Abstract
Anthocyanins, a class of flavonoids, are recognized for their diverse functions in plant development and beneficial effects on human health. Many of the genes encoding anthocyanin biosynthesis enzymes and the transcription factors that activate or repress them have been identified. Regulatory proteins that control anthocyanin biosynthesis by regulating the expression of different structural genes at the transcriptional and post-transcriptional levels are differentially modulated by environmental and biological factors such as light, temperature, sugar and hormones. This minireview summarizes the recent findings contributing to our understanding of the role of sugars and hormones in the modulation of the anthocyanin biosynthesis pathway with emphasis on the coordinated regulation of the critical transcriptional R2R3-MYB/bHLH/WD40 (MBW) complex in Arabidopsis.
Keywords: anthocyanin, hormone, light, MBW complex, sugar
INTRODUCTION
Anthocyanins are ubiquitous flavonoid pigments found in most plant organs that play important roles in attracting pollinators and seed distributors, and protect the plant from pathogens, herbivores and environmental stresses such as UV-B light (Steyn et al., 2002). Anthocyanin biosynthesis is mainly regulated at the transcriptional level via a set of transcription factors including basic helix-loop-helix (bHLH), Leu-zipper, MADS-box, R2R3-MYB, WD40, WIP and WRKY factors (Marles et al., 2003). In Arabidopsis, the phytochrome (PHY)-interacting transcription factor 3 (PIF3), a bHLH protein, interacts directly with PHYs and positively regulates anthocyanin biosynthesis (Kim et al., 2006; Shin et al., 2007). LONG HYPOCOTYL5 (HY5), a Leu-zipper transcription factor (TF), serves as a point of convergence for phytochrome (PHY) and cryptochrome (CRY) signalings (Gyula et al., 2003), functioning as a positive regulator of anthocyanin biosynthesis (Chattopadhyay et al., 1998). It binds directly to the promoters of early biosynthesis genes (EBGs) such as chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H) and flavonoid 3′-hydroxylase (F3′H), which are common to different flavonoid subpathways, and late biosynthesis genes (LBGs) such as dihydroflavonol 4-reductase (DFR), leucoanthocyanidin oxygenase (LDOX), anthocyanidin reductase (ANR) and UDP-glucose: flavonoid 3-O-glucosyltransferase (UF3GT) (Lee et al., 2007; Shin et al., 2007). Transcription factors from the R2R3-MYB, bHLH and WD40 classes interact to form MBW regulatory complexes (Borevitz et al., 2000; Gonzalez et al., 2008; Tohge et al., 2005; Zhang et al., 2003). For instance, TRANSPARENT TESTA 2 (TT2; MYB123) requires the cofactors TT8 (bHLH42) and TTG1 (WD40-repeat protein) to form a ternary MBW complex that regulates the expression of the DFR and ANR genes, which are involved in the accumulation of proanthocyanidins in the seed coat (Baudry et al., 2004; Nesi et al., 2000; 2001). Similarly, MYB75/PAP1 (PRODUCTION OF ANTHOCYANIN PIGMENT1) and MYB90/PAP2, which are members of the R2R3-MYB family, are responsible for bHLH co-factor-dependent transcriptional activation of phenylalanine-ammonia lyase (PAL), CHS, DFR and glutathione-S-transferase (GST), which are key enzymes of the anthocyanin biosynthesis pathway (Hichri et al., 2011). Contrary to the positive TFs, MYBL2, an R3 MYB protein, acts as a negative regulator of anthocyanin biosynthesis in response to environmental cues, such as high light and, presumably, nitrogen deficiency. It interacts with TT8 to form a transcriptional inhibitory complex, MYBL2/bHLH/TTG1 (L2BW). Anthocyanin biosynthesis in specific tissues and organs is therefore likely to be regulated by the balance between MBW and L2BW complexes (Dubos et al., 2008; Matsui et al., 2008).
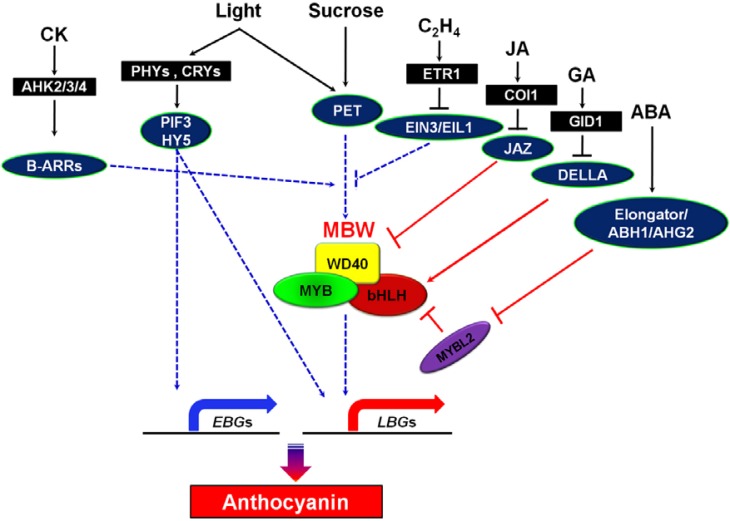
Application of exogenous sugars and hormones can affect the expression of anthocyanin biosynthesis genes that is dependent on the presence of light. However, the signaling pathways triggered by sugars, hormones and light and the interactions between these signals, if any, are not well understood. A recent finding that the photosynthetic electron transport (PET) chain, in addition to the well-characterized HY5-dependent signaling, plays an important role in light signaling pathways in green, vegetative leaf tissues (Das et al., 2011; Jeong et al., 2010) demonstrates the complexity of the cross-talk between various pathways. Sugar-induced anthocyanin biosynthesis is apparently modulated by the heterotrimeric M(L2)BW complexes that are under the regulation of hormones (Das et al., 2012; Jeong et al., 2010; Loreti et al., 2008; Shan et al., 2009), as schematically highlighted in Fig. 1. The present review summarizes the recent progress in our understanding of the interactions between sugars and hormones, primarily cytokinin (CK), ethylene, jasmonic acid (JA), abscisic acid (ABA) and gibberellic acid (GA), and their role in the light-dependent regulation of anthocyanin formation.
