Abstract
Circadian clock genes are regulated by a transcriptional-translational feedback loop. In Arabidopsis, LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) transcripts are highly expressed in the morning. Translated LHY and CCA1 proteins repress the expression of TIMING OF CAB EXPRESSION 1 (TOC1), which peaks in the evening. TOC1 protein induces expression of LHY and CCA1, forming a negative feedback loop which is believed to constitute the oscillatory mechanism of the clock. The rhythmic oscillation of mouse clock genes mPERIOD 1 (mPER1) and mPER2 has been correlated with regular alteration of chromatin structure through histone acetylation/deacetylation. However, little is known about the relationship between the transcriptional activity of Arabidopsis clock genes and their chromatin status. Here, we report that histone H3 acetylation (H3Ac) and H3 lysine 4 tri-methylation (H3K4me3) levels at LHY, CCA1, and TOC1 are positively correlated with the rhythmic transcript levels of these genes, whereas H3K36me2 level shows a negative correlation. Thus, our study suggests rhythmic transcription of Arabidopsis clock genes might be regulated by rhythmic histone modification, and it provides a platform for future identification of clock-controlling histone modifiers.
Keywords: Arabidopsis, chromatin, circadian clock, circadian rhythm, histone modification
INTRODUCTION
Circadian rhythm, controlled by biological circadian clocks, is a rhythmic variation with a 24-hour (h) period. It is found in diverse organisms ranging from prokaryotes to humans, allowing coordination with daily light and temperature changes. In animals and plants, circadian clocks regulate diverse physiological processes such as human sleep/wake cycles, Neurospora sporulation, plant flowering, and robust rhythmic gene expression (Covington et al., 2008; Dowson-Day and Millar, 1999; Dunlap, 1990; Imaizumi and Kay, 2006; Moore, 1997). Functional circadian clocks confer enhanced fitness and, therefore, allow organisms to be more adaptive (Dodd et al., 2005; Yerushalmi et al., 2011). According to experimental observations and mathematical modeling, circadian clocks are based on interlocking negative feedback loops of transcriptional activators and repressors (Locke et al., 2006; Song and Noh, 2007; Zhang and Kay, 2010). In Arabidopsis, the central loop operates through reciprocal regulation between the morning-phased, partially redundant MYB transcription factors, LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), and the evening-phased pseudo-response regulator TIMING OF CAB EXPRESSION 1 (TOC1; Harmer, 2009; McClung and Gutiérrez, 2010). LHY and CCA1 are transcriptional repressors that bind and inhibit expression from the TOC1 promoter (Alabadí et al., 2001; Schaffer et al., 1998; Wang and Tobin, 1998). TOC1 indirectly promotes expression of LHY and CCA1 partly through the CCA1-binding transcription factor, CCA1 HIKING EXPEDITION (CHE; Alabadí et al., 2001; Pruneda-Paz et al., 2009). Such daily interactions between transcriptional activators and inhibitors and their corresponding cis regulatory elements can achieve phase control over gene expression rhythms. More than 10% of the Arabidopsis transcriptome exhibits 24-h period rhythmicity (Covington et al., 2008; Harmer et al., 2000), suggesting a large portion of the genome is directly or indirectly controlled by the circadian clock.
Eukaryotic genomic DNA is packed around histone proteins into repeating nucleosome units, which form higher-order chromatin structures (Clapier et al., 2009). DNA methylation, covalent modification of histone proteins, and ATP-dependent chromatin remodeling are the best-known mechanisms influencing chromatin structure. The histone code or, more appropriately, histone language of various histone modifications and their combinations influences transcription (Cedar and Bergman, 2003; Lee et al., 2010). A number of different chemical modifications occur on histone proteins, especially at their N-terminal tails (Kouzarides, 2007; Tan et al., 2011). Some of these modifications lead to open chromatin status and active transcription, while others allow heterochromatin formation and transcriptional repression (Li et al., 2007). For example, acetylation on histone H3 (H3Ac) or H4 (H4Ac), tri-methylation on H3 lysine 4 (H3K4me3), and H3K36me2/me3 are well known activation markers, whereas H3K9me2/me3, H3K27me3, and symmetric di-methylation on H4 arginine 3 (H4R3me2s) are representative repressive markers (reviewed in Kouzarides, 2007; Li et al., 2007).
Recent studies have revealed a link between circadian-regulated gene expression and dynamic histone modifications. Circadian changes in histone modifications at the promoters of a few clock genes have been documented (Belden et al., 2007; Doi et al., 2006; Etchegaray et al., 2003). In Neurospora, an ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH (CSW) is required for clock function (Belden et al., 2007). CSW binds to the promoter of the central clock gene FREQUENCY (FRQ) and regulates FRQ expression by controlling the chromatin structure of its promoter. In mammals, the key clock genes, PERIOD 1 (PER1), PER2, and PER3, exhibit circadian rhythms in H3Ac levels, and histone acetyltransferase (HAT) p300 precipitates with CLOCK protein in this process in a time-dependent manner (Etchegaray et al., 2003). The transcription factor CLOCK, a central component of the circadian clock, possesses intrinsic HAT activity, suggesting that histone acetylation is important for core clock mechanisms (Doi et al., 2006). Although most studies have focused on histone acetylation in the circadian system, histone methylation is also crucial for clock function. Mammalian histone methyltransferase (HMT) Enhancer of Zeste Homolog 2 (EZH2), a polycomb group enzyme mediating H3K27me2 and H3K27me3 at the promoters of PER1 and PER2, is required for mammalian circadian clock function (Etchegaray et al., 2006). WDR5, a member of an HMT complex, directly interacts with PER1 and mediates rhythmic methylation of H3K4 and H3K9 at the promoters of PER1-regulated genes (Brown et al., 2005).
