Abstract
MicroRNAs are a class of small, endogenous, non-coding RNA molecules that negatively regulate gene expression at the transcriptional or the post-transcriptional level. Although a large number of miRNAs have been identified in many plant species, especially from model plants and crops, they remain largely unknown in peach. In this study, 110 potential miRNAs belonging to 37 families were identified using computational methods. A total of 43 potential targets were found for 21 families based on near-perfect or perfect complementarity between the plant miRNA and the target sequences. A majority of the targets were transcription factors which play important roles in peach development. qRT-PCR analysis of RNA samples prepared from different peach tissues for 25 miRNA families revealed that miRNAs were differentially expressed in different tissues. Furthermore, two target genes were experimentally verified by detection of the miRNA-mediated mRNA cleavage sites in peach using RNA ligase-mediated 5′ rapid amplification of cDNA ends (RLM-RACE). Finally, we studied the expression pattern of the two target genes in three different tissues of peach to further understand the mechanism of the interaction between miRNAs and their target genes.
Keywords: computational prediction, microRNA, peach, qRT-PCR, target
INTRODUCTION
MicroRNAs (miRNAs) are a large group of endogenous ∼21 nt small RNAs that play essential regulatory roles in various biological and metabolic processes, including development, signal transduction, cell fate identity, organ differentiation and stress responses by targeting messenger RNAs (mRNAs) for degradation or translational repression (Bartel, 2004; Carrington and Ambros, 2003; Jones-Rhoades et al., 2006). miRNA genes are transcribed by RNA polymerase II into long primary transcripts (pri-miRNAs) (Bartel, 2004; Carrington and Ambros, 2003). Following transcription and several post-transcriptional modifications using a set of Dicer-like enzymes, miRNA precursors (pre-miRNAs) and eventually mature miRNAs are generated (Schauer et al., 2002). Subsequently, mature miRNAs are incorporated into the RNA-induced silencing complex (RISC) to modulate the expression of target genes (Carrington and Ambros, 2003).
Recently, miRNAs have been identified by three methods: genetic screens, cloning and computational approaches. Although miRNAs are initially discovered using genetic screening technology (Lee et al., 1993; Wightman et al., 1993), this method is limited as it is expensive, time consuming and dominated by chance (Zhang et al., 2006b). Direct cloning of miRNAs, followed by construction and sequencing of a small RNA library, has been successfully employed to identify miRNAs in plants (Feng et al., 2009; Jian et al., 2010; Sunkar et al., 2005; Yao et al., 2007). However, only abundant miRNA genes can be easily detected by cloning or Northern blot. To find low-expression miRNA genes, computational prediction provides a convenient, valid strategy. A large number of miRNAs have been identified using computational approaches based on the high degree of conservation in plants (Dhandapani et al., 2011; Jones-Rhoades and Bartel, 2004; Lindow and Krogh, 2005; Zhang et al., 2006a).
miRNA regulation mechanisms include repression of translation (Aukerman and Sakai, 2003; Chen, 2004), cleavage of targeted mRNAs (Schwab et al., 2005; Sunkar et al., 2005) and chromatin modification (Jeong et al., 2011). The known miRNA targets have a high degree of complementarity to miRNAs and cleavage of the target mRNA typically occurs at the centre of the paired region, especially for plant miRNAs (German et al., 2008). This allows for the prediction of miRNA targets by computational approaches. Several studies have utilised this fact and identified many target genes in different plant species while allowing 1–3 nucleotide mismatches between the target mRNAs and miRNAs (Song et al., 2009; Wang et al., 2012; Zhang et al., 2008).
A growing number of miRNAs and their targets have been predicted and/or experimentally discovered in many plants; however, the miRNAs in peach are largely unknown despite a previous study reporting a few miRNAs in this species (Zhang et al., 2011). The systematic identification and characterisation of miRNAs in peach using the peach genome would help in identifying more miRNAs. We could better understand the genetic and morphological diversity of this species though analysing the functions of miRNAs. In addition, miRNA gene expression analyses have provided a good method to discern complex biological processes in plants. Peach is an economically important group of cultivated fruits and is considered one of the genetically most well-characterised species in the Rosaceae. The doubled haploid cultivar ‘Lovell’ Genome Sequencing Project was completed by employing the accurate Sanger methodology in 2010, which generated a 219 Mbp genome sequence and a 37.2 Mbp mRNAs sequence. There are a total of 202 scaffolds in this assembly of peach and are 26,938 CDS sequences (http://www.rosaceae.org/node/355). In this study, we predicted peach miRNAs and their targets by computational approaches using peach genome sequences. These potential miRNAs and their targets needed to be further validated and characterised by detecting and quantifying their expression in different tissues. A simple, accurate, special and sensitive method for miRNA detection and target expression profiling is in high demand. Here, we used a qRT-PCR-based method.
MATERIALS AND METHODS
Plant materials
Samples of young leaves, stems and flowers were collected from one-year old grafted ‘Lovell’ (Prunus persica) trees that were grown in the garden of Nanjing Agricultural University, China. After collection, all the samples were immediately frozen in liquid nitrogen and stored at −70°C for the following study.
Reference set of miRNA and peach genome sequences
A total of 2675 previously known mature plant miRNAs and their precursor sequences from 43 plant species were downloaded from the miRBase (Griffiths-Jones, 2006; Griffiths-Jones et al., 2008). Peach genome sequences and mRNA sequences were obtained from GDR (http://www.rosaceae.org/node/355).
Analysis software
A computer program, microHARVESTER (Dezulian et al., 2006), was used to identify potential miRNAs. The prediction of secondary structures and the stability of miRNA precursors were assessed by RNAfold (Hofacker et al., 1994) and Mfold-3.1.2 (Zuker, 2003).
Prediction of potential Prunus persica miRNAs (ppe-miRNAs) and targets
We used the microHARVESTER program to predict peach miRNAs. This approach, with excellent sensitivity and specificity, was based on a homology search followed by a set of structural filters. First, BLASTn (Altschul et al., 1997) and the Smith-Waterman pairwise alignment algorithm (Smith and Waterman, 1981) were employed to precisely determine the precursor and mature sequence candidates with a sensitive BLAST parameter setting (word-length 7 and E-value cutoff 10). We discarded those candidates whose aligned segments did not span most of the mature segment of the known precursor sequences and whose mature segments differed by more than four mistakes with a previously known mature miRNAs. Second, we used RNAfold to predict the minimal free energy structure of the candidate sequence. We discarded a candidate if more than six nucleotides of its miRNA* did not form bonds with its mature miRNA. Finally, Mfold was used to predict whether the remaining precursors had high negative minimal folding free energy (MFE), adjusted minimum folding free energy (AMFE) and a high minimal folding free energy index (MFEI) or not (Zhang et al., 2006c).
Previous studies have shown that all plant miRNAs mediate gene expression by targeting mRNA sequences at a perfect or near-perfect complementary site. This allowed the prediction of miRNA targets by computational approaches. To identify potential regulatory targets, we tested the 110 identified miRNAs against the peach mRNA sequence using a BLASTn search. Gaps were not allowed and G:U and other non-canonical pairs were treated as mismatches. The number of allowed mismatches at complementary sites between miRNA sequences and potential mRNA targets was no more than three. BLASTx was performed with the selected target transcripts to identify the functions of miRNAs.
Low molecular weight RNA extraction
Low molecular weight (LMW) RNA was independently isolated from different tissues by using CTAB reagent according to the procedure previously described by Wang et al. (2010). The concentration of RNA was measured by a UV-1800 spectro-photometer (Eppendorf, Germany) at 260 and 280 nm and visually ascertained on a 1.5% agarose gel.
Construction of small RNA cDNA libraries
Small RNAs were isolated from three peach tissues including young leaves, young stems and flowers. The small RNAs were polyadenylated using a poly(A) polymerase (NEB, USA) and then the poly(A)-tailed RNAs were recovered by phenol/chloroform extraction followed by ethanol precipitation. Reverse transcription was performed using MLV reverse transcriptase (Promega, USA), 1 μg of RT primers (Table 1) and 1 μg of poly(A)-tailed RNA to synthesise the small RNA cDNAs following the manufacturer’s instructions (Ro et al., 2006).
Table 1.
