a truncated form of the AtMYB44 protein, containing the R2R3 binding domain (AtMYB44 R2R3-domain protein), exhibits extensive binding to synthetic oligonucleotides containing the sequence pAACnG (Figs. 3 and 4). Thus, the N-terminal-side R2R3 domain is sufficient for the AtMYB44 binding activity and specificity. It was reported previously that C-terminal truncation of the mouse c-MYB protein reportedly had no effect on binding to DNA (Howe et al., 1990). In this experiment, the full-length AtMYB44 synthesized in E. coli did not show detectable binding activity for unknown reasons (data not shown). The inactivity of most of the whole AtMYB44 proteins expressed in bacteria might be due to, for example, misfolding. Similarly, the full-size AtMYB77 protein had a lower binding affinity than the truncated R2R3 domain protein (Romero et al., 1998), as is the case with other R2R3 MYB proteins, such as PhMYB3 and MmMYB (Ramsay et al., 1992; Solano et al., 1995). Binding activity was detected in PBM for AtMYB44 (Fig. 2) and AtMYB77 (Oh et al., 2012), demonstrating a higher degree of sensitivity for the PBM assay than EMSA.
The binding site selection assay using a pool of synthetic oligonucleotides with degenerate sequences showed that the binding consensus sequence of AtMYB77 (R2R3 domain) was pAACpGpC, in which the last C appeared with 82% probability at this position (Romero et al., 1998). By contrast, the quadruple 9-mer based PBM assay revealed that the full-size AtMYB77 protein binds specifically to the consensus sequence pAACnG (Oh et al., 2012). Similarly, the PBM assay performed in this study indicated that AtMYB44 binds to AACnG (Fig. 2), eliminating any additional nucleotide identified ambiguously in the binding site selection assay.
The PBM data from the AtMYB77 analysis revealed a nucleotide p (T or C) at the first position of the consensus sequence in addition to AACnG (Oh et al., 2012). The significance of this additional nucleotide in the binding between AtMYB77 and the promoter region is unknown. When we compared the top 50 nine-mer nucleotides that showed high affinity with AtMYB44 (Table 1) and AtMYB77 (Oh et al., 2012), no significant difference was found between the binding sites of AtMYB44 and AtMYB77. The two proteins may bind the same sites on the promoters, without exerting any specificity in terms of activation of transcription of their target genes. However, AtMYB44 and AtMYB77 have quite different biological roles. AtMYB77 is involved in auxin responses to control lateral root growth and development under changing environmental conditions (Shin et al., 2007). Double-knockout mutations of AtMYB44 and At-MYB77 (myb44myb77) did not change the auxin-responsive phenotypes of myb77. Thus, the difference of AtMYB44 and AtMYB77 in biological function may not due to their binding sequence specificity.
AtMYB44 recognizes and binds to AACnG (Fig. 2) and, thus, belongs to the Type I R2R3 MYB transcription factor family (Romero et al., 1998). The sequence AACnG appears every 256 (= 44) base pairs on average in the Arabidopsis genome. Assuming that a promoter region contains 2,000 base pairs, most promoters contain approximately eight AACnG sites. This implies that the binding of AtMYB44 in vitro to a promoter fragment does not mean that the corresponding gene is a target of AtMYB44. The randomly selected Arabidopsis genes in this study, including ABI5, ERA1, ERD1 and FCA, contain the consensus binding sequence in their promoter regions. An EMSA revealed that AtMYB44 proteins bind to the promoters (∼2 kb) of these genes (Fig. 4). However, we have no evidence that these genes are the real targets of the specific AtMYB44 function.
Transcriptional activation assays in yeast with the C-terminal side of AtMYB44 revealed that motifs 22.1 and 22.2 and a 41 amino acid region (aa 200–240) are critical for AtMYB44 to function as a transcriptional activator (Fig. 5). In this region, an asparagine-rich fragment, NINNTTSSRHNHNN (aa 215–228), is included. Such polyasparagine-and polyglutamine-containing proteins appear to be over-represented in protein kinases, lipid kinases, transcription factors, RNA helicases and messenger RNA-binding proteins, such as spliceosome components in Saccharomyces cerevisiae (Peters and Huang, 2007) and Dictyostelium discoideum (Elchinger et al., 2005). The homopolymer regions have been assumed to serve some functional role in these protein classes, without any experimental evidence. A sequence similar to NTSSSHNNNNN is found in AtMYB77, but not in AtMYB73 or AtMYB70 (Fig. 1), suggesting that the asparagine-rich regions probably do not contribute to the catalytic activity of these proteins. We speculate that the expected high polarity of this region may provide high affinity to some other interacting regulatory protein(s), serving a functional specificity for AtMYB44.