Fig. 1.
Schematic representation of the crosstalk between sucrose and hormone signaling pathways in the regulation of the expression of early (EBG) and late (LBG) anthocyanin biosynthesis genes through the activation of the MYB-bHLH-WD40 (MBW) transcription complex and down-regulation of MYBL2 expression. The point(s) of convergence of the photosynthetic electron transport (PET) dependent light, sucrose, CK, and ethylene signaling pathways leading to transcriptional regulation of the MBW complex remain elusive. DELLA-independent GA regulation of PAP1 expression is not shown. The HY5-mediated light signaling pathway directly regulates the expression of EBGs and LBGs at the transcriptional level. Blue and red lines pointing to anthocyanin biosynthesis genes (EBGs and LBGs) and MBW complex represent transcriptional and translational control, respectively.
SUGAR INDUCTION OF ANTHOCYANIN PIGMENTATION
In addition to their role as energy sources, sugars are important signaling molecules involved in the growth and development of higher plants (Finkelstein and Gibson, 2002; Smeekens, 2000), and their signaling pathways have been studied extensively in free-living microorganisms such as cyanobacteria (Lee et al., 2007; Ryu et al., 2004; 2008), bacteria (Stulke and Hillen, 1999), and yeast (Rolland et al., 2006). In Arabidopsis, anthocyanin production in cotyledons or leaves increases when seedlings are grown in a sugar-containing medium (Mita et al., 1997; Ohto et al., 2001). A similar phenomenon has been reported in radish hypocotyls (Hara et al., 2003) and grape cells (Larronde et al., 1998). In grape berry skin, sucrose (Suc) acts as an endogenous trigger, modulating the expression of anthocyanin biosynthesis genes (Boss et al., 1996). The role of Suc was analyzed in detail in a study that examined the transcript levels of sugar responsive genes in Arabidopsis (Solfanelli et al., 2006). Exogenous Suc increased the transcript levels of the LBGs DFR, LDOX and UF3GT by several hundred-fold, while the transcripts levels of EBGs acting upstream of the DFR in anthocyanin biosynthesis, including CHI, CHS, and C4H, showed lower induction by Suc. This Suc effect on the induction of anthocyanin biosynthetic genes may be attributed to the greater than 2-fold upregulation of positive TFs such as GL3, TT8 and PAP1 concurrent with the 3.3-fold downregulation of the negative transcription factor MYBL2 (Jeong et al., 2010). Under these conditions, the active MBW complex would be dominant over the negative L2BW complex.
In plants, sugar specific signaling pathways are activated by different disaccharides such as Suc and maltose (Mal), or breakdown products such as glucose (Glc) and fructose (Fru). The activation of a hexokinase (HXK)-dependent pathway by Glc in Arabidopsis is mediated by the AtHXK1 protein, although a separate, HXK-independent pathway regulates the expression of specific genes including CHS and PAL1 (Smeekens, 2000; Xiao et al., 2000). A Suc-specific sensing pathway has been proposed, although a putative Suc sensor protein has not been identified. Suc-specific pathways are not or only partially activated by the Suc breakdown products Glc and Fru, or by other sugars (Teng et al., 2005). Suc- or Mal-dependent anthocyanin accumulation (Solfanelli et al., 2006) is mediated either by Suc transporters (SUCs) or proteins closely associated with SUCs, rather than by a membrane-bound hexose transporter or the activity of HXK1, an internal glucose (Glc) sensor. This view was supported by a recent report showing that suc1-defective mutants grown in 3% but not in 5% Suc-containing growth medium had diminished anthocyanin accumulation (Sivitz et al., 2008). However, SUC1 expression and anthocyanin accumulation are spatially separated; SUC1 is expressed preferentially in the roots (Sivitz et al., 2008), while anthocyanin accumulates predominantly in the sub-epidermal cell layers of leaves (Jeong et al., 2010; Kubo et al., 1999). Furthermore, SUC1 expression and anthocyanin accumulation differ with respect to sugar specificity; anthocyanin synthesis is preferentially induced by the metabolizable disaccharides Suc and Mal, while SUC1 expression can be induced effectively by monosaccharide sugars such as Glc and Fru in addition to Suc and Mal (Jeong et al., 2010). There are no apparent differences between the induction of SUC1 expression by sugars and the osmoticums mannitol and palatinose (Jeong et al., 2010), indicating that SUC1 expression requires the presence of an osmotic sensor rather than membrane or intracellular receptors. AtSUC1 seems to play a role in Suc uptake rather than acting as a sugar sensor for anthocyanin production. How Suc is sensed in shoots therefore remains unclear.
Recently, Jeong et al. (2010) reported that light is required for Suc-induction of anthocyanin accumulation. Furthermore, the stimulatory effect of Suc was abolished in the light signaling mutants hy1, cry1/2, and hy5 treated with the photosynthetic electron transport inhibitor, 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), which functions at the QB binding site of photosystem II, but not by 2,5-dibromo-3-methyl-6-isopropyl-pbenzoquinone [DBMIB, an inhibitor of plastoquinone (PQ) oxidation]. These findings suggested that the PQ pool plays a role in sugar and light signaling associated with anthocyanin accumulation, as DCMU keeps the PQ pool oxidized and DBMIB reduces it. Therefore, sugar signaling is highly likely to be mediated by the redox state of the photosynthetic electron transport, which awaits further studies (Das et al., 2011).