Primers used for qRT-PCR
| Name | Sequences (5′ → 3′) |
|---|---|
| ppe-miR1511 | AACCTGGCTCTGATACCATA |
| ppe-miR172b | GGAATCTTGATGATGCTGCAG |
| ppe-miR171b | TGATTGAGCCACGCCAAC |
| ppe-miR171d | TGATTGAGCCGTGCCAA |
| ppe-miR2275 | TTTAGTTTCCTCCAATATCTCA |
| ppe-miR398b | TGTGATCTCAGGTCACCCC |
| ppe-miR164c | TGGAGAAGGGGAGCACG |
| ppe-miR169g | TAGCCAAGGATGACTTGCC |
| ppe-miR403 | TTAGATTCACGCACAAACTC |
| ppe-miR396c | TTCCACAGCTTTCTTGAACT |
| ppe-miR408 | ATGCACTGCCTCTTCCCT |
| ppe-miR319f | TTGGACTGAAGGGAGCTC |
| ppe-miR395g | CTGAAGTGTTTGGGGGAAC |
| ppe-miR390 | AAGCTCAGGAGGGATAGC |
| ppe-miR156i | TGACAGAAGATAGAGAGCACAA |
| ppe-miR447 | ACTCTCCCTCAAGGGCT |
| ppe-miR164a | TGGAGAAGCAGGGCACG |
| ppe-miR535a | TGACGACGAGAGAGAGCAC |
| ppe-miR477 | CTCTCCCTCAAAGGCTTC |
| 5.8s rRNA forward primer | CTCGGCAACGGATATCTCG |
| 5.8s rRNA reverse primer | CTAATGGCTTGGGGCG |
| ppa005013m forward primer | TTGCTGATGGAGTGGAAT |
| ppa005013m reverse primer | TCTGCTGGTTGTAACTTCT |
| ppa005230m forward primer | TTCAACTCCTCCTCCAAC |
| ppa005230m reverse prime | ATGACGACGAAGAAGAAGA |
| PRII forward primer | TGAAGCATACACCTATGATGATGAAG |
| PRII reverse primer | CTTTGACAGCACCAGTAGATTCC |
| RT-primer | CCAGTAGCGTATGATGAGCACAGAGTCTGAGATCACTCGTAGCGAGG-d(T)33-V(A/C/G)N(A/C/G/T) |
| ppe-miR172a | AGAATCTTGATGATGCTGCATAA |
| ppe-miR171a | TGATTGAGCCGCGTCAAT |
| ppe-miR171c | AGATTGAGCCGCGCC |
| ppe-miR167b | TGAAGCTGCCAGCATGATCT |
| ppe-miR398a | TGTGTTCTCAGGCATCACAC |
| ppe-miR398c | TGTGTTCTCAGGTCGCC |
| ppe-miR169d | CAGCCAAGGATGACTTGC |
| ppe-miR169i | TGAGCCAAGAATGACTTGCT |
| ppe-miR396b | TTTCACAGCTTTCTTGAACTG |
| ppe-miR166e | GCGAACCAGACAGCATTC |
| ppe-miR482a | TCTTTCCGAAACCTCCC |
| ppe-miR395d | ATGAAGTGTTCAAGGGAACTC |
| ppe-miR160a | TGCCTGGTTCCCTGTATG |
| ppe-miR156e | TTGACAGAAGATAGAGAGCACA |
| ppe-miR827 | GTAGATGACCATAAACAAACAA |
| ppe-miR2118 | CTACCGATTCCACCCATTC |
| ppe-miR393a | TCCCAAGGGATCGCATCG |
| ppe-miR397 | TCATTGAGTGCAGCGTTGAT |
| URP | CCAGTAGCGTATGATGAGCA |
qRT-PCR analysis of peach miRNAs
The templates used for qRT-PCR were the miRNA-enriched cDNA libraries generated from young leaves, stems and flowers. A miRNA-specific primer and a universal reverse primer, URP, were used for real-time quantitative PCR (Table 1). For real-time PCR, cDNA was mixed with 2× SYBR Green Mix (Takara, Japan) and each of the miRNA specific primers and a universal reverse primer in a final volume of 20 μl. PCR runs were 40 cycles each at 95°C for 10 s, 60°C for 20 s and 72°C for 45 s. Each reaction was repeated three times. The relative miRNA expression was quantified using the comparative ΔΔCT method (Livak and Schmittgen, 2001). All expression profiles were normalised to expression levels in the stem. 5.8S rRNAs (Design, 2005), was used as an internal control. The primer sequences are shown in Table 1.
Validation of miRNA target genes using RLM-RACE
To find the internal cleavage site in ppa005013m (targeted by miR156) and ppa005230m (targeted by miR172), RLM-RACE was performed using the 5′-Full Race Kit (Takara, Japan). A modified procedure for RLM-RACE was carried out following the instruction of the kit without calf intestinal phosphatase and tobacco acid pyrophosphatase treatment. Total RNA was extracted from young leaves, young stems and flowers using the CTAB method. Poly (A)+ mRNA was purified from pooled tissue RNA using the Oligotex™ -dT30<Super> mRNA Purification Kit (Takara) according to the manufacturer’s instructions. The adapter was directly ligated to the mRNA, then first strand cDNA was synthesised in a reverse transcription reaction. An amplification step, the same as the one used for gene-specific RACE, was recommended by the manufacturer and included a 5′ nest primer and a 3′ gene specific primer. The primer sequences were as follows: 5′ RACE outer primer (CATGGCTACATGCTGACAGCCTA), 5′ RACE inner primer (CGCGGA TCCACAGCCTACTGATGATCAGTCGATG), ppa005013m 3′ outer primer (GAATTGGTTGGTTTGAGAACCAAAC), ppa005013m 3′ inner primer (GGTTGCTGCCATTACTGTGTGAGTG), ppa 005230m 3′ outer primer (AGCTCCTGCAATAGAAACCGGG TAT) and ppa005230m 3′ inner primer (CGATGGTGAAAA ATGGTGGTGGAGA). After amplification, the 5′ RACE products were gel-purified and cloned, and at least 10 independent clones were randomly chosen and sequenced.
The expression analysis of miRNA targets
Reverse transcription of the total RNA that was extracted from young leaves, young stems and flowers was performed using the PrimeScript® RT reagent Kit. cDNA was mixed with 2× SYBR Green Mix (Takara, Japan) and each of the miRNA target-specific primers in a final volume of 20 μl for qRT-PCR. PRII (Tong et al., 2009) was used as an internal control. The primer sequences are shown in Table 1.
RESULTS
Computational prediction of potential ppe-miRNAs
Since the beginning of abundant miRNA identification and an-notation, it has been well-recognised that miRNAs are evolutionarily conserved in plants and animals. Based on the conserved sequences and secondary structures, 290 potential candidates were selected through a computer program, micro-HARVESTER. Of these, 207 candidates had fewer than four mismatches with known mature miRNA sequences. After carefully evaluating the secondary structures using the criteria described in the Methods section, there remained 110 potential miRNAs (Table 2).
Table 2.