In Arabidopsis, TOC1 expression is affected by clock-controlled cycles of histone acetylation (Perales and Más, 2007), although the enzymes responsible remain unknown. Except for this single report, few researchers have addressed the relationship between clock gene expression and changes in chromatin structure. Here we show that H3Ac and H3K4me3 levels at the LHY, CCA1, and TOC1 loci positively correlate with transcription from these loci, whereas the level of H3K36me2 inversely correlates with transcription. Furthermore, the correlations between clock gene expression and histone modifications are consistently observed during free run under constant light conditions. Based on these results, we propose that the rhythmic activity of the Arabidopsis circadian clock might be regulated by rhythmic histone modifications.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis thaliana (ecotype Columbia-0) seeds were sown on Murashige and Skoog (MS) agar supplemented with 1% sucrose and refrigerated for at least 3–4 days before growing at 22°C under 100 μmol·m−2·s−1 cool white fluorescent light. Photoperiods (12 h of light/12 h of dark: 12L12D or constant light: LL) were programmed according to the purpose of each experiment.
RT-PCR analysis
Total RNA was isolated from the seedlings by using TRI Re-agent (Molecular Research Center) according to the manufacturer’s instructions. Reverse transcription (RT) was performed with M-MuLV Reverse Transcriptase (Fermentas) and an oligod (T) primer according to the manufacturer’s instructions using 3 μg of total RNA. Semi-quantitative polymerase chain reaction (PCR) was performed using first-strand cDNA with i-Taq DNA polymerase (iNtRON Biotechnology). Primer sequences for each PCR reaction were as follows: LHY (5′-CAAAGCAGC GAGAGCGATGG-3′ and 5′-GCGTGCCCGTGAGTTTCTTC-3′), CCA1 (5′-GGTAGAGAAAGAGGCTGAAGC-3′ and 5′-GG AAACGACTGATAATCTCCTGC-3′), TOC1 (5′-GGATTTGAAC GGTGAGTG-3′ and 5′-CACTTGAAACTTCTCCGCC-3′), and ubiquitin 10 (UBQ10; 5′-GATCTTTGCCGGAAAACAATTGGA GGATGGT-3′ and 5′-CGACTTGTCATTAGAAAGAAAGAGA TAACAGG-3′). Transcript levels were quantified from gel images using the Metamorph™ software (Universal Imaging Corp.) and normalized to the level of UBQ10 before plotting.
ChIP assay
Chromatin immunoprecipitation (ChIP) was performed as described by Han et al. (2007) using 14- to 16-day-old seedlings. Seedlings were vacuum infiltrated with 1% formaldehyde for cross-linking and ground in liquid nitrogen after quenching the cross-linking process. Chromatin was isolated and sonicated into ∼0.5–1 kb fragments. Each modification-specific antibody (H3Ac, Millipore 06–599; H4Ac, Upstate 06–946; H3K4me3, Abcam ab8580; H3K36me2, Upstate 07–369) was added to the chromatin solution, which had been pre-cleared with salmon sperm DNA/ProteinA agarose beads (Millipore). Then, the immunocomplexes were precipitated and eluted. The cross-links were reversed and residual proteins were removed by incubation with proteinase K followed by phenol/chloroform extraction. DNA was recovered by ethanol precipitation. The amount of immunoprecipitated chromatin was determined by semi-quantitative PCR. Primer sequences for each PCR reaction were as follows: LP1 (5′-GTGGCTGAGATTGCTTCTGG-3′ and 5′-CTTGAGAGTAGCCATGGAGG-3′), LP2 (5′-TGCGGT GGATTCGTTTGGG-3′ and 5′-AGCCGTTGTGATAACCTC-3′), LX1 (5′-CCGGTCCTGTTATGGATAC-3′ and 5′-TCAGTCCA TCGCTCTCGC-3′), LX4 (5′-TAGCAGGTAAGTGGCGAC-3′ and 5′-GAGATACCATACCTGAGGG-3′), L3U (5′-CTACATG ACAGACTTGGAGG-3′ and 5′-GTACAGAACCTGACATGACC-3′), CP1 (5′-CAGGTAGTCCCGGAACTCGTGG-3′ and 5′-CGGAAATGGAGAAATCTCAGCC-3′), CP2 (5′-CTCCATTTC CGTAGCTTCTGG-3′ and 5′-CAAACAATAAGAAAGACCAT GAC-3′), CX1 (5′-GGGGTCATGGTCTTTCTTATTG-3′ and 5′-GTCGCAAATATGATGGACGC-3′), CX6 (5′-GGGAAGTCAG AATAACAGGG-3′ and 5′-GGAAACGACTGATAATCTCCTGC-3′), C3U (5′-CGGATGCGGTTGGAAACTC-3′ and 5′-CAAGA GCCCCTTGAGTGAAG-3′), TP1 (5′-CTCTTCTTGTCGGAAAT TCACC-3′ and 5′-CCCCAACCTAATCCTTTATCC-3′), TP2 (5′-AGGGGATAAATTAGGCGAC-3′ and 5′-CCATCTCCTCCTT TACACTC-3′), TX1 (5′-GAACGGTGAGTGTAAAGG-3′ and 5′-AAGCCAGACGACAAGAACC-3′), TX5 (5′-GCTGGTAGTGG TCTGTTGC-3′ and 5′-CAAAGAGAAACAATCACCTGG-3′), T3U (5′-TACACCAAGAACTGAAAACCG-3′ and 5′-CCAATG AGAGCTTTTCAATGC-3′), and ACTIN1 (5′-CGTTTCGCTTT CCTTAGTGTTAGCT-3′ and 5′-AGCGAACGGATCTAGAGA CTCACCTTG-3′). The amounts of PCR products were quantified from gel images using the Metamorph™ software (Universal Imaging Corp.) and normalized to the amounts of the corresponding inputs before plotting.