For transgenic Arabidopsis, microarray analysis showed that in the absence of salt treatment, the overall gene expression patterns were not significantly different from those of wild-type plants (Jung et al., 2008). Thus, AtMYB44 overproduction in itself does not appear to be sufficient to regulate the expression of a group of specific target genes. Instead, this transcription factor may function either by working cooperatively with other transcription factors or through stimulus-induced structural modification. Indeed, the C-terminus activation domain of At-MYB77 interacts with auxin response factors (ARFs) in vitro, resulting in a strong reduction in lateral root numbers (Shin et al., 2007). Similarly, it appears that the functional specificity of AtMYB44 results from its interaction with other protein(s), not from the specificity of the binding sequence on the promoter regions of target genes. In relation to structural modification, it was recently reported that AtMYB44 is phosphorylated by the MAP kinases MPK3 and MPK6 to regulate seed germination (Nguyen et al., 2012).
The AtMYB44 gene is upregulated by a variety of hormone treatments, abiotic stresses and microbial infections (Jung et al., 2010; Kranz et al., 1998; Yanhui et al., 2006). AtMYB44 regulates diverse cellular responses mediated by abscisic acid (Jung et al., 2008), jasmonate (Jung et al., 2010) and ethylene (Liu et al., 2010; 2011; Lü et al., 2011). Our data suggest that functional diversity of AtMYB44 among the type I MYB transcription factors is not due to their binding specificity on a particular promoter sequence. Taken together, our data suggest that AtMYB44 is induced by diverse stimuli and acts as a common regulator that interacts directly or indirectly with other regulator(s) generated by each signal or stimulus, thus determining the target genes to be transcribed.
Acknowledgments
This work was supported by the Basic Science Research Program through the National Research Foundation (grant number 2012-0007030), by the Next Generation BioGreen 21 Research Program through the National Center for GM Crops of the Rural Development Administration (grant number PJ008073), and by the Technology Development Program for Life Industry through the Korea Institute of Planning and Evaluation for Technology of Food, Agriculture, Forestry and Fisheries (grant number 111076-5).
REFERENCES
- Das P.K., Shin D.H., Choi S.-B., Park Y.-I. Sugarhormone cross-talk in anthocyanin biosynthesis. Mol Cells. 2012;34:501–507. doi: 10.1007/s10059-012-0151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elchinger L., Pachebat J.A., Glöckner G., Rajandream M.-A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R.A., Schiestl R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K.M., Watson R.J. Nucleotide preferences in sequence-specific recognition of DNA by c-myc protein. Nucleic Acids Res. 1991;19:3913–3919. doi: 10.1093/nar/19.14.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K.M., Reakes C.F.L., Watson R.J. Characterization of the sequence-specific interaction of mouse c-myb protein with DNA. EMBO J. 1990;9:161–169. doi: 10.1002/j.1460-2075.1990.tb08092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999;41:577–585. doi: 10.1023/a:1006319732410. [DOI] [PubMed] [Google Scholar]
- Jung C., Seo J.S., Han S.W., Koo Y.J., Kim C.H., Song S.I., Nahm B.H., Choi Y.D., Cheong J.-J. Overexpression of AtMYB44 enhances stomata closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008;146:623–635. doi: 10.1104/pp.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C., Shim J.S., Seo J.S., Lee H.Y., Kim C.H., Choi Y.D., Cheong J.-J. Non-specific phytohormonal induction of AtMYB44 and suppression of jasmonate-responsive gene activation in Arabidopsis thaliana. Mol Cells. 2010;29:71–76. doi: 10.1007/s10059-010-0009-z. [DOI] [PubMed] [Google Scholar]
- Kim M.-J., Lee T.-H., Pahk Y.-M., Kim Y.-H., Park H.-Mi, Choi Y.D., Nahm B.H., Kim Y.-K. Quadruple 9-mer-based protein binding microarray with DsRed-monomer fusion protein. Methods Mol. Biol. 2009;786:65–77. doi: 10.1007/978-1-61779-292-2_4. [DOI] [PubMed] [Google Scholar]
- Kim M.-J., Chung P.J., Lee T.-H., Kim T.-H., Nahm B.H., Kim Y.-K. Convenient determination of protein-binding DNA sequences using quadruple 9-mer-based microarray and DsRed fusion protein. BMC Mol. Biol. 2012;10:91. doi: 10.1007/978-1-61779-292-2_4. [DOI] [PubMed] [Google Scholar]