Details of identified ppe-miRNAs and structural information
| miRNA | Location | Mature miRNA | MAS | MN (nt) | ML (nt) | PL (nt) | (A+U)% | MFEs | AMFEs | MFEIs |
|---|---|---|---|---|---|---|---|---|---|---|
| ppe-miR156a | scaffold_7:22103897-22104005 | UGACAGAAGAGAGUGAGCAC | 5′ | 0 | 20 | 109 | 50.46 | 47.70 | 43.76 | 0.810 |
| ppe-miR156b | scaffold_1:33986381-33986484 | UGACAGAAGAGAGUGAGCAC | 5′ | 0 | 20 | 104 | 53.85 | 53.70 | 51.63 | 1.076 |
| ppe-miR156c | scaffold_7:19399614-19399720 | UGACAGAUAGAGAGAGAGCAC | 5′ | 2 | 21 | 107 | 57.94 | 52.20 | 48.79 | 1.084 |
| ppe-miR156d | scaffold_1:34819687-34819794 | UGACAGAUAGAGAGUAAGCAC | 5′ | 1 | 21 | 108 | 52.78 | 47.40 | 43.89 | 0.861 |
| ppe-miR156e | scaffold_3:10599623-10599738 | UUGACAGAAGAUAGAGAGCAC | 5′ | 0 | 21 | 116 | 52.59 | 48.30 | 41.64 | 0.757 |
| ppe-miR156f | scaffold_5:17453185-17453288 | UUGACAGAAGAUAGAGAGCAC | 5′ | 0 | 21 | 104 | 57.69 | 47.70 | 45.87 | 1.040 |
| ppe-miR156g | scaffold_3:21012691-21012815 | UUGGCAGAAGAAAAGAGAGCAC | 5′ | 3 | 22 | 125 | 60.80 | 49.00 | 39.2 | 0.800 |
| ppe-miR156h | scaffold_6:3280057-3280163 | UUGACAGAAGAAAGAGAGCAC | 5′ | 1 | 21 | 107 | 52.34 | 53.30 | 49.81 | 0.977 |
| ppe-miR156i | scaffold_3:10599079-10599182 | UGACAGAAGAUAGAGAGCACA | 5′ | 1 | 21 | 104 | 55.77 | 47.40 | 45.58 | 0.991 |
| ppe-miR159a | scaffold_2:18908762-18908950 | AUUGGAGUGAAGGGAGCUCC | 3′ | 2 | 20 | 189 | 52.91 | 92.10 | 48.73 | 0.548 |
| ppe-miR159b | scaffold_5:17562391-17562575 | UUUGGAUUGAAGGGAGCUCUA | 3′ | 1 | 21 | 185 | 51.89 | 78.70 | 42.54 | 0.478 |
| ppe-miR159c | scaffold_4:13455964-13456050 | CUUGGCUUGAAGGGAGCUCCG | 3′ | 2 | 21 | 87 | 48.28 | 24.40 | 28.05 | 0.623 |
| ppe-miR160a | scaffold_6:21902556-21902657 | UGCCUGGUUCCCUGUAUGCCA | 5′ | 1 | 21 | 102 | 50.00 | 45.30 | 44.41 | 0.871 |
| ppe-miR160b | scaffold_4:5325323-5325427 | UGCCUGGCUCCCUGUAUGCCA | 5′ | 0 | 21 | 105 | 46.67 | 63.80 | 60.76 | 1.085 |
| ppe-miR162 | scaffold_5:16885784-16885886 | UCGAUGAACCGCUGCCUCCAG | 3′ | 3 | 21 | 103 | 52.43 | 43.30 | 42.04 | 0.858 |
| ppe-miR164a | scaffold_6:26465850-26465969 | UGGAGAAGCAGGGCACGUGCA | 5′ | 0 | 21 | 120 | 53.33 | 60.00 | 50.00 | 0.893 |
| ppe-miR164b | scaffold_6:24710611-24710792 | UGGAGAAGCAGGGCACGUGCA | 5′ | 0 | 21 | 182 | 59.89 | 56.60 | 31.10 | 0.426 |
| ppe-miR164c | scaffold_8:21367800-21367915 | UGGAGAAGGGGAGCACGUGCA | 5′ | 3 | 21 | 116 | 50.86 | 53.80 | 46.38 | 0.814 |
| ppe-miR164d | scaffold_3:1784022-1784116 | UGGAGAGCUAGAGCACAUGCA | 5′ | 4 | 21 | 95 | 54.74 | 41.30 | 43.47 | 1.011 |
| ppe-miR166a | scaffold_8:19800532-19800631 | UCGGACCAGGCUUCAUUCCC | 3′ | 0 | 20 | 100 | 51.00 | 55.40 | 55.40 | 1.131 |
| ppe-miR166b | scaffold_5:12581627-12581786 | UCGGACCAGGCUUCAUUCCC | 3′ | 0 | 20 | 160 | 59.38 | 59.60 | 37.25 | 0.573 |
| ppe-miR166c | scaffold_2:26094634-26094793 | UCGGACCAGGCUUCAUUCCC | 3′ | 0 | 20 | 160 | 53.13 | 63.40 | 39.63 | 0.528 |
| ppe-miR166d | scaffold_2:19692915-19693064 | UCGGACCAGGCUUCAUUCCC | 3′ | 0 | 20 | 150 | 61.33 | 64.20 | 42.80 | 0.738 |
| ppe-miR166e | scaffold_1:2213219-2213318 | UCGAACCAGACAGCAUUCCC | 3′ | 4 | 20 | 100 | 53.00 | 49.50 | 49.50 | 1.053 |
| ppe-miR167a | scaffold_4:5610366-5610741 | UGAAGCUGCAAGAUGACCUG | 5′ | 4 | 20 | 376 | 69.95 | 102.20 | 27.18 | 0.241 |
| ppe-miR167b | scaffold_6:27656223-27656309 | UGAAGCUGCCAGCAUGAUCUG | 5′ | 0 | 21 | 87 | 58.62 | 43.00 | 49.43 | 1.373 |
| ppe-miR167c | scaffold_1:1563822-1563911 | UGAAGCUGCCAGCAUGAUCUU | 5′ | 1 | 21 | 90 | 56.67 | 45.60 | 50.67 | 1.299 |
| ppe-miR167d | scaffold_8:20014988-20015080 | UGAAGCUACCACAUGAUCUG | 5′ | 3 | 20 | 93 | 51.61 | 42.80 | 46.02 | 1.023 |
| ppe-miR169a | scaffold_3:19573376-19573494 | UAGCCAGAGACGACUUGCCGA | 5′ | 4 | 21 | 119 | 50.42 | 46.80 | 39.33 | 0.667 |
| ppe-miR169b | scaffold_4:16645761-16645853 | GAGCCAAGGAUGACUUGCCA | 5′ | 2 | 20 | 93 | 53.76 | 47.30 | 50.86 | 1.183 |
| ppe-miR169c | scaffold_4:16664507-16664599 | GAGCCAAGGAUGACUUGCCA | 5′ | 2 | 20 | 93 | 53.76 | 45.10 | 48.49 | 1.128 |
| ppe-miR169d | scaffold_4:16676120-16676220 | CAGCCAAGGAUGACUUGCCGG | 5′ | 3 | 21 | 101 | 53.47 | 47.00 | 46.53 | 0.990 |
| ppe-miR169e | scaffold_4:16648995-16649095 | CAGCCAAGGAUGACUUGCCGG | 5′ | 3 | 21 | 101 | 52.48 | 48.60 | 48.12 | 1.002 |
| ppe-miR169f | scaffold_3:21677982-21678091 | UAGCCAAGGAUGACUUGCCUG | 5′ | 0 | 21 | 110 | 50.91 | 43.60 | 39.64 | 0.734 |
| ppe-miR169g | scaffold_4:10137181-10137297 | UAGCCAAGGAUGACUUGCCUGC | 5′ | 0 | 22 | 117 | 53.85 | 45.20 | 38.63 | 0.715 |
| ppe-miR169h | scaffold_1:22425535-22425679 | UAGCCAAGGAGACUGCCUGU | 5′ | 3 | 20 | 145 | 51.03 | 55.80 | 38.48 | 0.542 |
| ppe-miR169i | scaffold_4:16691539-16691653 | UGAGCCAAGAAUGACUUGCUG | 5′ | 2 | 21 | 115 | 58.26 | 46.90 | 40.78 | 0.850 |
| ppe-miR171a | scaffold_3:16525609-16525709 | UGAUUGAGCCGCGUCAAUAUC | 3′ | 1 | 21 | 101 | 57.43 | 41.90 | 41.49 | 0.965 |
| ppe-miR171b | scaffold_3:21505900-21505996 | UGAUUGAGCCACGCCAACAUC | 3′ | 2 | 21 | 97 | 61.86 | 37.50 | 38.66 | 1.045 |
| ppe-miR171c | scaffold_7:21451283-21451383 | AGAUUGAGCCGCGCCAAUAUC | 3′ | 1 | 21 | 101 | 53.47 | 48.60 | 48.12 | 1.024 |
| ppe-miR171d | scaffold_3:16557260-16557354 | UGAUUGAGCCGUGCCAAUAUC | 3′ | 1 | 21 | 95 | 54.74 | 49.60 | 52.21 | 1.214 |
| ppe-miR171e | scaffold_1:32200679-32200763 | UGAUUGAGCCGUGCCAAUAUC | 3′ | 1 | 21 | 85 | 52.94 | 38.30 | 45.06 | 1.126 |
| ppe-miR172a | scaffold_2:20686356-20686484 | AGAAUCUUGAUGAUGCUGCAU | 3′ | 0 | 21 | 129 | 59.69 | 53.10 | 41.16 | 0.792 |