INTERACTION OF SUGAR WITH CK
The stimulatory effect of CK on anthocyanin accumulation has been reported in several plants including carrot, artichoke, Haplopappus graciliss, garden balsam, rose and rape (Nakamura et al., 1980). In Arabidopsis, CK is a positive regulator of the anthocyanin biosynthesis pathway and functions by modulating the expression of the anthocyanin biosynthesis genes PAL1, CHI, CHS and DFR (Deikman and Hammer, 1995). Transgenic plants overexpressing the isopentenyltransferase (IPT) gene, which is associated with CK biosynthesis, showed a greater accumulation of anthocyanin than wild-type plants in the presence of light and Suc (Guo et al., 2005). Interestingly, CK- and sugar-induced anthocyanin accumulation is strictly linked to light signaling pathways (Cominelli et al., 2008). Anthocyanin accumulation is not detected in dark grown seedlings, although the Arabidopsis mutant increased chalcone synthase expression 1 (icx1) exhibited higher accumulation of anthocyanin than wild-type plants in the presence of light (Wade et al., 2003). However, several studies have questioned the role of CK as a positive regulator of anthocyanin production. Indeed, Loreti et al. (2008) showed that treatment with 10 μM 6-benzylaminopurine (BA), a synthetic CK, does not affect sugar-induced anthocyanin accumulation. The addition of BA to non-chlorophyllous corn (Zea mays L.) failed to influence Suc induction of anthocyanin accumulation (Kim et al., 2006).
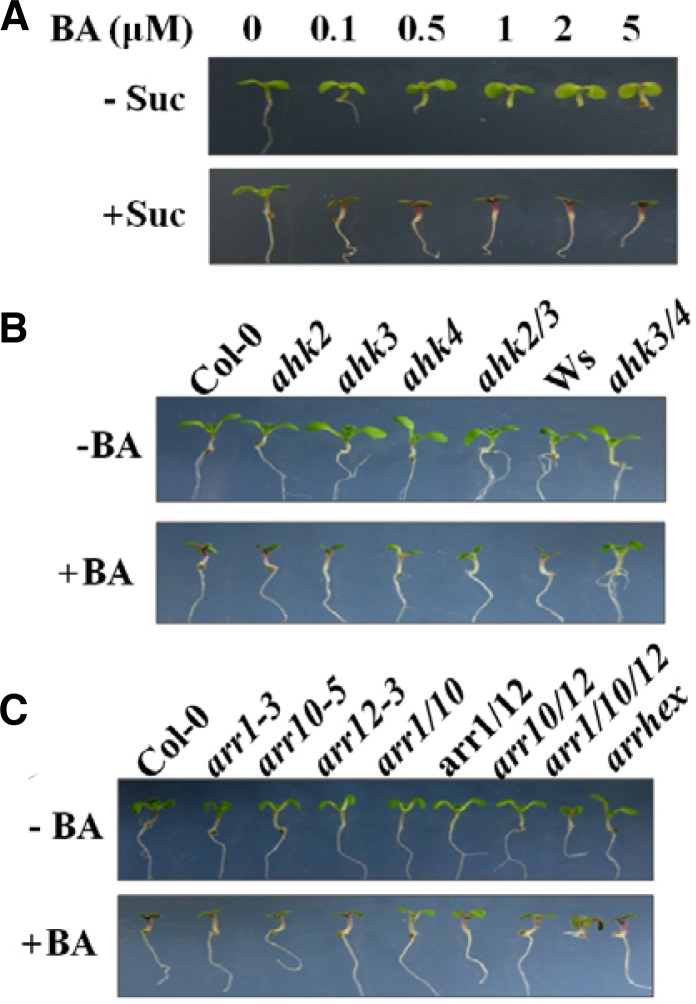
CK signaling, which involves a phosphorelay mechanism similar to the bacterial two-component system (Ferreira and Kieber, 2005), is mediated by sensor His kinases (AHKs), His-containing phosphotransfer proteins (AHPs), and type-B and -A response regulators (ARRs) (Hwang and Sheen, 2001). AHKs are positive and functionally overlapping regulators of CK signaling (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006). Several Arabidopsis CK signaling pathway mutants have been used to investigate the role of CK in the regulation of anthocyanin accumulation. The double mutants ahk2/3 and ahk3/4 showed significant reduction of anthocyanin accumulation in response to treatment with Suc and CK (Das et al., 2012; Figs. 2A and 2B). The type-B ARRs are redundant positive regulators of CK signaling and are the immediate upstream activators of type-A ARR gene expression (Argyros et al., 2008; Ishida et al., 2008). Of 11 known type-B ARRs in Arabidopsis, the subfamily 1 contains 7 ARRs that are associated with CK signaling (Mason et al., 2005). Suc-induced anthocyanin accumulation was significantly reduced in the single mutants arr1, arr10, and arr12, the double mutants arr1/10, arr1/12, and arr10/12, and the triple mutant arr1/10/12 treated with CK, indicating that ARR1, 10 and 12 act as positive regulators of CK signaling (Fig. 2C; Das et al., 2012). Furthermore, it turns out that CK enhances Suc induction of anthocyanin accumulation by affecting the PET signaling pathway via transcriptional activation of the positive regulators PAP1, (E)GL3 and TT8, and by repressing the MYBL2 transcript level (Fig. 1) (Das et al., 2012).
Fig. 2.
CK enhances sucrose-mediated anthocyanin pigmentation predominantly via AHK2/3/4 and type-B ARRs. (A) Col-0 seedlings were grown in 1/2-strength MS media supplemented with various 6-benzylaminopurine (BA) concentrations (0, 0.1, 0.5, 1, 2, and 5 μM) with (+Suc) or without (-Suc) 90 mM sucrose (Suc). (B) Wild-type Col-0 or Ws or single and double CK receptor mutants (ahk2, ahk3, ahk4, ahk2/3, and ahk3/4) or (C) ARR (arr1-3, arr10-5, arr12-1, arr1/10, arr1/12, arr10/12, arr1/10/12, and arrhex (arr3/4/5/6/8/9)) defective mutant seedlings were grown in 1/2 strength MS media supplemented with 90 mM Suc alone (−BA) or with 0.5 μM BA (+BA). Arabidopsis seedlings were grown for 7 days at a light intensity of 140 μmol m−2 s−1.