RESULTS
Rhythmic expression of LHY, CCA1, and TOC1 under light-dark cycling conditions correlates with histone modification
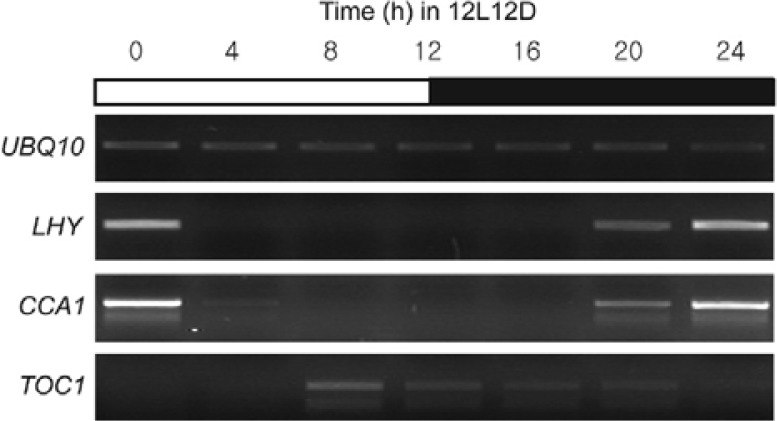
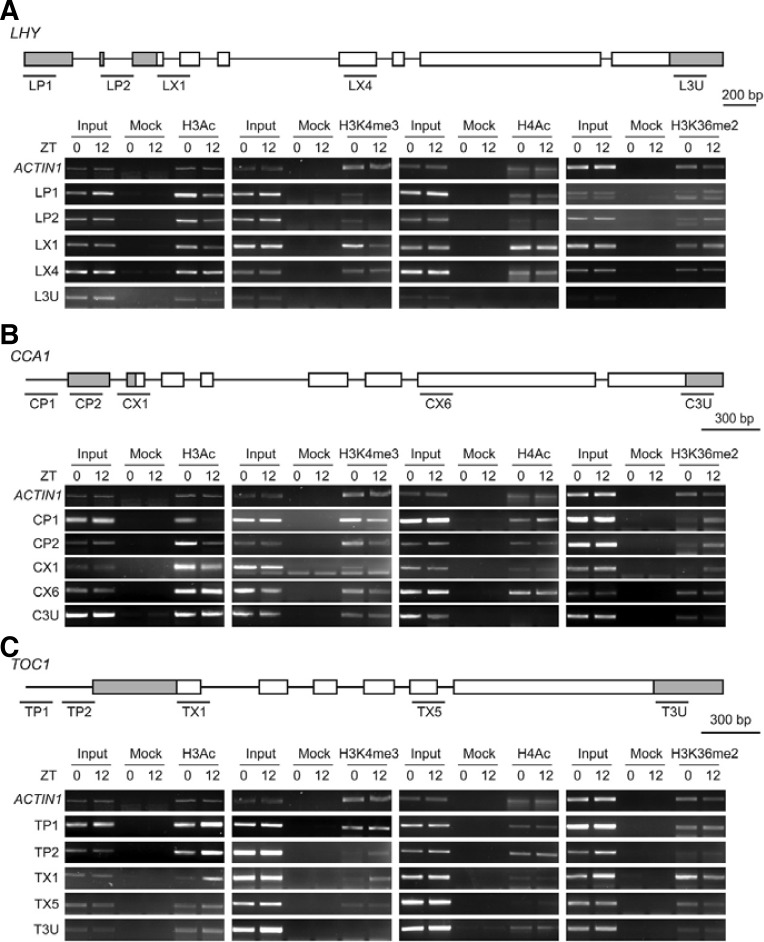
Transcript levels of LHY, CCA1, and TOC1, which compose the central oscillator of the Arabidopsis circadian clock, exhibit high-amplitude rhythms under 12-h light/dark photocycles (12L12D; Niwa et al., 2007; Yamashino et al., 2008). LHY, CCA1, and TOC1 transcripts showed expected rhythmicity under these conditions, with LHY and CCA1 transcript levels peaking in the early morning (zeitgeber 0; ZT0) and TOC1 transcription peaking in the late afternoon (ZT8-12; Fig. 1). Because chromatin modification is important for clock-regulated gene expression in Neurospora and mammals (Belden et al., 2007; Doi et al., 2006), we sought to determine whether a similar correlation exists between the rhythmic oscillation of LHY, CCA1, and TOC1 and chromatin structure in Arabidopsis. Wild-type (WT) Arabidopsis seedlings were grown in 12L12D and ChIP assays were performed with antibodies specific to H3Ac, H4Ac, H3K4 me3, and H3K36me2. H3Ac and H3K4me3 levels in LP1, LP2, and LX1 but not the LX4 and L3U regions of LHY were higher at ZT0 than at ZT12 (Fig. 2A). The levels of H4Ac in all regions of LHY did not significantly differ between ZT0 and ZT12 (Fig. 2A). However, the pattern of H3K36me2 enrichment within LHY chromatin contrasted those of H3Ac and H3K4me3: H3K36me2 levels in LP1, LP2, and LX1 were higher at ZT12 than at ZT0 (Fig. 2A). No change in H3K36me2 was observed in LX4 and H3K36me2 enrichment in L3U was minimal (Fig. 2A). The change in H3Ac, H3K4me3, H4Ac, and H3K36me2 patterns at LHY was similar at the CCA1 locus (Fig. 2B). Thus, at LHY and CCA1, H3Ac and H3K4me3 positively correlated with LHY and CCA1 transcription, whereas the level of H3K36me2 showed a negative correlation.
Fig. 1.
Expression of LHY, CCA1, and TOC1 transcripts in wild-type (WT) Arabidopsis grown in 12L12D. WT seedlings were grown under 12L12D conditions for 7 days and harvested every 4 h for 1 day. Expression was analyzed by RT-PCR. UBQ10 was included as an expression control.
Fig. 2.