| ppe-miR172b | scaffold_2:22285766-22285872 | GGAAUCUUGAUGAUGCUGCAG | 3′ | 2 | 21 | 107 | 55.14 | 49.50 | 46.26 | 0.964 |
| ppe-miR172c | scaffold_6:4912768-4912951 | UGAAUCUUGAUGAUGCCGCAC | 3′ | 3 | 21 | 184 | 59.24 | 58.50 | 31.79 | 0.424 |
| ppe-miR319a | scaffold_1:29856933-29857147 | UUGGACUGAAGGGAGCUCCU | 3′ | 1 | 20 | 215 | 51.63 | 102.90 | 47.86 | 0.460 |
| ppe-miR319b | scaffold_2:23738870-23739096 | UUGGACUGAAGGGAGCUCCUC | 3′ | 1 | 20 | 227 | 57.71 | 88.40 | 38.94 | 0.406 |
| ppe-miR319c | scaffold_2:18914576-18914767 | UUGGAUUGAAGGGAGCUCCA | 3′ | 2 | 20 | 192 | 48.96 | 100.40 | 52.29 | 0.534 |
| ppe-miR319d | scaffold_5:17220685-17220922 | UUGGACUGAAGGGAGCUCCC | 3′ | 0 | 20 | 238 | 49.16 | 111.20 | 46.72 | 0.386 |
| ppe-miR319e | scaffold_6:2304460-2304649 | UUGGACUGAAGGGAGCUCCC | 3′ | 0 | 20 | 190 | 56.84 | 78.30 | 41.21 | 0.503 |
| ppe-miR319f | scaffold_8:17944925-17945032 | UUGGACUGAAGGGAGCUCUCA | 3′ | 2 | 21 | 108 | 50.93 | 40.40 | 37.41 | 0.706 |
| ppe-miR390a | scaffold_6:24551362-24551453 | AAGCUCAGGAGGGAUAGCGCC | 5′ | 0 | 21 | 92 | 54.35 | 44.30 | 48.15 | 1.146 |
| ppe-miR390b | scaffold_6:1728631-1728735 | AAGCUCAGGAGGGAUAGCGCC | 5′ | 0 | 21 | 105 | 61.90 | 48.50 | 46.19 | 1.155 |
| ppe-miR393a | scaffold_2:25056675-25056781 | UCCCAAGGGAUCGCAUCGAUCC | 5′ | 2 | 22 | 107 | 57.94 | 39.50 | 36.92 | 0.820 |
| ppe-miR393b | scaffold_2:22650086-22650180 | UCCAAAGGGAUCGCAUUGAUCC | 5′ | 0 | 22 | 95 | 56.84 | 40.80 | 42.95 | 1.047 |
| ppe-miR394a | scaffold_1:43600159-43600272 | UUGGCAUUCUGUCCACCUCCAU | 5′ | 0 | 22 | 114 | 58.77 | 49.60 | 43.51 | 0.926 |
| ppe-miR394b | scaffold_1:32136128-32136229 | UUGGCAGUAUGCCCACCUCCAC | 5′ | 3 | 22 | 102 | 53.92 | 37.50 | 36.76 | 0.782 |
| ppe-miR395a | scaffold_1:26767775-26767862 | CUGAAGUGUUUGGGGGGACC | 3′ | 1 | 20 | 88 | 52.27 | 37.80 | 42.95 | 1.023 |
| ppe-miR395b | scaffold_1:26799377-26799487 | CUGAAGUGUUUGGGGGGACC | 3′ | 1 | 20 | 111 | 55.86 | 43.80 | 39.46 | 0.805 |
| ppe-miR395c | scaffold_1:26765063-26765158 | AUGAAGUGAGUGAGGGAACUC | 3′ | 4 | 21 | 96 | 57.29 | 42.60 | 44.38 | 1.082 |
| ppe-miR395d | scaffold_1:26805384-26805488 | AUGAAGUGUUCAAGGGAACUC | 3′ | 4 | 21 | 105 | 59.05 | 39.40 | 37.52 | 0.873 |
| ppe-miR395e | scaffold_1:26780279-26780383 | AUGAAGUGUUCAAGGGAACUC | 3′ | 4 | 21 | 105 | 59.05 | 35.20 | 33.52 | 0.780 |
| ppe-miR395f | scaffold_1:26764880-26764975 | AUGAAGUGUUCAAGGGAACUC | 3′ | 4 | 21 | 96 | 55.21 | 43.40 | 45.21 | 1.051 |
| ppe-miR395g | scaffold_1:26806562-26806681 | CUGAAGUGUUUGGGGGAACUC | 3′ | 1 | 21 | 120 | 60.00 | 50.60 | 42.17 | 0.878 |
| ppe-miR395h | scaffold_1:26748442-26748561 | CUGAAGUGUUUGGGGGAACUC | 3′ | 1 | 21 | 120 | 60.00 | 38.30 | 31.92 | 0.665 |
| ppe-miR396a | scaffold_7:21479083-21479229 | UUCCCACAGCUUUAUUGAACCG | 5′ | 4 | 22 | 147 | 54.42 | 51.60 | 35.10 | 0.524 |
| ppe-miR396b | scaffold_1:39280926-39281045 | UUUCACAGCUUUCUUGAACUGU | 5′ | 2 | 22 | 120 | 58.33 | 55.90 | 46.58 | 0.932 |
| ppe-miR396c | scaffold_7:21474174-21474288 | UUCCACAGCUUUCUUGAACUU | 5′ | 0 | 21 | 115 | 56.52 | 51.30 | 44.61 | 0.892 |
| ppe-miR397 | scaffold_4:1619240-1619325 | UCAUUGAGUGCAGCGUUGAUG | 5′ | 1 | 21 | 86 | 62.79 | 42.60 | 49.53 | 1.548 |
| ppe-miR398a | scaffold_1:27542748-27542857 | UGUGUUCUCAGGCAUCACACCUU | 3′ | 4 | 23 | 110 | 57.27 | 55.40 | 50.36 | 1.072 |
| ppe-miR398b | scaffold_4:22864986-22865110 | UGUGAUCUCAGGUCACCCCUGU | 3′ | 3 | 22 | 125 | 42.40 | 76.50 | 61.20 | 0.85 |
| ppe-miR398c | scaffold_4:23714196-23714310 | UGUGUUCUCAGGUCGCCCCUG | 3′ | 2 | 21 | 115 | 46.09 | 51.20 | 44.52 | 0.718 |
| ppe-miR399a | scaffold_4:3161730-3161872 | UGCCAAAGGAGUAAUUGCCCAG | 3′ | 2 | 22 | 143 | 55.24 | 50.50 | 35.31 | 0.552 |
| ppe-miR399b | scaffold_4:3186380-3186502 | UGCCAAAGGAGAAUUGCCCUG | 3′ | 0 | 21 | 123 | 60.98 | 60.50 | 49.19 | 1.025 |
| ppe-miR399c | scaffold_4:3187835-3187944 | UGCCACUAGAGAAUUGCCCUG | 3′ | 3 | 21 | 110 | 51.82 | 49.80 | 45.27 | 0.854 |
| ppe-miR399d | scaffold_4:3192972-3193111 | UGCCAGAGGAGACUUUGCCCUG | 3′ | 3 | 22 | 140 | 56.43 | 51.40 | 36.71 | 0.602 |
| ppe-miR399e | scaffold_3:4392605-4392744 | UGCCAGAGGAGACUUUGCCCUG | 3′ | 3 | 22 | 140 | 55.00 | 55.20 | 39.43 | 0.626 |
| ppe-miR399f | scaffold_3:541261-541369 | UGCCAAAGAAGAGUUGCCCUA | 3′ | 2 | 21 | 109 | 54.13 | 51.30 | 47.06 | 0.941 |
| ppe-miR399g | scaffold_3:584777-584885 | UGCCAAAGAAGAGUUGCCCUA | 3′ | 2 | 21 | 109 | 53.21 | 48.50 | 44.50 | 0.872 |
| ppe-miR399h | scaffold_3:574913-575021 | UGCCAAAGAAGAGUUGCCCUA | 3′ | 2 | 21 | 109 | 54.13 | 50.00 | 45.87 | 0.917 |
| ppe-miR399i | scaffold_1:46720492-46720600 | UGCCAAAGAAGAGUUGCCCUA | 3′ | 2 | 21 | 109 | 54.13 | 54.20 | 49.72 | 0.994 |
| ppe-miR399j | scaffold_5:9980708-9980803 | UGCCAAUGGAGAGACGCCCUA | 3′ | 4 | 21 | 96 | 53.13 | 35.90 | 37.40 | 0.831 |
| ppe-miR399k | scaffold_4:3179684-3179797 | UGCCAAAGGAGAAUUGCCGUG | 3′ | 1 | 21 | 114 | 58.77 | 34.30 | 30.09 | 0.640 |
| ppe-miR403 | scaffold_1:8677734-8677847 | UUAGAUUCACGCACAAACUCG | 3′ | 0 | 21 | 114 | 54.39 | 44.80 | 39.30 | 0.756 |
| ppe-miR408 | scaffold_10:245026-245130 | AUGCACUGCCUCUUCCCUGGC | 3′ | 2 | 21 | 105 | 48.57 | 46.60 | 44.38 | 0.822 |
| ppe-miR414 | scaffold_7:21519316-21519594 | UCAUCAUCAUCAUCAUCGUCU | 5′ | 2 | 21 | 279 | 47.67 | 121.50 | 43.55 | 0.298 |
| ppe-miR447 | scaffold_5:11892465-11892595 | ACUCUCCCUCAAGGGCUUCUCAG | 5′ | 3 | 23 | 131 | 54.96 | 55.90 | 42.67 | 0.723 |
| ppe-miR477 | scaffold_5:11892655-11892736 | CUCUCCCUCAAAGGCUUCUA | 5′ | 1 | 20 | 82 | 54.88 | 43.30 | 52.80 | 1.427 |