INTERACTION OF SUGAR WITH ETHYLENE
Ethylene has been associated with the increase in the soluble sugar contents of ripening fruit and the synthesis of flavonoids (Lelievre et al., 1997). Treatment of grape berries with the ethylene-releasing compound 2-chloroethylphosphonic acid activated the transcription of structural genes encoding the key enzymes of anthocyanin biosynthesis and increased anthocyanin accumulation (El-Kereamy et al., 2003). Ethylene also plays a negative regulatory role in anthocyanin biosynthesis. Anthocyanin accumulation in red cabbage grown in the dark and etiolated cabbage was markedly suppressed by ethylene (Craker and Wetherbee, 1973; Kang and Burg, 1973) and the inhibition of ethylene biosynthesis by cobalt increased anthocyanin accumulation in corn (Rengel and Kordan, 1987). Transgenic tobacco transformed with the ethylene receptor gene was used to demonstrate the role of ethylene as a negative regulator of anthocyanin biosynthesis. Anthocyanin accumulation was increased in the petals of tobacco plants over-expressing the mutated ethylene receptor gene, ethylene response 1 (ETR1H69A) (Takada et al., 2005). In contrast, constitutive triple response (ctr1) mutants grown in the presence of high levels of Suc had similar levels of anthocyanin than wild-type plants (Gibson et al., 2001).
In Arabidopsis, ethylene suppresses anthocyanin accumulation by binding to redundant receptors such as ETR1, ETR2, ERS1 and ERS2, with ETR1 possibly playing a dominant regulatory role. Slightly reduced anthocyanin levels in ctr1 loss-of-function mutants suggest that CTR1 may function immediately downstream of or within the ethylene receptor complex. EIN2, which acts downstream of CTR1 and functions in a wide range of ethylene responses in plants, is involved in the ethylene signaling pathway. EIN3 and its homologue EIL1 appear to function redundantly in the ethylene pathway, as anthocyanin accumulation is increased significantly in the ein3 eil1 double mutant but not in the ein3-1 or eil1-3 single mutants. Therefore, the effect of ethylene on the suppression of sugar-induced anthocyanin biosynthesis likely involves the ethylene triple response (Jeong et al., 2010).
In light-grown Arabidopsis, ethylene repression of anthocyanin accumulation is regulated at the transcriptional level, as demonstrated by the down-regulation of positive TFs (bHLHs such as GL3, EGL3 and TT8 and MYBs including PAP1 and PAP2) and the up-regulation of the negative TF MYBL2 when ethylene signaling was inhibited. In contrast, ethylene signaling has little effect on the mRNA levels of TTG1, which encodes an essential regulator of anthocyanin accumulation (Das et al., 2011; Jeong et al., 2010). Thus, ethylene seems to suppress the activity of the MBW complex through the transcriptional regulation of bHLHs and MYBs (Fig. 1). However, whether the ethylene sensitive components of the M(L2)BW complex are directly regulated by EIN3/EIL1 transcription factors needs further study.
INTERACTION OF SUGAR WITH JA
JAs, which include jasmonic acid and its cyclopentane precursors as well as cyclopentenones, modulate anthocyanin accumulation (Loreti et al., 2008). In the absence of sugar, JA treatment failed to induce the accumulation of anthocyanin and the DFR transcript, indicating that JA synergistically modulates sugar-induced anthocyanin biosynthesis by regulating the expression of the DFR gene (Loreti et al., 2008). The F-box protein Coronatine Insensitive-1 (COI1), which forms the SCFCOI1 complex with ASK1/ASK2, Cullin1, and Rbx1, is involved in diverse JA responses (Xu et al., 2002), most of which are disrupted in the Arabidopsis mutant coronatine insensitive 1 (coi1). A coi1-1 mutant with a premature stop codon at W467 failed to accumulate anthocyanin in response to methyl JA (Reymond and Farmer, 1998; Xie et al., 1998). Analysis of several transcript levels in coi1-1 mutants showed reduced expression of three anthocyanin regulatory factors, PAP1, PAP2 and GL3, which transcriptionally regulate the LBGs DFR, LDOX and UF3GT (Devoto et al., 2005).
The JA-ZIM-domain (JAZ) family proteins, which are substrates of the SCFCOI1 complex, consist of 12 members and function as negative regulators of JA responses, probably by directly inhibiting various transcriptional regulators (Xie et al., 1998; Xu et al., 2002). Protein-protein interaction studies revealed that JAZs directly interact with bHLHs (TT8, GL3, and EGL3) and R2R3-MYB TFs (PAP1 and GL1) in the MBW complex (Fig. 1; Qi et al., 2011). Recently, a model describing the COI1 regulation of JAZ proteins was proposed. Upon perception of the JA signal, COI1 recruits JAZs to the SCFCOI1 complex for ubiquitination and subsequent degradation by the 26S proteasome, which triggers the release of the MBW complexes to activate JA-induced anthocyanin biosynthesis.
INTERACTION OF SUGAR WITH GA
GA acts as an antagonist of Suc-induced anthocyanin biosynthesis. The R2R3-MYB transcription factors PAP1 and PAP2, which are Suc-dependent modulators of anthocyanin biosynthesis, were repressed by GA3 (Loretti et al., 2008). The genes encoding enzymes that function downstream of naringenin were up-regulated in ga1-5 mutant seedlings. The ga1-5 mutant has a deficiency in the step catalyzing the conversion of geranylgeranyl pyrophosphate (GGPP) to copalyl pyrophosphate (CPP), and therefore low endogenous GA levels. The low endogenous GA concentrations in ga1-5 activated the expression of genes involved in anthocyanin biosynthesis, thus enhancing the accumulation of anthocyanin in the mutant plants (Loreti et al., 2008).