Histone modifications within LHY, CCA1, and TOC1 chromatin in 12L12D. (A) ChIP assay of LHY chromatin with H3Ac-, H4K4me3-, H4Ac-, and H3K36me2-specific antibodies. WT seedlings were grown in 12L12D for 7 days and harvested at ZT0 and ZT12 (A-C). ZT means time (h) after lights on. “Input” indicates chromatin before immunoprecipitation. “Mock” refers to control samples lacking antibody. ACTIN 1 was used as a control chromatin. The gray boxes in the front indicate exonic 5′ untranslated regions (UTRs), and the rear gray boxes represent 3′ UTRs (AC). Labeled lines indicate regions amplified in ChIP-PCRs using specific primers (see “Materials and Methods”; A-C). (B) ChIP assay of CCA1 chromatin. (C) ChIP assay of TOC1 chromatin.
In the case of TOC1, H3Ac and H3K4me3 levels were higher at ZT12 than at ZT0 in the TP1, TP2, and TX1 regions, whereas the differences were minor or undetectable in T3U and TX5 (Fig. 2C). The levels of H4Ac within TOC1 chromatin did not significantly differ between ZT0 and ZT12 samples in all regions tested (Fig. 2C). Unlike H3Ac and H3K4me3, the levels of H3K36me2 at TOC1 were higher at ZT0 than at ZT12 in the TX1 and TX5 regions (Fig. 2C). Thus, enrichment of H3Ac and H3K4me3 or H3K36me2 within TOC1 chromatin positively or negatively correlates, respectively, with TOC1 transcript levels. These results indicate that the transcriptional activity of all Arabidopsis central clock genes, LHY, CCA1, and TOC1, in 12L12D is positively correlated with H3Ac and H3K4me3 or negatively correlated with H3K36me2 within their chromatin.
Rhythmic expression of LHY, CCA1, and TOC1 during free run is correlated with histone modification
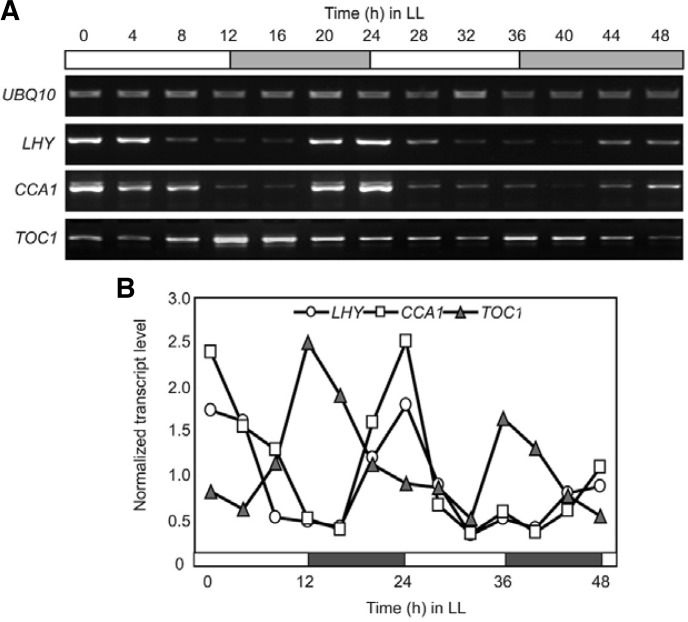
An important property of circadian rhythms is their ability to persist in the absence of initial input signals. This property reflects the regulation by an endogenous and self-sustaining oscillator, the circadian clock. Expression of LHY, CCA1, and TOC1 maintains oscillation in constant light (LL) and constant dark (DD) conditions (Niwa et al., 2007; Wenden et al., 2011; Yamashino et al., 2008). To determine whether the free-running rhythm also correlates with changes in chromatin structure, we analyzed H3Ac, H3K4me3, and H3K36me2 levels at LHY, CCA1, and TOC1 in LL, because these chromatin markers were positively or negatively correlated with transcription of LHY, CCA1, and TOC1 in 12L12D (Figs. 1 and 2). For free-run experiments, WT Arabidopsis seedlings were grown and entrained under 12L12D for 7 days, transferred to LL at ZT0, and harvested every 4 h for 2 days. Before performing ChIP experiments, transcript levels of LHY, CCA1, and TOC1 were assessed by RT-PCR to see if the condition of the prepared samples was consistent with previous reports and adequate for ChIP assays. LHY and CCA1 transcripts oscillated with peaks at subjective dawns ZT0, ZT24, and ZT48, while TOC1 transcript peaked at late afternoons ZT12 and ZT36 (Figs. 3A and 3B). These patterns were consistent with previous reports (e.g., Niwa et al., 2007; Wenden et al., 2011; Yamashino et al., 2008); thus, the samples were judged adequate for subsequent ChIP assays.
Fig. 3.
Expression of LHY, CCA1, and TOC1 transcripts in WT seedlings grown in LL. (A) RT-PCR analysis of LHY, CCA1, and TOC1 expression in LL. WT seedlings were grown under 12L12D conditions for 7 days and then released to constant light at Time 0. Plants were harvested every 4 h for 2 days in LL and used for RNA extraction and RT-PCR. UBQ10 was included as an expression control. (B) Quantification of LHY, CCA1, and TOC1 expression in (A). Each transcript level was normalized by the corresponding transcript level of UBQ10 before plotting.