| ppe-miR482a | scaffold_1:29646074-29646169 | UCUUUCCGAAACCUCCCAUUCC | 3′ | 3 | 22 | 96 | 65.63 | 32.20 | 33.54 | 1.016 |
| ppe-miR482b | scaffold_1:29651603-29651699 | CCUACUCCACCCAUUCC | 3′ | 1 | 17 | 97 | 55.67 | 43.70 | 45.05 | 1.048 |
| ppe-miR482c | scaffold_3:10579299-10579395 | UCUUCCCAAGCCCGCCCAUUCC | 3′ | 1 | 22 | 97 | 62.89 | 42.50 | 43.81 | 1.217 |
| ppe-miR535a | scaffold_8:17685868-17685968 | UGACGACGAGAGAGAGCACGC | 5′ | 1 | 21 | 101 | 48.51 | 61.90 | 61.29 | 1.179 |
| ppe-miR535b | scaffold_8:17689597-17689697 | UGACAACGAGAGAGAGCACGC | 5′ | 0 | 21 | 101 | 48.51 | 62.30 | 61.68 | 1.186 |
| ppe-miR538 | scaffold_4:4636437-4636570 | UUGCAUGCAGUCUAUGUCUGGG | 5′ | 2 | 22 | 134 | 61.19 | 36.70 | 27.39 | 0.527 |
| ppe-miR827 | scaffold_7:22580971-22581076 | GUAGAUGACCAUAAACAAACA | 3′ | 2 | 21 | 106 | 70.75 | 35.20 | 33.21 | 1.071 |
| ppe-miR858 | scaffold_5:17626942-17627247 | UCUCGUUGUCUGUUCGACCUU | 5′ | 1 | 21 | 306 | 66.67 | 67.30 | 21.99 | 0.216 |
| ppe-miR1030 | scaffold_6:7773855-7774079 | UCUGCAUUUGCACCUGCACUU | 5′ | 3 | 21 | 225 | 59.11 | 76.30 | 33.91 | 0.369 |
| ppe-miR1446 | scaffold_3:21872535-21872639 | UUCUUAACUCUCUCCCUCAUA | 5′ | 2 | 21 | 105 | 59.05 | 44.00 | 41.90 | 0.975 |
| ppe-miR1511 | scaffold_3:8575412-8575509 | AACCUGGCUCUGAUACCAUA | 3′ | 2 | 20 | 98 | 63.27 | 41.30 | 42.14 | 1.17 |
| ppe-miR2111 | scaffold_4:5129206-5129292 | UAAUCUGCAUCCUGAGGUUUA | 5′ | 0 | 21 | 87 | 51.72 | 52.80 | 60.69 | 1.445 |
| ppe-miR2118 | scaffold_1:29644637-29644733 | CUACCGAUUCCACCCAUUCCGA | 3′ | 2 | 22 | 97 | 58.76 | 39.90 | 41.13 | 1.028 |
| ppe-miR2275 | scaffold_3:19748539-19748641 | UUUAGUUUCCUCCAAUAUCUCA | 3′ | 1 | 22 | 103 | 60.19 | 47.00 | 45.63 | 1.113 |
| ppe-miR3627 | scaffold_3:19985286-19985440 | UGGUCGCAUAGCGACGGCACU | 5′ | 4 | 21 | 155 | 43.23 | 55.20 | 35.61 | 0.405 |
| ppe-miR3629a | scaffold_4:14155746-14155846 | GGCUGCCGAGAAAGUGUGGGA | 5′ | 3 | 21 | 101 | 53.47 | 27.00 | 26.73 | 0.659 |
| ppe-miR3629a | scaffold_6:6832326-6832628 | GGUUGCUGAGAAAAUGCAGGA | 5′ | 2 | 21 | 303 | 61.72 | 84.30 | 27.82 | 0.240 |
| ppe-miR3629a | scaffold_6:6555645-6555802 | GGUUGAUGAGAAAAUGAAGGA | 5′ | 3 | 21 | 158 | 70.25 | 39.10 | 24.75 | 0.527 |
| ppe-miR3629a | scaffold_2:19091880-19092047 | GGCUGCUGAGAAAUCUGGGA | 5′ | 3 | 20 | 168 | 64.29 | 48.80 | 29.05 | 0.484 |
| ppe-miR3629a | scaffold_5:12208693-12208905 | GGUUACUGAGAAAAUGAAGGA | 5′ | 3 | 21 | 213 | 63.38 | 71.40 | 33.52 | 0.430 |
| ppe-miR3629a | scaffold_8:14952639-14952764 | GGUUGCUGAGAAAAUGGAGGA | 3′ | 2 | 21 | 126 | 57.94 | 27.20 | 21.59 | 0.407 |
| ppe-miR3629a | scaffold_7:14342466-14342736 | GGUUGCUGAGAAAGUGUGGGA | 3′ | 3 | 21 | 271 | 53.14 | 88.00 | 30.66 | 0.241 |
MAS, mature miRNA arm sided in hairpin secondary structure; MN, mismatch number; ML, mature miRNA length; PL, precursor miRNA length; MFE, minimum folding free energy; AMFE, adjusted minimum folding free energy; MFEI, minimum folding free energy index.
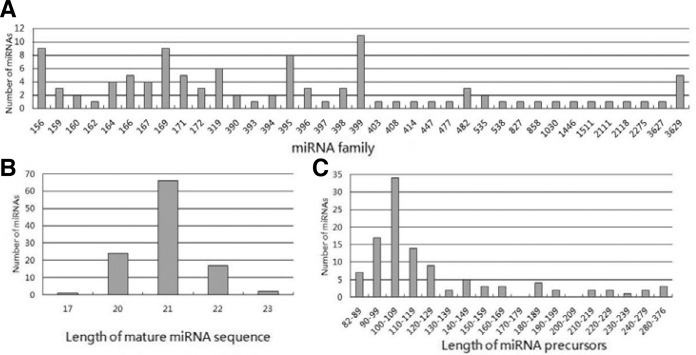
The 110 predicted peach miRNAs belong to 37 miRNA families. The largest miRNA family size identified was miR399 that consisted of 11 members. miR156, miR169 and miR395 possessed nine, nine and eight members, respectively, whereas, 17 miRNA families had only one member identified in this study (Fig. 1A). The length of the mature miRNAs ranged from 17 to 23 nt (Table 2 and Fig. 1B). The majority of miRNAs were 21 nt long (60.0%) followed by 20 (21.8%), 22 (15.5%), 23 (1.8%) and 17 (0.9%) (Fig. 1B).
Fig. 1.
Analysis of potential miRNAs in peach (A) miRNA family in peach (B) Size distribution of miRNAs in peach (C) Number of peach miRNAs against different lengths of mature miRNAs.
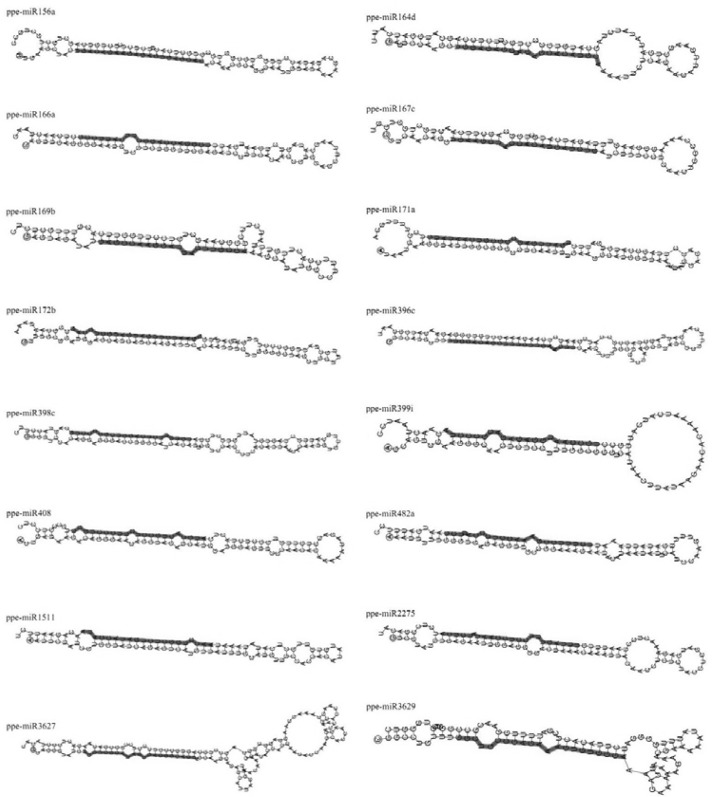
Following peach miRNA identification, diversity could be found not only in the numbers within each miRNA family, but also in other aspects, such as the location of the mature miRNAs and the length of miRNA precursors. The 49.1% of the mature miRNA sequences were located at the 5′-end of the miRNA precursor sequence, with the others at the 3′-end. The length of the miRNA precursors varied from 82 to 376 nucleotides with an average of 128.6. However, the majority of the identified miRNAs (73.6%) had 82–129 nucleotides, with more than half of the miRNAs (59.1%) at 90–119 nucleotides (Fig. 1C). Compared with animal miRNAs, which have a consistent nucleotide length (∼70–80 nt) (Ambros, 2004; Bartel, 2004), the length of plant (including peach) miRNA precursors varies. Although all identified ppe-miRNAs had similar predicted stem-loop hairpin structures, their hairpin shapes varied due to differences in length (Fig. 2). The percentage AU content ranged from 42.4% to 70.75% with an average of 55.68% (Table 2). Several studies have found that miRNA precursors have low folding free energy, and have suggested that low free energy is an important characteristic of miRNAs (Dhandapani et al., 2011; Zhang et al., 2006b). The MFE of the 110 identified ppe-miRNAs ranged from −121.5 to −24.4 kcal/mol with an average of −52.82 kcal/mol. However, minimal folding free energy depends on the length of the RNA sequence (Seffens and Digby, 1999). Thus, to avoid the effect of using minimal folding free energy as the one and only criterion to identify new miRNAs (Adai et al., 2005), MFEI was used to distinguish miRNAs from other non-coding and coding RNAs. The MFEI values in our study for precursor miRNA sequences ranged from 0.216 to 1.548 with an average of 0.835.