The DELLA proteins GAI, RGA, RGL1, RGL2 and RGL3 are negative regulators of GA signaling that act immediately downstream of the GA receptor in Arabidopsis. DELLA proteins repress stem elongation and are important negative regulators of seed germination. Binding of GA to its soluble receptor GID1 causes binding of GID1-GA to DELLAs and leads to their degradation via the ubiquitin-proteasome pathway (Sun and Gubler, 2004). The function of DELLA proteins in anthocyanin accumulation was investigated in gai mutants, which are characterized by a dark green and dwarf phenotype (Jiang et al., 2007). The gai mutation affects GA sensing or subsequent signal transduction, and does not result in GA deficiency. The transcript levels of the DFR gene in gai mutants were similar to those of the wild type in plants treated with GA in the presence of Suc (Loreti et al., 2008), suggesting that GA modulates sugar-induced anthocyanin biosynthesis by regulating the activity of DELLA proteins (Fig. 1). In DELLA protein quadruple mutants (gai-t6 rga-t2 rgl1-1 rgl2-1), the expression of anthocyanin biosynthesis genes such as LDOX and F3’H is GA-DELLA dependent, while the expression of UF3GT and PAP1 is independent of DELLA proteins (Jiang et al., 2007). Because GA signaling is dependent on the activity of the SCFSLY1 E3 ubiquitin ligase, the negative effects of GAs on anthocyanin biosynthesis may be mediated by the degradation of the DELLA proteins.
GA has also been implicated in the low temperature induction of anthocyanin biosynthesis, as GA2ox1 transcript upregulation significantly inactivated GA in a HY5/HYH-dependent manner (Zhang et al., 2011), thereby establishing the prime role of light in GA regulation of anthocyanin accumulation.
INTERACTION OF SUGAR WITH ABA
ABA induces anthocyanin accumulation in many plants including maize kernels and grape (Kim et al., 2006; Mori et al., 2005; Paek et al., 1997). In maize, viviparous-1 (vp1), an ABA insensitive mutant that fails to express the C1 gene (a MYB transcription factor similar to PAP1 and PAP2), shows no accumulation of anthocyanin (McCarty et al., 1989). In Arabidopsis seedlings, although ABA alone could induce PAP2, the hormone showed a synergistic effect with Suc on anthocyanin accumulation and caused a significant increase in the transcript levels of biosynthesis genes including CHS, DFR, LDOX, UF3GT, and C4H, among others (Loreti et al., 2008; Tonelli et al., 2007). However, the expression of the DFR gene in the ABA-deficient mutant, aba1-3, which is impaired in the oxidation of zeaxanthin and antheraxanthin to violaxanthin, and in the ABA-insensitive abi1-1 mutant did not affect anthocyanin accumulation, irrespective of the presence of sugar. This result suggests that ABA is not strictly required for Suc-induced anthocyanin biosynthesis (Loreti et al., 2008).
ABA signaling is also associated with Elongator, a histone acetyl-transferase complex that was first identified by its direct association with RNA polymerase-II (Chen et al., 2006). Elongator consists of six subunits and its mutant phenotype is characterized by narrow leaves, reduced root growth, ABA hypersensitivity and increased accumulation of anthocyanins (Zhou et al., 2009). Elongator mutations have a negative effect on the expression of anthocyanin biosynthesis pathway genes, probably by the direct regulation of the expression of the MYBL2 gene during transcription elongation (Fig. 1; Zhou et al., 2009). Thus, ABA may exert its effect by regulating mRNA stability, as evident in the mutants defective in the mRNA-cap-binding protein ABA hypersensitive-1 (ABH1) (Hugouvieux et al., 2001) and poly(A)-specific ribonuclease (AtPARN) AHG2 (Arabidopsis hypersensitive germination 2) (Nishimura et al., 2005).
CONCLUSIONS
Sugar signaling associated with anthocyanin accumulation is linked to several signaling pathways such as those triggered by light, CK, ethylene, JA, and ABA, which might be interconnected with Ca2+ signaling by as yet unknown mechanisms (Vitrac et al., 2000). Although light is known to be a crucial factor for the hormonal regulation of anthocyanin biosynthesis, the details of the interactions between sugar, light and hormones remain to be elucidated. For instance, epistatic interactions between the MBW complex and the photoreceptor signaling component HY5 should be addressed, as some LBGs are regulated by both PAP1 and HY5 in the presence of light, and this is further enhanced by sugars (Jeong et al., 2010). Anthocyanin accumulation is a cell autonomous process; anthocyanin accumulates in non-photosynthetic organs and tissues, including the lower epidermis and trichomes (Lee and Collins, 2001), while light signals for anthocyanin induction seem to originate in photosynthetic cells in response to exogenously supplied sugars (Das et al., 2011; Jeong et al., 2010). The identification of the entities involved in cell to cell communication between acyanic and cyanic cells is therefore challenging (Das et al., 2011; Jeong et al., 2010). Substantial progress has been made regarding the regulation of anthocyanin genes by the MBW complex, and the physical interaction with repressors such as MYBL2 has been proposed. However, recent results indicated that miR156-regulated SPL9 may function in the regulation of the MWB complex by binding to PAP1 (Guo et al., 2011). Future studies should be aimed at the identification of such additional factors that are under the influence of sugars or other stimuli and could be involved in the transcriptional and post-translational regulation of PAP1. Furthermore, the interaction between different hormones also needs to be considered, and several genetic mutants have been isolated that may help clarify the effect of hormones on anthocyanin biosynthesis. For example, high ABA levels in the ein2 mutant might be responsible for the high anthocyanin accumulation in these plants, demonstrating the dual regulation of ABA and ethylene signaling (Wang et al., 2007). New screening strategies coupled with proteomic and genomic techniques might further clarify the regulation of anthocyanin biosynthesis.