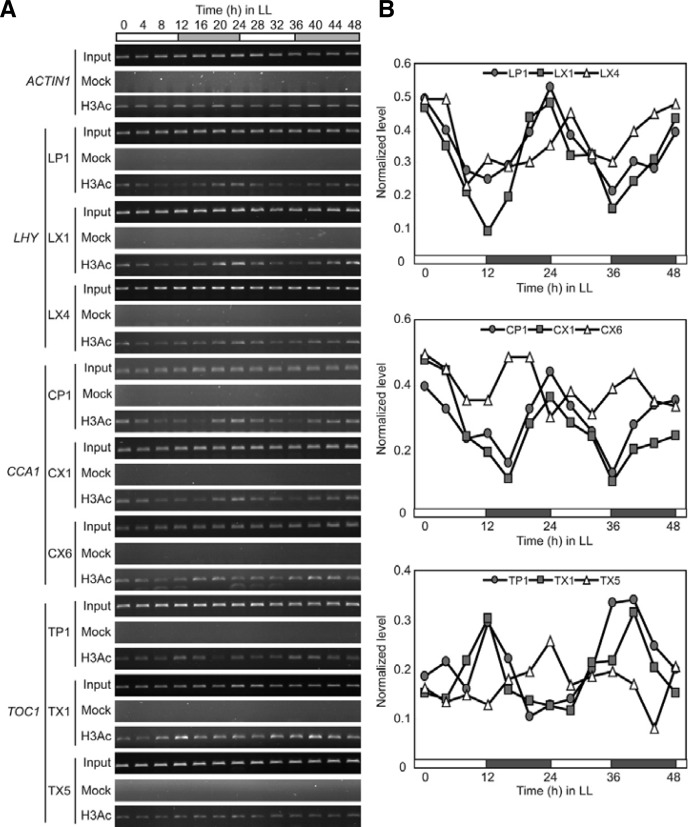
First, H3Ac levels at the LHY, CCA1, and TOC1 loci showed robust oscillations in LL (Figs. 4A and 4B). H3Ac levels peaked at ZT0, ZT24, and ZT48 in the LP1 and LX1 regions of LHY and reached minimum at ZT12 and ZT36, indicating that the H3Ac enrichment pattern correlates with LHY expression during free run. H3Ac enrichment of CCA1 chromatin was similar to that of LHY chromatin (Figs. 4A and 4B). High levels of H3Ac were observed at ZT0, ZT24, and ZT48 in the CP1 and CX1 regions of CCA1, while the lowest levels were detected at ZT16 and ZT36. However, H3Ac oscillation in CX6 was less clear, and H3Ac levels peaked earlier in CX6 than in CP1 and CX1 (Figs. 4A and 4B). H3Ac levels oscillated robustly within TOC1 chromatin (Fig. 4A and 4B). High levels of H3Ac were observed at ZT12 and ZT36 in TP1 and TX1, while levels were low at ZT0, ZT24, and ZT48. Therefore, H3Ac levels correlated positively with LHY, CCA1, and TOC1 transcription in LL.
Fig. 4.
Oscillation of H3Ac within LHY, CCA1, and TOC1 chromatin in LL. (A) ChIP-PCR analysis of H3Ac within LHY, CCA1, and TOC1 chromatin. WT seedlings were entrained under 12L12D conditions for 7 days and subsequenttly released to LL conditions. Plants were harvested every 4 h for 2 days in LL and used for ChIP using H3Ac-specific antibody. ACTIN 1 was used as a control chromatin. Regions tested and primers used for PCR are those shown in Fig. 2. (B) Quantification of the H3Ac levels in (A). Each H3Ac level was normalized by the level of corresponding input before plotting.
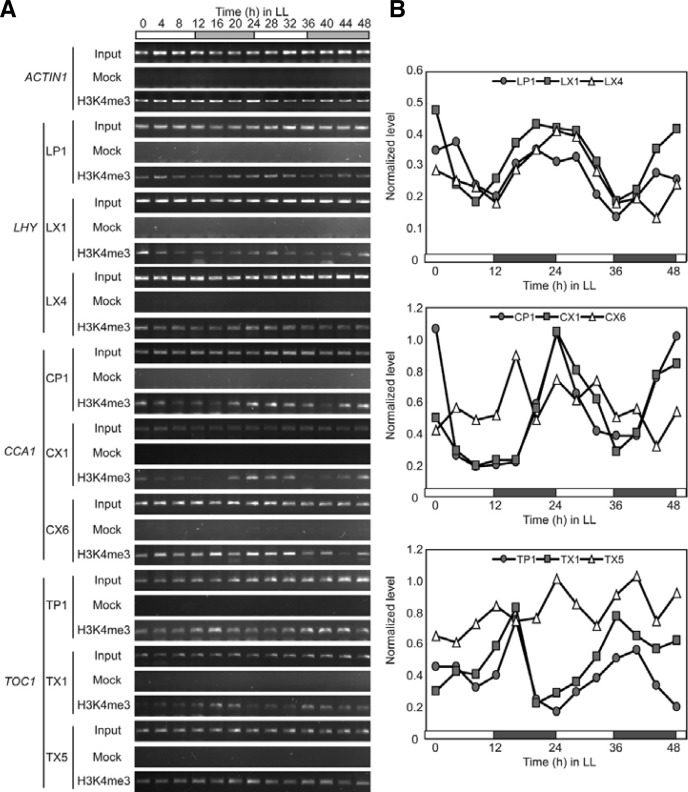
Second, H3K4me3 levels at the LHY, CCA1, and TOC1 loci also showed robust oscillations with an approximately 24-h period in LL (Figs. 5A and 5B). The highest enrichments of H3K4me3 were detected at ZT0-4, ZT24-28, and ZT44-48 in the LP1 and LX1 regions of LHY, and enrichment levels were the lowest at ZT8-12 and ZT36. Thus, the free-run rhythm of LHY in LL correlated with H3K4me3 levels within LHY chromatin. A similar correlation between rhythm and H3K4me3 enrichment was observed at the CCA1 locus (Figs. 5A and 5B). H3K4me3 levels were the highest at ZT0, ZT24, and ZT48 and the lowest around ZT12 and ZT36 in CP1 and CX1. However, the rhythmic oscillation of H3K4me3 in CP1 and CX1 was barely detectable in CX6 (Figs. 5A and 5B). The rhythmic pattern of H3K4me3 enrichment within TOC1 chromatin was the reverse of those within LHY and CCA1 chromatin (Figs. 5A and 5B). H3K4me3 levels were the highest at ZT16 and ZT36-40 and the lowest at ZT0, ZT20-24, and ZT44-48 in TP1 and TX1. In sum, H3K4me3 levels were positively correlated with the transcript levels of LHY, CCA1, and TOC1 in LL.