Fig. 2.
Stem-loop hairpin structures of representative miRNA families. Red indicates mature miRNA sequences.
Potential targets of peach miRNA
In this study, we identified a total of 43 potential target genes, involved in different biological functions, for the 21 identified miRNA families in peach based on the fact that miRNAs perfectly or near-perfectly complement their target sequences in peach. We could not identify the target genes for the following 16 miRNA families: ppe-miR 2275, 398, 169, 403, 3627, 396, 482, 390, 827, 1446, 414, 447, 535, 162, 538, 477 and 858. The majority of these miRNA targets were various transcription factor genes including SBP, MYB, ARF, NAC, AP2, PHB, F-box, GRAS and PPR that are known to regulate plant development. Some miRNA targets included the inorganic phosphate transporter (miR399), the sulphate transporter (miR395) and laccase (miR397). Other targets were uncharacterised (miR408) and hypothetical proteins (miR1511, miR3629, miR1030).
The identified target genes were conserved in several plants, including the squamosa promoter-binding-like (SPL) genes of miR156, the MYB domain containing gene of miR159, the NAC-domain containing gene of miR164, HD-ZipIII transcription factors of miR166, auxin responsive factors (ARFs) of miR160, scarecrow-like transcription factor of miR171, AP2 domain-containing transcription factor of miR172 and laccase of miR397 (Table 3). This analysis revealed that the majority of target transcripts were highly correlated with plant development and metabolic processes. Several of the well-annotated target transcripts such as MYB, NAC1, PHB and ARFs have putative functions involved in floral organ formation. miR164 and miR166 are root-associated miRNAs that regulate the NAC and HD-ZIP transcription factor genes, respectively. HD-ZIP proteins also regulate vascular development as well as lateral organ polarity and meristem formation. miR156, which targets eight squamosa promoter binding protein-like (SPL) transcription factor genes, is involved in flowering time modulation and leaf morphogenesis. miR159 and miR319 both target MYB which is involved in flower development. Auxin responsive factors (ARFs) were also a class of targets of miRNA160. ARFs are important components of auxin signal transduction. Unfortunately, the function of the targets of miR1511, miR3629, miR408, miR2118 and miR1030 are currently unknown.
Table 3.
Potential targets of identified peach miRNAs
| miRNA family | Targeted genes | Targeted protein | Target function |
|---|---|---|---|
| miR156 | ppa024285m | Probable receptor-like protein kinase | Adjust protein kinase activity |
| ppa005013m | Squamosa promoter-binding-like protein 12-like | Transcription factor (TF) | |
| ppa023657m | Squamosa promoter-binding-like protein 16 | TF | |
| ppa007056m | SPL domain class transcription factor | TF | |
| ppa006611m | SPL domain class transcription factor | TF | |
| ppa021582m | SPL domain class transcription factor | TF | |
| ppa017695m | Squamosa promoter-binding-like protein 6-like | TF | |
| ppa003644m | Squamosa promoter-binding-like protein 6-like | TF | |
| ppa007202m | Squamosa promoter-binding-like protein 16-like | TF | |
| miR159 | ppa003628m | Transcription factor GAMYB | TF |
| miR160 | ppa002710m | Auxin response factor 18 | TF |
| ppa002082m | Auxin response factor 18-like | TF | |
| ppa002195m | Auxin response factor 16 | TF | |
| ppa003136m | Auxin response factor | TF | |
| miR164 | ppa007653m | NAC domain-containing protein | TF |
| miR166 | ppa001405m | HB15 HD-ZipIII transcription factors | TF |
| miR167 | ppa017885m | Pentatricopeptide repeat-containing protein (PPR) | TF |
| miR171 | ppa001781m | GRAS family transcription factor (SCARECROW-like) | TF |
| ppa001561m | GRAS family transcription factor (SCARECROW-like) | TF | |
| miR172 | ppa021782m | Ethylene-responsive transcription factor RAP2-7-like | TF |
| ppa005230m | AP2 domain-containing transcription factor | TF | |
| ppa003783m | AP2 domain class transcription factor | TF | |
| ppa018704m | AP2 domain class transcription factor | TF | |
| miR319 | ppa003628m | Transcription factor GAMYB | TF |
| miR393 | ppa003465m | Protein auxin signalling F-box 3 | TF |
| ppa003344m | Transport inhibitor response 1 protein | TF | |
| miR394 | ppa004699m | F-box family protein | TF |
| miR395 | ppa002425m | Sulphate transporter 2.1-like | Adjusts nutrient balance |
| miR397 | ppa015544m | Laccase-11-like | Oxidoreductase |
| ppa017222m | Laccase-11-like | Oxidoreductase | |
| ppa003590m | Laccase-11-like | Oxidoreductase | |
| ppa003714m | Laccase-17 | Oxidoreductase | |
| ppa003296m | Laccase-2-like | Oxidoreductase | |
| miR399 | ppa025234m | Probable inorganic phosphate transporter 1–3 | Adjusts nutrient balance |
| miR408 | ppa018507m | Uncharacterised protein LOC100305588 precursor | Unknown |
| miR477 | ppa016418m | GRAS family transcription factor | TF |
| ppa026722m | GRAS family transcription factor | TF | |
| ppa025123m | Hypothetical protein | Unknown | |
| miR1030 | ppa000744m | Hypothetical protein | Unknown |
| miR1511 | ppb023395m | Hypothetical protein | Unknown |
| miR2111 | ppa023821m | F-box family protein | TF |
| miR2118 | ppa013258m | Unknown | Unknown |
| miR3629 | ppa018289m | Hypothetical protein | Unknown |
Expression analysis of peach miRNAs by qRT-PCR
To validate the existence and spatiotemporal expression of miRNAs in organisms and to assess their potential roles in regulating the expression of the genes, we analysed the expression of a sample of 37 miRNA sequences belonging to 25 families using qRT-PCR in young leaves, stems and flowers of ‘Lovell’. The qRT-PCR analyses demonstrated that all miRNAs were expressed in all the three tissues tested. However, while analysing the results from qRT-PCR, we observed that the expression level of miRNAs differed from each other in the three peach tissues tested (Fig. 3). The qRT-PCR results showed that miR160a, miR166e, miR169d/g, miR171a/b, miR172a/b, miR390, miR393a, miR395d/g, miR396b/c, miR397, miR398a//b/c, miR403, miR482a, miR827 and miR1511 expression levels were not significantly different in all tested tissues. Several miRNAs had different expression patterns in leaf, stem and flower tissues. miR319f and miR2275 accumulated in young leaves, while miR156a, miR164, miR408, miR447, miR477 and miR2118 were expressed predominantly in flowers. miR156i, miR167b, miR169i, miR171c/d and miR535 were all expressed more abundantly in young leaves and flowers in peach compared with the expression in stems, whereas the expression of miR166e, miR169g, miR393a and miR1511 in young leaves was lower than in young stems and flowers.
Fig. 3.
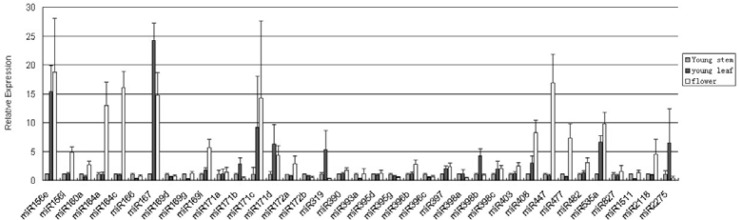
Relative expression levels of peach miRNAs in different tissues.
The results show that different family members, even different members of the same family, display drastically different expression levels. Abundance comparisons of different members in one miRNA family in various peach tissues may provide valuable information on the role played by miRNAs in plant growth.