Acknowledgments
This work was supported by Grant PJ8205 from the Next-Generation BioGreen 21 Program, Rural Development Administration and Grant 2011-0031344 from the Advanced Biomass Research and Development Center, Republic of Korea.
REFERENCES
- Ahmad M., Cashmore A.R. The blue-light receptor cryptochrome 1 shows functional dependence on phytochrome A or phytochrome B in Arabidopsis thaliana. Plant J. 1997;11:421–427. doi: 10.1046/j.1365-313x.1997.11030421.x. [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero F., Arroyo A., Zhou L., Sheen J., Leon P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 2000;14:2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Argyros R.D., Mathews D.E., Chiang Y.H., Palmer C.M., Thibault D.M., Etheridge N., Argyros D.A., Mason M.G., Kieber J.J., Schaller G.E. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell. 2008;20:2102–2116. doi: 10.1105/tpc.108.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E., Rolland F., Thevelein J.M., Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- Borevitz J.O., Xia Y., Blount J., Dixon R.A., Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss P.K., Davies C., Robinson S.P. Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol. Biol. 1996;32:565–569. doi: 10.1007/BF00019111. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S., Puente P., Deng X.W., Wei N. Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 1998;15:69–77. doi: 10.1046/j.1365-313x.1998.00180.x. [DOI] [PubMed] [Google Scholar]
- Chen Z., Zhang H., Jablonowski D., Zhou X., Ren X., Hong X., Schaffrath R., Zhu J.K., Gong Z. Mutations in ABO1/ELO2, a subunit of holo-Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol. Cell. Biol. 2006;26:6902–6912. doi: 10.1128/MCB.00433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Cominelli E., Gusmaroli G., Allegra D., Galbiati M., Wade H.K., Jenkins G.I., Tonelli C. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J. Plant Physiol. 2008;165:886–894. doi: 10.1016/j.jplph.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Craker L.E., Wetherbee P.J. Ethylene, light, and anthocyanin synthesis. Plant Physiol. 1973;51:436–438. doi: 10.1104/pp.51.3.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P.K., Bang G., Choi S.-B., Yoo S.D., Park Y.-I. Photosynthesis-dependent anthocyanin pigmentation in Arabidopsis. Plant Signal. Behav. 2011;6:1–4. doi: 10.4161/psb.6.1.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P.K., Shin D.H., Choi S.-B., Yoo S.D., Choi G., Park Y.-I. Cytokinins enhance sugar-induced anthocyanin biosynthesis in Arabidopsis. Mol Cells. 2012;34:93–101. doi: 10.1007/s10059-012-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deikman J., Hammer P.E. Induction of anthocyanin accumulation by cytokinins in Arabidopsis thaliana. Plant Physiol. 1995;108:47–57. doi: 10.1104/pp.108.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A., Ellis C., Magusin A., Chang H.S., Chilcott C., Zhu T., Turner J.G. Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol. Biol. 2005;58:497–513. doi: 10.1007/s11103-005-7306-5. [DOI] [PubMed] [Google Scholar]
- Dubos C., Le Gourrierec J., Baudry A., Huep G., Lanet E., Debeaujon I., Routaboul J.M., Alboresi A., Weisshaar B., Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008;55:940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- El-Kereamy A., Chervin C., Roustan J.P., Cheyhnier V., Souquet J.M., Moutounet M., Raynal J., Ford C., Latché A., Pech J.C., et al. Exogenous ethylene stimulates the long term expression of genes related to anthocyanin biosynthesis in grape berries. Physiol. Plant. 2003;119:175–282. [Google Scholar]
- Ferreira F.J., Kieber J.J. Cytokinin signaling. Curr. Opin. Plant Biol. 2005;8:518–525. doi: 10.1016/j.pbi.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R., Gibson S.I. ABA and sugar interactions regulating development: Cross-talk or voices in a crowd? Curr. Opin. Plant Biol. 2002;5:26–32. doi: 10.1016/s1369-5266(01)00225-4. [DOI] [PubMed] [Google Scholar]
- Gibson S.I., Laby R.J., Kim D. The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem. Biophys. Res. Commun. 2001;280:196–203. doi: 10.1006/bbrc.2000.4062. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- Gray J., Picton S., Shabbeer J., Schuch W., Grierson D. Molecular biology of fruit ripening and its manipulation with antisense genes. Plant Mol. Biol. 1992;19:69–87. doi: 10.1007/BF00015607. [DOI] [PubMed] [Google Scholar]
- Guo J.C., Hu X.W., Duan R.J. Interactive effects of CKs, light and sucrose on the phenotypes and the syntheses of anthocyanins, lignins in cytokinin over-producing transgenic Arabidopsis. J. Plant Growth Regul. 2005;24:93–101. [Google Scholar]
- Guo J.-Y., Felippes F.F., Liu C.-J., Weigel D., Wang J.-W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23:1512–1522. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyula P., Schafer E., Nagy F. Light perception and signaling in higher plants. Curr. Opin. Plant Biol. 2003;6:446–452. doi: 10.1016/s1369-5266(03)00082-7. [DOI] [PubMed] [Google Scholar]
- Hara M., Oki K., Hoshino K., Kuboi T. Enhancement of anthocyanin biosynthesis by sugar in radish (Raphanus sativus) hypocotyl. Plant Sci. 2003;164:259–265. [Google Scholar]
- Hichri I., Barrieu F., Bogs J., Kappel C., Delrot S., Lauvergeat V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011;62:2465–2483. doi: 10.1093/jxb/erq442. [DOI] [PubMed] [Google Scholar]