Fig. 5.
Oscillation of H3K4me3 within LHY, CCA1, and TOC1 chromatin in LL. (A) ChIP-PCR analysis of H3K4me3 levels within LHY, CCA1, and TOC1 chromatin. (B) Quantification of the H3K4me3 levels in (A). Experimental conditions were as described in Fig. 4, with H3K4me3-specific antibody in ChIP.
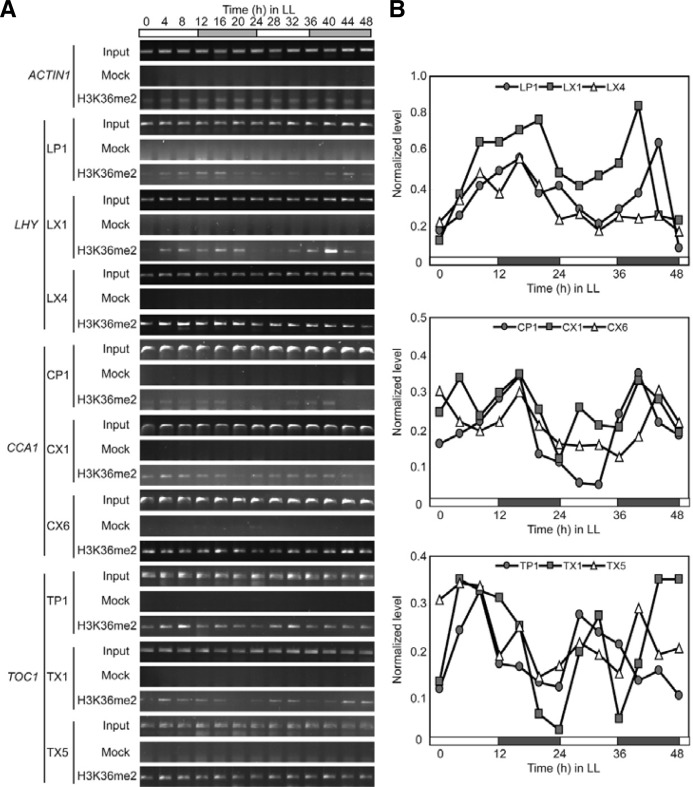
Third, H3K36me2 levels at LHY, CCA1, and TOC1 inversely correlated with the transcript levels in LL (Figs. 6A and 6B). The highest levels of H3K36me2 within LHY chromatin were detected in LP1 and LX1 around ZT16 and ZT40-44, when LHY expression was in the trough phase (Fig. 3B); the lowest levels were observed at ZT0, around ZT28, and ZT48. Rhythmic oscillation of H3K36me2 was less clear in LX4 than in LP1 and LX1 (Figs. 6A and 6B). H3K36me2 levels within CCA1 chromatin oscillated similarly to those of LHY (Figs. 6A and 6B). The highest enrichments of H3K36me2 were found in CP1 and CX1 at ZT16 and ZT40, while the lowest enrichments were observed at ZT0, ZT24-32, and ZT48. A similar lower-amplitude oscillation pattern was observed in CX6 (Figs. 6A and 6B). In the case of TOC1, the inverse correlation between transcript and H3K36me2 was less clear, although the highest H3K36me2 enrichments were consistently observed at ZT4-8 and ZT28-32, when TOC1 expression was low (Fig. 3), in TP1 and TX1 (Figs. 6A and 6B). Together, the results in Fig. 6 show that H3K36me2 inversely correlates with transcriptional activity of LHY, CCA1, and TOC1 in LL.
Fig. 6.
Oscillation of H3K36me2 within LHY, CCA1, and TOC1 chromatin in LL. (A) ChIP-PCR analysis of H3K36me2 levels within LHY, CCA1, and TOC1 chromatin. (B) Quantification of the H3K36me2 levels in (A). Experimental conditions were as described in Fig. 4, with H3K36me2-specific antibody in ChIP.
DISCUSSION
Epigenetic changes, such as DNA methylation and histone modifications, cause structural changes in chromatin and affect gene expression. Chromatin modification also provides an efficient mechanism for the relay of environmental signals, such as light or nutrient availability, to differential gene expression (Cho et al., 2012; Masri and Sassone-Corsi, 2010). Exposure to light causes rapid phosphorylation on H3 serine 10 within SCN chromatin. This phosphorylation parallels the induction of immediate early genes such as c-fos and Per1, indicating that light-mediated signaling can regulate circadian gene expression through chromatin modification (Crosio et al., 2000). Interplay between DNA methylation and histone modifications has also been well documented. For example, in a genome-wide mapping analysis, the Arabidopsis thaliana H3K9 methyltransferase KRYPTONITE was shown to direct some CNG and CNN DNA methylation (Sasaki et al., 2012). The Arabidopsis thaliana histone deacetylase (HDAC) HDA6 is required for enhanced RNA-directed DNA methylation (To et al., 2011). Genome-wide mapping of EMF1, which shares no sequence homology with known polycomb group proteins, showed that the EMF1-binding pattern is similar to that of H3K27me3 on the chromosomal and genic level (Kim et al., 2012). Two contrasting studies have demonstrated that intragenic DNA methylation induces a closed chromatin structure that excludes H3K4me2, H3K4me3, H3K9Ac, and H3K14Ac (Lorincz et al., 2004; Okitsu and Hsieh, 2007).
Recent studies have shown that activation of a number of clock-regulated genes in mammals, flies, and plants is coupled with changes in histone modifications such as acetylation (Hirayama et al., 2007; Naruse et al., 2004; Perales and Mas, 2007; Ripperger and Schibler, 2006). Thus, despite divergences in oscillator components, mechanisms by which the expression of oscillator genes is regulated through chromatin modification are common to the circadian systems of mammals, flies, and plants. Consistent with these observations, we demonstrated that the levels of H3Ac, H3K4me3, and H3K36me2 but not H4Ac at the LHY, CCA1, and TOC1 loci are correlated with the expression of these central clock genes in Arabidopsis under both 12L12D and LL conditions (Figs. 2–6).