Identification of miRNA-guided cleavage and expression analysis of miRNA targets in peach
To verify the potential miRNA targets and better comprehend how ppe-miRNAs regulate their target genes, the cleavage sites of the miRNA target and its expression in three tissues were identified and analysed. The RLM-RACE procedure was successfully used to map the cleavage sites in two predicted ppe-miRNA target genes. Ppa005013m and ppa005230m were confirmed as the real targets of ppe-miR156 and ppe-miR172, respectively, since all the 5′-ends in the mRNA fragments mapped to the nucleotide that pairs to the tenth nucleotide of each miRNA with higher frequencies than depicted for each pairing oligo (Fig. 4). The two predicted targets were found to have specific cleavage sites corresponding to the miRNA complementary sequences (Fig. 4) and might be regulated by the two ppe-miRNAs. Ppa005013m and ppa005230m are similar to Arabidopsis proteins coded by the SPL domain class and AP2 domain-containing transcription factors, respectively (Table 3). To further assess the regulatory action between the miRNA and its target, we analysed the expression of two miRNA target genes, ppa005013m and ppa005230m, using qRT-PCR. The qRT-PCR analysis demonstrated that all miRNA target genes were expressed in all the three tissues tested. However, the expression levels of miRNA target genes were different from each other in the three peach tissues tested (Fig. 5). The two genes were both expressed more abundantly in young stems and were lower in young leaves. Their expression levels were not significantly different in the three tested tissues.
Fig. 4.
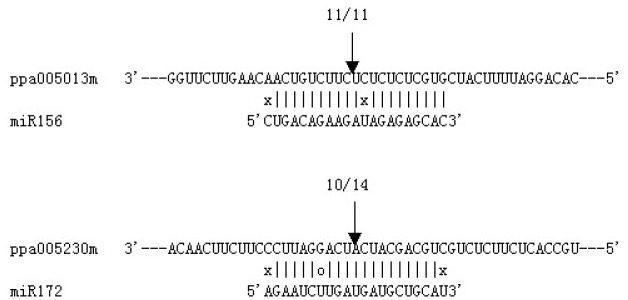
Mapping of mRNA cleavage sites by RNA ligase-mediated 5′ RACE. Each top strand depicts a miRNA-complementary site in the target mRNA, and each bottom strand depicts the miRNA. Watson-Crick pairing (vertical dashes), G:U wobble pairing (circles) and mismatched base pairing (X) are indicated. Arrows indicate the 5′ termini of mRNA fragments isolated from peach. Numbers indicate the fraction of terminating cloned PCR products.
Fig. 5.
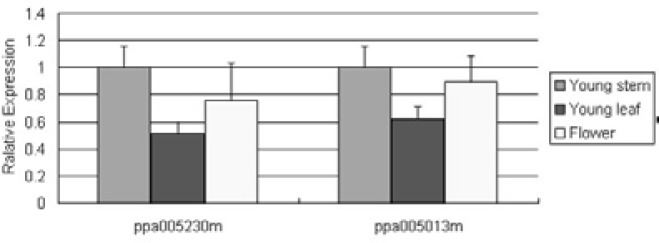
Relative expression levels of peach miRNA target genes in different tissues.
DISCUSSION
miRNAs have been extensively studied in recent years, and thousands of miRNA genes in the plant kingdom, from mosses and ferns to higher flowering plants, have been computationally predicted and/or experimentally cloned either by traditional genetic approaches or by the recently developed next-generation sequencing (NGS) strategy (Meng et al., 2011). However, only 22 miRNAs belonging to seven miRNA families were computationally predicted in peach using peach EST sequences (Zhang et al., 2011). A systematic study of miRNAs has not been completely performed in peach using peach genome sequence. The identification of entire sets of peach miRNA genes and, subsequently, their targets will lay the foundation for unravelling the complex miRNA-mediated regulatory networks controlling development and other physiological processes. Computational approach and high-throughput sequencing approach are the two main methods used to identify miRNAs (Song et al., 2009; 2010b; Yu et al., 2011; Zhao et al., 2010). In our study, we used the peach genome sequence to predict miRNAs and their targets and found 110 potential miRNAs and 43 presumed miRNA targets. Of these predicted miRNAs, 23 miRNA families were conserved, often over broad evolutionary distances, while 19 miRNAs belonging to 14 miRNA families were not conserved, as they exist in only a small number of species. Furthermore, miR3629 and miR3627 have been found only in Vitis vinifera. miR1511, miR2275, mi1446, miR1030, miR538, miR414, miR447 and miR858 were also very rare miRNAs, only found in one or two species. Our results indicated that many miRNAs were specific to small groups of related species and we speculated that they could play a part in speciation. The high-throughput sequencing approach was also employed to identify peach miRNAs. The 631 known miRNA families and 341 potential novel miRNAs were identified (unpublished data). In the known miRNAs, 34 (31%) miRNAs predicted by computational method were the same as the miRNAs identified using high-throughput sequencing approach. In the remaining different miRNAs, 21 miRNAs (miR160a, miR164c, miR169d/i, miR171a/b/c, miR393a, miR395d/g, miR396b/c, miR398a/b/c, miR408, miR447, miR482, miR827, miR1511, miR2275) were verified by qRT-PCR methods. That is to say 55 (50%) miRNA sequences were valid at least. In addition, four miRNA families, miR538/1030/1446/3629, were not found in miRNAs that produces by high-throughput sequencing, while these miRNA families were predicted by computational methods. It is confirmed that our computational method is efficient and reliable and can help identify miRNAs irrespective of expression conditions.
The identification of target genes for miRNAs is an important step in understanding the regulation of miRNA via structural genes. Although thousands of miRNAs have been identified in plants, the targets for these miRNAs have not been tested and verified due to the fact that there has been no large-scale experimental method available (Zhang et al., 2006b). We first predicted miRNA targets, then verified two miRNA target genes by RLM-RACE. We searched candidate targets of peach miRNAs using a BLASTn search with 110 identified miRNAs against peach mRNA sequences. Our analysis revealed that most of the predicted targets in peach have a conserved function with miRNA targets in Arabidopsis (Rhoades et al., 2002) and a wide variety of plant species (Dhandapani et al., 2011; Jones-Rhoades and Bartel, 2004). Consistent with previous reports, most of these targets in peach were plant-specific transcription factors, such as AP2, NAC, SBP, MYB and the ARF family. Nonetheless, the discovery that miRNAs regulate genes such as the sulphate transporter, the inorganic phosphate transporter and laccases showed that miRNAs also have a crucial role in regulating other aspects of plant biology. Upregulation of miR395 could suppress the corresponding target genes during sulphate starvation and miR399 may control Pi homeostasis by regulating the expression of a ubiquitin-conjugating E2 enzyme in Arabidopsis (Chiou, 2006). These miRNAs may play important roles in plant nutrient homeostasis and responses to environmental biotic and abiotic stresses. Finding a cleavage site supposedly located in the sequence of the target gene complementary to the miRNA is necessary to verify the cleavage of target mRNAs. Among the methods used to observe miRNA-dependent cleavage of targets, RLM-RACE is the most useful (Llave et al., 2002; Song et al., 2009). Our results show that two potential target genes for the two ppe-miRNAs had specific cleavage sites corresponding to their miRNA complementary sequences. Furthermore, it was also observed that, consistent with previous reports (Debernardi et al., 2012; Song et al., 2010b; Wang et al., 2012), ppe-miRNA targets have an miRNA-complementary site located in their coding regions.
Currently, the major outlines of the functional interactions of plant miRNAs are gradually becoming known and the functions of miRNAs are being generally investigated by altering miRNA expression or by analysing mutant target genes lacking miRNA binding sites (Sun et al., 2011). The expression of miRNAs and their target gene pattern might provide clues about miRNA functions. Previous reports have demonstrated that several Arabidopsis, Oryza and Populus miRNAs are expressed ubiquitously while the expression of others is regulated by development and show preferential accumulation in certain tissues, while some others are regulated in response to stress (Yao et al., 2007). In this study, we used qRT-PCR to validate the existence and spatiotemporal expression of miRNAs and their target genes. In our work, 37 miRNAs in 25 families were identified. miR156, miR171 and miR408 have been tested and verified in peach (Zhang et al., 2011), and the expression of these miRNAs in different tissues corresponded with our study with the exception of miR156. Zhang et al. (2011) reported that the expression of miR156 is higher in young leaves than in flowers; however, our results were exactly the opposite. It is apparent that the expression of miRNAs may differ in diverse varieties. In trifoliate orange, miR156 was accumulated more in flowers than leaves and stems while miR156 was expressed more abundantly in leaves compared with flowers and young shoots in citrus (Song et al., 2009; 2010a). In apple, miR156 were high expression in stems and low in flowers and leaves (Yu et al., 2011). It is revealed that the same miRNA family have variations expression patterns to facilitate functional specialization in different plant. To further understand the mechanism of the interaction between miRNAs and their target genes, we studied the expression of miRNA target genes (ppa005013 and ppa 005230) in three tissues. The expression levels of the target genes of miR156 and miR172, ppa005013m and ppa005230m, were not significantly different in the three tested tissues. However, it is interesting that the expression level tendency of ppe-miRNAs (miR156 and miR172) and their target genes (ppa 005013m and ppa005230m) were not opposite in three tissues. miR156 accumulated more in flowers and less in young stems, while the expression of the miR156 target gene, ppa005013m, was higher in young stems and lower in young leaves. The expression of miR172a and miR172b in the same family was different in the three peach tissues. miR172a was expressed more abundantly in flowers than young stems and leaves, while miR172b was expressed more abundantly in young stems than young leaves and flowers. Their common target gene, ppa 005230m, was expressed more abundantly in young stems than in young leaves and flowers. It seems that ppa005013m and ppa005230m are not just regulated by miR156 and miR172, respectively. They may regulated by other miRNAs or genes. The emerging picture of miRNA regulation is a complex and comprehensive gene regulatory network. Comprehensive characterisation of all the identified peach miRNAs and their target genes in different tissues would be helpful to understand the tissue-specific expression of all the miRNAs as well as their regulatory roles with respect to different tissues, organs, and conditions.