- Higuchi M., Pischke M.S., Mahonen A.P., Miyawaki K., Hashimoto Y., Seki M., Kobayashi M., Shinozaki K., Kato T., Tabata S., et al. In planta functions of the Arabidopsis CK receptor family. Proc. Natl. Acad. Sci USA. 2004;101:8821–8826. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V., Kwak J.M., Schroeder J.I. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106:477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- Hwang I., Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- Ishida K., Yamashino T., Yokoyama A., Mizuno T. Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 2008;49:47–57. doi: 10.1093/pcp/pcm165. [DOI] [PubMed] [Google Scholar]
- Jeong S.W., Das P.K., Jeoung S.C., Song J.Y., Lee H.K., Kim Y.K., Kim W.J., Park Y.I., Yoo S.D., Choi S.B., et al. Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis thaliana. Plant Physiol. 2010;154:1515–1531. doi: 10.1104/pp.110.161869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Gao X., Liao L., Harberd N.P., Fu X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signalling pathway in Arabidopsis. Plant Physiol. 2007;145:1460–1470. doi: 10.1104/pp.107.103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B.G., Burg S.P. Role of ethylene in phytochrome induced anthocyanin biosynthesis. Planta. 1973;110:227–235. doi: 10.1007/BF00387635. [DOI] [PubMed] [Google Scholar]
- Kim J.S., Lee B.H., Kim S.H., Ok K.H., Cho K.Y. Response to environmental and chemical signals for anthocyanin biosynthesis in non-chlorophyllous corn (Zea mays L.) leaf. J. Plant Biol. 2006;49:16–25. [Google Scholar]
- Kubo H., Peeters A.J.M., Aarts M.G.M., Pereira A., Koornneef M. ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell. 1999;11:1217–1226. doi: 10.1105/tpc.11.7.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larronde F., Krisa S., Decendit A., Cheze C., Merillon J.M. Regulation of polyphenol production in Vitis vinifera cell suspension cultures by sugars. Plant Cell Rep. 1998;17:946–950. doi: 10.1007/s002990050515. [DOI] [PubMed] [Google Scholar]
- Lee D.W., Collins T.M. Phylogenetic and ontogenetic influences on the distribution of anthocyanins and betacyanins in leaves of tropical plants. Int. J. Plant Sci. 2001;162:1141–1153. [Google Scholar]
- Lee S., Ryu J.Y., Kim S.Y., Jeon J.H., Song J.Y., Cho H.T., Choi S.B., Marsac N.T., Park Y.-I. Transcriptional regulation of the respiratory genes in the cyanobacterium Synechocystis sp. PCC 6803 during the early response to glucose feeding. Plant Physiol. 2007;145:1019–1030. doi: 10.1104/pp.107.105023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelievre J.M., Tichit L., Dao P., Fillion L., Nam Y.W., Pech J.L., Latche A. Effects of chilling on the expression of ethylene biosynthetic gene in ‘Passe-crassane’ pears (Pyrus communis) fruits. Plant Mol. Biol. 1997;33:847–855. doi: 10.1023/a:1005750324531. [DOI] [PubMed] [Google Scholar]
- Loreti E., Povero G., Novi G., Solfanelli C., Alpi A., Perata P. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 2008;179:1004–1016. doi: 10.1111/j.1469-8137.2008.02511.x. [DOI] [PubMed] [Google Scholar]
- Marles M.A., Ray H., Gruber M.Y. New perspectives on proanthocyanidin biochemistry and molecular regulation. Phytochemistry. 2003;64:367–383. doi: 10.1016/s0031-9422(03)00377-7. [DOI] [PubMed] [Google Scholar]
- Mason M.G., Mathews D.E., Argyros D.A., Maxwell B.B., Kieber J.J., Alonso J.M., Ecker J.R., Schaller G.E. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell. 2005;17:3007–3018. doi: 10.1105/tpc.105.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K., Umemura Y., Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008;55:954–967. doi: 10.1111/j.1365-313X.2008.03565.x. [DOI] [PubMed] [Google Scholar]
- McCarty D.R., Carson C.B., Stinard P.S., Robertson D.S. Molecular analysis of viviparous-1: an abscisic acid-insensitive mutant of maize. Plant Cell. 1989;1:523–532. doi: 10.1105/tpc.1.5.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita S., Hirano H., Nakamura K. Negative regulation in the expression of a sugar-inducible gene in Arabidopsis thaliana-a recessive mutation causing enhanced expression of a gene for β-amylase. Plant Physiol. 1997;114:575–582. doi: 10.1104/pp.114.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Saito H., Goto-Yamamoto N., Kitayama M., Kobayashi S., Sugaya S., Gemma H., Hashizume K. Effects of abscisic acid treatment and night temperatures on anthocyanin composition in Pinot noir grapes. Vitis. 2005;44:161–165. [Google Scholar]
- Nakamura N., Nakamae H., Maekawa L. Effects of light and kinetin on anthocyanin accumulation in the petals of Rosa hybrid Hort cv. Ehigasa. Z Pflanzenphysiol. 1980;98:263–270. [Google Scholar]
- Nesi N., Debeaujon I., Jond C., Pelletier G., Caboche M., Lepiniec L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N., Jond C., Debeaujon I., Caboche M., Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura C., Ohashi Y., Sato S., Kato T., Tabata S., Ueguchi C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell. 2004;16:1365–1377. doi: 10.1105/tpc.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Kitahata N., Seki M., Narusaka Y., Narusaka M., Kuromori T., Asami T., Shinozaki K., Hirayama T. Analysis of ABA hypersensitive germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J. 2005;44:972–984. doi: 10.1111/j.1365-313X.2005.02589.x. [DOI] [PubMed] [Google Scholar]