H3 within LHY and CCA1 chromatin was highly acetylated at ZT0 and deacetylated at ZT12, while H3 within TOC1 chromatin was highly acetylated at ZT12 and deacetylated at ZT0 (Fig. 4). Therefore, H3Ac levels positively correlate with the transcript levels of all Arabidopsis central clock genes, consistent with the general role of H3Ac as a marker for active transcription. The Arabidopsis genome encodes 18 HDACs divided into 3 types: Reduced Potassium Deficiency 3, HD-tuin, and Sirtuin (Hollender and Liu, 2008). Twelve HATs encoded by the Arabidopsis genome are grouped into 4 families based on sequence homology and mode of action: GNAT, MYST, p300/CBP, and TAFII250 (Chen and Tian, 2007). However, the HDACs and HATs responsible for dynamic acetylation of H3 within LHY, CCA1, and TOC1 chromatin are yet to be identified. Considering the known redundancy in the activity of Arabidopsis HATs (Han et al., 2007), it is possible that multiple redundant HATs and HDACs affect histone acetylation levels at the clock gene loci.
H3K4me3 is another well-known marker for active transcription (Li et al., 2007). H3K4me3 is generally enriched in the transcription initiation regions of actively transcribed genes (Li et al., 2007) and can be recognized directly by the basal transcription factor TFIID via the plant homeodomain finger of TAF3 (Vermeulen et al., 2007), providing a platform for engagement of RNA Polymerase II. Curiously, the role of H3K36me is still ambiguous. H3K36me2 or H3K36me3 is generally enriched in the transcription elongation regions of actively transcribed genes; however, it exerts a negative effect on transcription when it is found in promoter regions (Landry et al., 2003; Li et al., 2007; Strahl et al., 2002). Our results show that the positive or negative correlation of H3K4me3 or H3K36me2 levels, respectively, with the transcription activity at the LHY, CCA1, and TOC1 loci is observed in regions around the transcription start sites but not in the transcription elongation regions (Fig. 2). Therefore, it is reasonable to suggest H3K4me3 and H3K36me2 might have positive and negative effects, respectively, on transcription of Arabidopsis central clock genes. Identification of the catalytic components (either HMTs or histone demethylases) responsible for the dynamic oscillation of H3K4me3 and H3K36me2 within LHY, CCA1, and TOC1 chromatin would be a challenging and important future task.
A number of cross-talk relationships among different histone modifications are known (reviewed in Latham and Dent, 2007). For example, high levels of H3Ac and H3K4me3 are detected at the transcriptional start sites of active promoters, and a human HMT, MLL4, links increased H3K9Ac and H3K14Ac to increased H3K4me3 (Pokholok et al., 2005). In addition, H3K4 me3 in budding yeast is thought to promote nucleosome acetylation and remodeling near promoters, while H3K4me2 leads to reduced histone acetylation levels (Kim and Buratowski, 2009). In Arabidopsis, high-resolution mapping of H3K9Ac, H3K27Ac, H3K9me3, and H3K27me3 in dark-grown and light-treated seedlings provided comprehensive insights into a combinatorial interplay between histone modifications and light-regulated gene expression (Charron et al., 2009). More H3K9Ac and H3K27me3 enriched regions were detected in light-treated seedlings, while more H3K27Ac and H3K9me3 enriched regions were detected in dark-grown seedlings. These results imply an adjustment of histone modification patterns in response to light. Cross-talk among different histone modifications at the Arabidopsis central clock loci is yet to be studied, although our understanding would shed light on the type of histone modification that has a primary influence on clock-gene activity.
We demonstrated correlations between histone modifications and clock-gene expression in Arabidopsis. A central question is whether these histone modifications have regulatory roles in clock-gene expression. This question might be answered by identification and functional studies of the catalytic components for histone modification. Considering the numerous functional redundancies among histone modifiers, reverse genetic approaches rather than forward genetic mutant screens might provide more opportunities for success.
Acknowledgments
This work was supported by grants from the National Research Foundation (2011-0015177) and by the Next-Generation BioGreen21 Program (TAGC with grant # PJ008202 and SSAC with grant # PJ008141) of the Rural Development Administration.