Acknowledgments
We gratefully acknowledge support for this research from project #948, with funds from the National Science Foundation of China (31102516), the Natural Science Foundation of Jiangsu Province (BK2011642) and a project funded by the Priority Academic Programme Development of the Jiangsu Higher Education Institutions (PAPD). The authors also thank Doctor Reiqhard from the Genetics and Biochemistry Department at Clemson University for providing the plant materials.
REFERENCES
- Adai A., Johnson C., Mlotshwa S., Archer-Evans S., Manocha V., Vance V., Sundaresan V. Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 2005;15:78–91. doi: 10.1101/gr.2908205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell Online. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs:: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Carrington J.C., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T.J. Regulation of Phosphate Homeostasis by MicroRNA in Arabidopsis. Plant Cell Online. 2006;18:412–421. doi: 10.1105/tpc.105.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi J.M., Rodriguez R.E., Mecchia M.A., Palatnik J.F. Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet. 2012;8:e1002419. doi: 10.1371/journal.pgen.1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Design R.T.P.C.R.P. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- Dezulian T., Remmert M., Palatnik J.F., Weigel D., Huson D.H. Identification of plant microRNA homologs. Bioinformatics. 2006;22:359–360. doi: 10.1093/bioinformatics/bti802. [DOI] [PubMed] [Google Scholar]
- Dhandapani V., Ramchiary N., Paul P., Kim J., Choi S.H., Lee J., Hur Y., Lim Y.P. Identification of potential microRNAs and their targets in Brassica rapa L. Mol. Cells. 2011;32:21–37. doi: 10.1007/s10059-011-2313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Wang K., Liu X., Chen S., Chen J. The quantification of tomato microRNAs response to viral infection by stem-loop real-time RT-PCR. Gene. 2009;437:14–21. doi: 10.1016/j.gene.2009.01.017. [DOI] [PubMed] [Google Scholar]
- German M.A., Pillay M., Jeong D.H., Hetawal A., Luo S., Janardhanan P., Kannan V., Rymarquis L.A., Nobuta K., German R., et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotech. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S.M. The microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., Van Dongen S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer L.S., Tacker M., Schuster P. Fast folding and comparison of RNA secondary structures. Monatshefte für Chemie/Chemical Monthly. 1994;125:167–188. [Google Scholar]
- Jeong D.H., Park S., Zhai J., Gurazada S.G., De Paoli E., Meyers B.C., Green P.J. Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell. 2011;23:4185–4207. doi: 10.1105/tpc.111.089045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian X., Zhang L., Li G., Wang X., Cao X., Fang X., Chen F. Identification of novel stress-regulated microRNAs from Oryza sativa L. Genomics. 2010;95:47–55. doi: 10.1016/j.ygeno.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lindow M., Krogh A. Computational evidence for hundreds of non-conserved plant microRNAs. BMC Genomics. 2005;6:119. doi: 10.1186/1471-2164-6-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta] CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K.D., Carrington J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Meng Y., Wu P., Chen M. MicroRNAs in plant roots: current understanding and future perspectives. Non Coding RNAs in Plants. 2011. pp. 269–284.
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Ro S., Park C., Jin J., Sanders K.M., Yan W. A PCR-based method for detection and quantification of small RNAs. Biochem. Biophys. Res. Commun. 2006;351:756–763. doi: 10.1016/j.bbrc.2006.10.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer S.E., Jacobsen S.E., Meinke D.W., Ray A. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002;7:487–491. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Seffens W., Digby D. mRNAs have greater negative folding free energies than shuffled or codon choice randomized sequences. Nucleic Acids Res. 1999;27:1578. doi: 10.1093/nar/27.7.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.F., Waterman M.S. Identification of common molecular subsequences. J. Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Song C., Fang J., Li X., Liu H., Thomas Chao C. Identification and characterization of 27 conserved microRNAs in citrus. Planta. 2009;230:671–685. doi: 10.1007/s00425-009-0971-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Fang J., Wang C., Guo L., Nicholas K.K., Ma Z. MiR-RACE, a new efficient approach to determine the precise sequences of computationally identified trifoliate orange (Poncirus trifoliata) microRNAs. PloS One. 2010a;5:e10861. doi: 10.1371/journal.pone.0010861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Wang C., Zhang C., Korir N.K., Yu H., Ma Z., Fang J. Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata) BMC Genomics. 2010b;11:431. doi: 10.1186/1471-2164-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.H., Shi R., Zhang X.H., Chiang V.L., Sederoff R.R. MicroRNAs in trees. Plant Mol. Biol. 2011:1–17. doi: 10.1007/s11103-011-9864-z. [DOI] [PubMed] [Google Scholar]
- Sunkar R., Girke T., Jain P.K., Zhu J.K. Cloning and characterization of microRNAs from rice. Plant Cell Online. 2005;17:1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z., Gao Z., Wang F., Zhou J., Zhang Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009;10:71. doi: 10.1186/1471-2199-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Xiong A.S., Yao Q.H., Zhang Z., Qiao Y.S. Direct isolation of high-quality low molecular weight RNA of pear peel from the extraction mixture containing nucleic acid. Mol. Biotechnol. 2010;44:61–65. doi: 10.1007/s12033-009-9204-6. [DOI] [PubMed] [Google Scholar]
- Wang C., Han J., Liu C., Kibet K.N., Kayesh E., Shangguan L., Li X., Fang J. Identification of microRNAs from Amur grape (vitis amurensis Rupr.) by deep sequencing and analysis of microRNA variations with bioinformatics. BMC Genomics. 2012;13:122. doi: 10.1186/1471-2164-13-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Yao Y., Guo G., Ni Z., Sunkar R., Du J., Zhu J.K., Sun Q. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.) Genome Biol. 2007;8:R96. doi: 10.1186/gb-2007-8-6-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Song C., Jia Q., Wang C., Li F., Nicholas K.K., Zhang X., Fang J. Computational identification of microRNAs in apple expressed sequence tags and validation of their precise sequences by miR-RACE. Physiol. Plant. 2011;141:56–70. doi: 10.1111/j.1399-3054.2010.01411.x. [DOI] [PubMed] [Google Scholar]
- Zhang B., Pan X., Anderson T.A. Identification of 188 conserved maize microRNAs and their targets. FEBS Lett. 2006a;580:3753–3762. doi: 10.1016/j.febslet.2006.05.063. [DOI] [PubMed] [Google Scholar]
- Zhang B., Pan X., Wang Q., Cobb G.P., Anderson T.A. Computational identification of microRNAs and their targets. Comput. Biol. Chem. 2006b;30:395–407. doi: 10.1016/j.compbiolchem.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Zhang B.H., Pan X.P., Cox S.B., Cobb G.P., Anderson T.A. Evidence that miRNAs are different from other RNAs. Cellular and molecular life sciences. Cell. Mol. Life Sci. 2006c;63:246–254. doi: 10.1007/s00018-005-5467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Pan X., Stellwag E.J. Identification of soybean microRNAs and their targets. Planta. 2008;229:161–182. doi: 10.1007/s00425-008-0818-x. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yu M., Yu H., Han J., Song C., Ma R., Fang J. Computational identification of microRNAs in peach expressed sequence tags and validation of their precise sequences by miR-RACE. Mol. Biol. Rep. 2011;39:1975–1987. doi: 10.1007/s11033-011-0944-6. [DOI] [PubMed] [Google Scholar]
- Zhao C.Z., Xia H., Frazier T.P., Yao Y.Y., Bi Y.P., Li A.Q., Li M.J., Li C.S., Zhang B.H., Wang X.J. Deep sequencing identifies novel and conserved microRNAs in peanuts (Arachis hypogaea L.) BMC Plant Biol. 2010;10:3. doi: 10.1186/1471-2229-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]