- Ohto M., Onai K., Furukawa Y., Aoki E., Araki T., Nakamura K. Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol. 2001;127:252–261. doi: 10.1104/pp.127.1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek N.C., Lee B.M., Bai D.G., Cobb B.G., Magill C.W., Smith J.D. Regulatory roles of abscisic acid for anthocyanin synthesis in maize kernels. Maydica. 1997;42:385–391. [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell. 2011;23:1795–1814. doi: 10.1105/tpc.111.083261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengel Z., Kordan H.A. Effects of growth regulators on light-dependent anthocyanin production in Zea mays seedlings. Physiol Plant. 1987;69:511–519. [Google Scholar]
- Reymond P., Farmer E.E. Jasmonate and salicylate as global signals for defence gene expression. Curr. Opin. Plant Biol. 1998;1:404–411. doi: 10.1016/s1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- Riefler M., Novak O., Strnad M., Schmülling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development and cytokinin metabolism. Plant Cell. 2006;18:40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F., Baena-Gonzalez E., Sheen J. Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- Ryu J.Y., Song J.Y., Lee J.M., Jeong S.W., Chow W.S., Choi S.B., Pogson B.J., Park Y.-I. Glucose-induced expression of carotenoid biosynthesis genes in the dark is mediated by cytosolic pH in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 2004;279:25320–25325. doi: 10.1074/jbc.M402541200. [DOI] [PubMed] [Google Scholar]
- Ryu J.Y., Jeong S.W., Lim A.Y., Ko Y., Yoon S., Choi A.B., Park Y.-I. Cyanobacterial glucokinase complements the glucose sensing role of Arabidopsis thaliana hexokinase 1. Biochem. Biophys. Res. Commun. 2008;374:454–459. doi: 10.1016/j.bbrc.2008.07.041. [DOI] [PubMed] [Google Scholar]
- Shan X., Zhang Y., Peng W., Wang Z., Xie D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 2009;13:3849–3860. doi: 10.1093/jxb/erp223. [DOI] [PubMed] [Google Scholar]
- Shin J., Park E., Choi G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 2007;49:981–994. doi: 10.1111/j.1365-313X.2006.03021.x. [DOI] [PubMed] [Google Scholar]
- Sivitz A.B., Reinders A., Ward J.M. Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiol. 2009;147:92–100. doi: 10.1104/pp.108.118992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S. Sugar-induced signal transduction in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:49–81. doi: 10.1146/annurev.arplant.51.1.49. [DOI] [PubMed] [Google Scholar]
- Solfanelli C., Poggi A., Loreti E., Alpi A., Perata P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 2006;140:637–646. doi: 10.1104/pp.105.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn W.J., Wand S.J.E., Holcroft D.M., Jacobs G. Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol. 2002;155:349–361. doi: 10.1046/j.1469-8137.2002.00482.x. [DOI] [PubMed] [Google Scholar]
- Stulke J., Hillen W. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 1999;2:195–201. doi: 10.1016/S1369-5274(99)80034-4. [DOI] [PubMed] [Google Scholar]
- Sun T., Gubler F. Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 2004;55:197–223. doi: 10.1146/annurev.arplant.55.031903.141753. [DOI] [PubMed] [Google Scholar]
- Takada K., Ishimaru K., Minamisawa K., Kamada H., Ezura H. Expression of a mutated melon ethylene receptor gene Cm-ETR1/H69A affects stamen development in Nicotiana tabacum. Plant Sci. 2005;169:935–942. [Google Scholar]
- Teng S., Keurentjes J., Bentsink L., Koornneef M., Smeekens S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005;139:1840–1852. doi: 10.1104/pp.105.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- Tohge T., Nishiyama Y., Hirai M.Y., Yano M., Nakajima J., Awazuhara M., Inoue E., Takahashi H., Goodenowe D.B., Kitayama M., et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- Tonelli C., Cominelli E., Allegra D., Galbiati M. Plant tolerance to drought and salinity: modulation of transcription factors. Proceedings of the 18th International Conference on Arabidopsis Research. 2007;176 [Google Scholar]
- Vitrac X., Larronde F., Krisa S., Decendit A., Deffieux G., Merillon J.M. Sugar sensing and Ca2+-calmodulin requirement in Vitis vinifera cells producing anthocyanins. Phytochemistry. 2000;53:659–665. doi: 10.1016/s0031-9422(99)00620-2. [DOI] [PubMed] [Google Scholar]
- Wade H.K., Sohal A.K., Jenkins G.I. Arabidopsis ICX1 is a negative regulator of several pathways regulating flavonoid biosynthesis genes. Plant Physiol. 2003;131:707–715. doi: 10.1104/pp.012377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu C., Li K., Sun F., Hu H., Li X., Zhao Y., Han C., Zhang W., Duan Y., et al. Arabidopsis EIN2 modulates stress response through abscisic acid response pathway. Plant Mol. Biol. 2007;64:633–644. doi: 10.1007/s11103-007-9182-7. [DOI] [PubMed] [Google Scholar]
- Xiao W.Y., Sheen J., Jang J.C. The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol. Biol. 2000;44:451–461. doi: 10.1023/a:1026501430422. [DOI] [PubMed] [Google Scholar]
- Xie D., Feys B.F, James S., Nieto-Rostro M., Turner J.G. COI1: an Arabidopsis gene required for jasmonate-regulated defence and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. The SCF(COI1) ubiq-uitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell. 2002;14:1919–1935. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu Z., Liu R., Hao H., Bi Y. Gibberellins negatively regulate low temperature-induced anthocyanin accumulation in a HY5/HYH-dependent manner. Plant Signal. Behav. 2011;6:632–634. doi: 10.4161/psb.6.5.14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Qi J., Ren N., Cheng J., Erb M., Mao B., Lou Y. Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J. 2009;60:638–648. doi: 10.1111/j.1365-313X.2009.03988.x. [DOI] [PubMed] [Google Scholar]