REFERENCES
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Belden W.J., Loros J.J., Dunlap J.C. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell. 2007;25:587–600. doi: 10.1016/j.molcel.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Brown S.A., Ripperger J., Kadener S., Fleury-Olela F., Vilbois F., Rosbash M., Schibler U. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Charron J.B., He H., Elling A.A., Deng X.W. Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell. 2009;21:3732–3748. doi: 10.1105/tpc.109.066845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., Tian L. Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim. Biophys Acta. 2007;1769:295–307. doi: 10.1016/j.bbaexp.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J.-N., Ryu J.-Y., Jeong Y.-M., Park J., Song J.-J., Amasino R.M., Noh B., Noh Y.-S. Control of seed germination by light-induced histone arginine demethylation activity. Dev Cell. 2012;22:736–748. doi: 10.1016/j.devcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- Clapier C.R., Cairns B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Covington M.F., Maloof J.N., Straume M., Kay S.A., Harmer S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9:R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosio C., Cermakian N., Allis C.D., Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat. Neurosci. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kevei E., Toth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A. Plant circadian clocks increase photosynthesis, growth, survival and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Doi M., Hirayama J., Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Dowson-Day M.J., Millar A.J. Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 1999;17:63–71. doi: 10.1046/j.1365-313x.1999.00353.x. [DOI] [PubMed] [Google Scholar]
- Dunlap J.C. Closely watched clocks: molecular analysis of circadian rhythms in Neurospora and Drosophila. Trends Genet. 1990;6:159–165. doi: 10.1016/0168-9525(90)90151-u. [DOI] [PubMed] [Google Scholar]
- Etchegaray J.P., Lee C., Wade P.A., Reppert S.M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Etchegaray J.P., Yang X., DeBruyne J.P., Peters A.H., Weaver D.R., Jenuwein T., Reppert S.M. The polycomb group protein EZH2 is required for mammalian circadian clock function. J. Biol. Chem. 2006;281:21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- Han S.K., Song J.D., Noh Y.S., Noh B. Role of plant CBP/p300-like genes in the regulation of flowering time. Plant J. 2007;49:103–114. doi: 10.1111/j.1365-313X.2006.02939.x. [DOI] [PubMed] [Google Scholar]
- Harmer S.L. The circadian system in higher plants. Annu. Rev. Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- Harmer S.L., Hogenesch J.B., Straume M., Chang H.-S., Han B., Zhu T., Wang X., Kreps J.A., Kay S.A. Orches-trated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Hirayama J., Sahar S., Grimaldi B., Tamaru T., Takamatsu K., Nakahata Y., Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- Hollender C., Liu Z. Histone deacetylase genes in Arabidopsis development. Integr. Plant Biol. 2008;50:875–885. doi: 10.1111/j.1744-7909.2008.00704.x. [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Kay S.A. Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci. 2006;11:550–558. doi: 10.1016/j.tplants.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Kim T., Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Lee J., Eshed-Williams L., Zilberman D., Sung Z.R. EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet. 2012;3:e1002512. doi: 10.1371/journal.pgen.1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Landry J., Sutton A., Hesman T., Min J., Xu R.M., Johnston M., Sternglanz R. Set2-catalyzed methylation of histone H3 represses basal expression of GAL4 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:5972–5978. doi: 10.1128/MCB.23.17.5972-5978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham J.A., Dent S.Y.R. Cross-regulation of histone modifications. Nat. Struct. Mol. Biol. 2007;14:1017–1024. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Smith E., Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Locke J.C., Kozma-Bognar L., Gould P.D., Feher B., Kevei E., Nagy F., Turner M.S., Hall A., Millar A.J. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2006;2:59. doi: 10.1038/msb4100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz M.C., Dickerson D.R., Schmitt M., Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- Masri S., Sassone-Corsi P. Plasticity and specificity of the circadian epigenome. Nat. Neurosci. 2010;13:1324–1329. doi: 10.1038/nn.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung C.R., Gutiérrez R.A. Network news: prime time for systems biology of the plant circadian clock. Curr. Opin. Genet. Dev. 2010;20:588–598. doi: 10.1016/j.gde.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R.Y. Circadian rhythms: basic neurobiology and clinical applications. Annu. Rev. Med. 1997;48:253–266. doi: 10.1146/annurev.med.48.1.253. [DOI] [PubMed] [Google Scholar]
- Naruse Y., Oh-hashi K., Iijima N., Naruse M., Yoshioka H., Tanaka M. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol. Cell. Biol. 2004;24:6278–6287. doi: 10.1128/MCB.24.14.6278-6287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y., Ito S., Nakamichi N., Mizoguchi T., Niinuma K., Yamashino T., Mizuno T. Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol. 2007;48:925–937. doi: 10.1093/pcp/pcm067. [DOI] [PubMed] [Google Scholar]
- Okitsu C.Y., Hsieh C.-L. DNA methylation dictates histone H3K4 methylation. Mol. Cell. Biol. 2007;27:2746–2757. doi: 10.1128/MCB.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M., Más P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell. 2007;19:2111–2123. doi: 10.1105/tpc.107.050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger J.A., Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Kobayashi A., Saze H., Kakutani T. RNAi-independent de novo DNA methylation revealed in Arabidopsis mutants of chromatin remodeling gene DDM1. Plant J. 2012;70:750–758. doi: 10.1111/j.1365-313X.2012.04911.x. [DOI] [PubMed] [Google Scholar]
- Schaffer R., Ramsay N., Samach A., Corden S., Putterill J., Carré I.A., Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Song H.-R., Noh Y.-S. Plants measure the time. J. Plant Biol. 2007;50:257–265. [Google Scholar]
- Strahl B.D., Grant P.A., Briggs S.D., Sun Z.W., Bone J.R., Caldwell J.A., Mollah S., Cook R.G., Shabanowitz J., Hunt D.F., et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M., Luo H., Lee S., Jin F., Yang J.S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N., et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To T.K., Kim J.M., Matsui A., Kurihara Y., Morosawa T., Ishida J., Tanaka M., Endo T., Kakutani T., Toyoda T., et al. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 2011;4:e1002055. doi: 10.1371/journal.pgen.1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M., Mulder K.W., Denissov S., Pijnappel W.W., van Schaik F.M., Varier R.A., Baltissen M.P., Stunnenberg H.G., Mann M., Timmers H.T. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Wang Z.-Y., Tobin E.M. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Wenden B., Kozma-Bognar L., Edwards K.D., Hall A.J., Locke J.C., Millar A.J. Light inputs shape the Arabidopsis circadian system. Plant J. 2011;66:480–491. doi: 10.1111/j.1365-313X.2011.04505.x. [DOI] [PubMed] [Google Scholar]
- Yamashino T., Ito S., Niwa Y., Kunihiro A., Nakamichi N., Mizuno T. Involvement of Arabidopsis clock-associated pseudo-response regulators in diurnal oscillations of gene expression in the presence of environmental time cues. Plant Cell Physiol. 2008;49:1839–1850. doi: 10.1093/pcp/pcn165. [DOI] [PubMed] [Google Scholar]
- Yerushalmi S., Yakir E., Green R.M. Circadian clocks and adaptation in Arabidopsis. Mol. Ecol. 2011;20:1155–1165. doi: 10.1111/j.1365-294X.2010.04962.x. [DOI] [PubMed] [Google Scholar]
- Zhang E.E., Kay S.A. Clocks not winding down: unraveling circadian networks. Nat. Rev. Mol. Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]