- Kranz H.D., Denekamp M., Greco R., Jin H., Leyva A., Meissner R.C., Petroni K., Urzainqui A., Bevan M., Martin C., et al. To-wards functional characterization of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- Liu R., Lü B., Wang X., Zhang C., Zhang S., Qian J., Chen L., Shi H., Dong H. Thirty-seven transcription factor genes differentially respond to a harpin protein and affect resistance to the green peach aphid in Arabidopsis. J. Biosci. 2010;35:435–450. doi: 10.1007/s12038-010-0049-8. [DOI] [PubMed] [Google Scholar]
- Liu R., Chen L., Jia Z., Lü B., Shi H., Shao W., Dong H. Transcription factor AtMYB44 regulates induced expression of the ETHYLENE INSENSITIVE2 gene in Arabidopsis responding to a harpin protein. Mol. Plant Microbe Interact. 2011;24:377–389. doi: 10.1094/MPMI-07-10-0170. [DOI] [PubMed] [Google Scholar]
- Lü B., Sun W., Zhang S., Zhang C., Qian J., Wang X., Gao R., Dong H. HrpN Ea-induced deterrent effect on phloem feeding of the green peach aphid Myzus persicae requires AtGSL5 and AtMYB44 genes in Arabidopsis thaliana. J. Biosci. 2011;36:123–137. doi: 10.1007/s12038-011-9016-2. [DOI] [PubMed] [Google Scholar]
- Martin C., Paz-Ares J. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/s0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- Meissner R.C., Jin H., Cominelli E., Denekamp M., Fuertes A., Greco R., Kranz H.D., Penfield S., Petroni K., Urzainqui A., et al. Function search in a large transcription factor gene family in Arabidopsis: assessing the potential of reverse genetics to identify insertional mutations in R2R3 MYB genes. Plant Cell. 1999;11:1827–1840. doi: 10.1105/tpc.11.10.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen X.C., Hoang M.H.T., Kim H.S., Lee K., Liu X.-M., Kim S.H., Bahk S., Park H.C., Chung W.S. Phosphorylation of the transcriptional regulator MYB44 by mitogen activated protein kinase regulates Arabidopsis seed germination. Biochem. Biophys. Res. Commun. 2012;423:703–708. doi: 10.1016/j.bbrc.2012.06.019. [DOI] [PubMed] [Google Scholar]
- Ogata K., Morikawa S., Nakamura H., Sekikawa A., Inoue T., Kanai H., Sarai A., Ishii S., Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- Oh N.I., Kim Y.K., Nahm B.H., Cheong J.-J. Quadruple 9-mer-based protein binding microarray analysis of the Arabidopsis transcription factor AtMYB77. J. Korean Soc. Appl. Biol. Chem. 2012;55 (in press). [Google Scholar]
- Peters T.W., Huang M. Protein aggregation and polyasparagine-mediated cellular toxicity in Saccharomyces cerevisiae. Prion. 2007;1:144–153. doi: 10.4161/pri.1.2.4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C.W.B., Sippel A.E., Vingron M., Klempnauer K.-H. Drosophila and vertebrate myb proteins share two conserved regions, one of which functions as a DNA-binding domain. EMBO J. 1987;6:3085–3090. doi: 10.1002/j.1460-2075.1987.tb02616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay R.G., Ishii S., Gonda T.J. Interaction of the Myb protein with specific DNA binding sites. J. Biol. Chem. 1992;267:5656–5662. [PubMed] [Google Scholar]
- Romero I., Fuertes A., Benito M.J., Malpica J.M., Leyva A., Paz-Ares J. More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 1998;14:273–284. doi: 10.1046/j.1365-313x.1998.00113.x. [DOI] [PubMed] [Google Scholar]
- Rosinsky J.A., Atchley W.R. Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J. Mol. Evol. 1998;46:74–83. doi: 10.1007/pl00006285. [DOI] [PubMed] [Google Scholar]
- Seo J.S., Sohn H.B., Noh K., Jung C., An J.H., Donovan C.M., Somers D.A., Kim D.I., Jeong S.-C., Kim C.-G., et al. Expression of the Arabidopsis AtMYB44 gene confers drought/salt-stress tolerance in transgenic soybean. Mol. Breed. 2012;29:601–608. [Google Scholar]
- Shin R., Burch A.Y., Huppert K.A., Tiwari S.B., Murphy A.S., Guilfoyle T.J., Schachtman D.P. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19:2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R., Nieto C., Avila J., Cañas L., Diaz I., Paz-Ares J. Dual DNA-binding specificity of petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J. 1995;14:1773–1784. doi: 10.1002/j.1460-2075.1995.tb07166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stober-Grässer U., Brydolf B., Bin X., Grässer F., Firtel R.A., Lipsick J.S. The Myb DNA-binding domain is highly conserved in Dictyostelium discoideum. Oncogene. 1992;7:589–596. [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Yanhui C., Xiaoyuan Y., Kun H., Meihua L., Jigang L., Zhaofeng G., Zhiqiang L., Yunfei Z., Xiaoxiao W., Xiaoming Q., et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]