Abstract
Primary lung tumors, breast tumors, and melanoma metastasize mainly in the brain where therapy is limited to surgery and radiation. To investigate the molecular basis of brain metastases, we isolated brain-trophic metastatic MDA-MB-435-LvBr2 (LvBr2) cells via left ventricle (LV) injection of MDA-MB-435 cells into immunodeficiency (NOD/SCID) mice. Whereas parent MDA-MB-435 cells displayed an elongated morphology, LvBr2 cells were round and displayed an aggregated distribution. LvBr2 cells expressed lower β-catenin levels and higher heterogeneous nuclear ribonucleoprotein C1/C2 (hnRNPC) levels than parental cells. Since microRNAs are known to play an important role in cancer progression including metastasis, we screened microRNAs expressed specifically in brain metastases. MicroRNA-146a was almost undetectable in LvBr2 cells and highly expressed in the parental cells. Overexpression of miR-146a increased β-catenin expression and suppressed the migratory and invasive activity of LvBr2 cells. The miR-146a-elicited decrease in hnRNPC in turn lowered the expression of MMP-1, uPA, and uPAR and inhibited the migratory and invasive activity of LvBr2 cells. Taken together, our findings indicate that miR-146a is virtually absent from brain metastases and can suppress their metastatic potential including their migratory and invasive activities associated with upregulation of β-catenin and downregulation of hnRNPC.
Keywords: β-catenin, brain metastasis, HnRNPC, MicroRNA-146a
INTRODUCTION
Despite great efforts to develop cancer therapies, cancer mortality has been on the rise, which is due to metastasis rather than to primary tumor itself (Nguyen et al., 2009). Metastasis has been recognized as the main cause of recurrence and poor prognosis. The metastatic process involves multiple genes which enable cells to enter the bloodstream, disseminate, adapt to new miroenvironments, and proliferate at distant organs (Chambers et al., 2002). Although brain metastases are highly aggressive, they can only be treated using surgery and radiation. Metastasis of melanoma cells to lymph nodes, liver, lung, and brain constitutes a major cause of death (Akslen et al., 1987). In the case of brain metastasis, the prognosis of melanoma patients is very poor and median survival is less than 6 months. Moreover, despite of advanced therapies, treatment options for brain metastatic melanoma are restricted and their efficiency is very low.
With the discovery of oncogenic and tumor suppressive miRNAs, aberrant expression of miRNAs has been implicated in the malignancy of cancer cells. Many miRNAs are found to regulate metastatic processes including epithelial-to-mesenchymal transition (EMT), adhesion, migration, invasion, apoptosis, angiogenesis, and colonization (Hurst et al., 2009). Metastasis-associated miRNAs (Metastamir) are categorized into metastasis-suppressing and metastasis-promoting miRNAs (Aigner, 2011; Hurst et al., 2009; Nicoloso et al., 2009) and their functions have been investigated in variety of cancer cells. MiR-146 is reported to enhance proliferation and inhibit apoptosis of gastric cancer cells (Xiao et al., 2012) and to diminish the metastatic potential of breast cancer cells by targeting IRAK1 and TRAF6, which suppress invasiveness of pancreatic cancer cells via positive regulation of NF-κB (Bhaumik et al., 2008; Li et al., 2010).
In the present report, we studied brain metastasis-specific miRNAs using a xenograft model. We found that brain-trophic metastatic LvBr2 cells expressed higher level of proteolytic enzymes than parent MDA-MB-435 cells. Of 97 miRNAs whose expression was significantly changed in brain metastasis, miR-146a showed potently decreased expression in brain metastasis and its overexpression suppressed the metastatic potential of LvBr2 cells, which is linked to the upregulation of β-catenin and the downregulation of hnRNPC.
MATERIALS AND METHODS
Xenograft model for brain metastasis and miRNA microarray
Brain-trophic metastatic MDA-MB-435 cells were isolated previously after serial injection of MDA-MB-435 cells through the left ventricle (LV) of heart (Nam et al., 2008). We termed these brain-trophic metastatic cells as MDA-MB-435-LvBr1 (LvBr1, first cycle) and MDA-MB-435-LvBr2 (LvBr2, second cycle). To rule out the effect of the brain microenvironment, MDA-MB-435-IcBr cells (IcBr) were isolated from brain lesions performed by intracranial (IC) injection of MDA-MB-435 cells. Since LvBr1 and LvBr2 cells showed very similar results, here we represented the results obtained using LvBr2. Total RNA was isolated using Trizol (Invitrogen) and used for microRNA microarray (Affymetrix GeneChip® microRNA microarray).
Cell culture and transfection
MDA-MB-435, LvBr1, LvBr2, and IcBr cells were maintained at 37°C, 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Hyclone) supplemented with 10% fetal bovine serum (GIBCO-BRL) and 1% antibiotic-antimycotic solution (GIBCO-BRL). Cells were transfected using lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol. HnRNPC siRNA was from Santa Cruz Biotechnology and control siRNA (CTRL) from Genolution (Korea). Precursor miR-146a (pre-miR-146a, Ambion) was used to overexpress miR-146a.
Reagents and antibodies
Cycloheximide was from Sigma Aldrich. The antibodies were as follows: anti-hnRNPC and anti-EGFR were from Santa Cruz Biotechnology; anti-Flag was from Sigma Aldrich, and antiphospho-Akt, anti-Akt, anti-phospho-ERK, and anti-ERK from Cell Signaling Technology. Anti-GAPDH, anti-ST6GALNAC5, and anti-E-cadherin were from Abcam.
Western blot and quantitative real-time PCR
For Western blot analysis, proteins were fractionated and then transferred to PVDF membrane (Millipore). Membranes were incubated with primary antibodies and HRP-conjugated secondary antibodies, and visualized using enhanced chemiluminiscence (ECL, Amersham). For reverse transcription (RT) and quantitative (q) real-time PCR analysis, total RNA was isolated using Trizol (Invitrogen) according to the manufacturer's procedure. The level of mRNA expression was determined using power SYBR® Green PCR Master Mix (Applied Biosystems, USA). Primer sequences are listed in Supplementary Table 1. The level of GAPDH mRNA was used as internal control for normalization.
Migration and invasion assay
Migratory activity was determined by measuring cells with the ability to cross the pores of migration chambers in the presence of chemoattractants (10% FBS). Briefly, equal numbers of starved cells were added onto the upper chambers of 24-well Falcon Cell Culture Transwells (8 μm pore, QCM Colorimetric Cell Migration Assay kit, Millipore). Migration of cells was initiated by adding serum-containing media into the lower chamber; 24 h later, the migrated cells were stained with crystal violet and the migratory activity was determined by measuring the optical density at 560 nm. For invasion assay, Matrigel Transwells (BD Biocoat Matrigel Invasion Chamber) were used. Equal numbers of cells were added to the upper chamber and cells were allowed to invade by adding fresh DMEM containing 10% fetal bovine serum into the lower compartment. Invading cells were fixed and stained with hematoxylin/eosin (H/E) and the number of invading cells on the lower surface of the membrane was counted under a microscope in several fields.
RESULTS
Brain-trophic metastatic LvBr2 cells showed higher metastatic activity than parent MDA-MB-435 cells
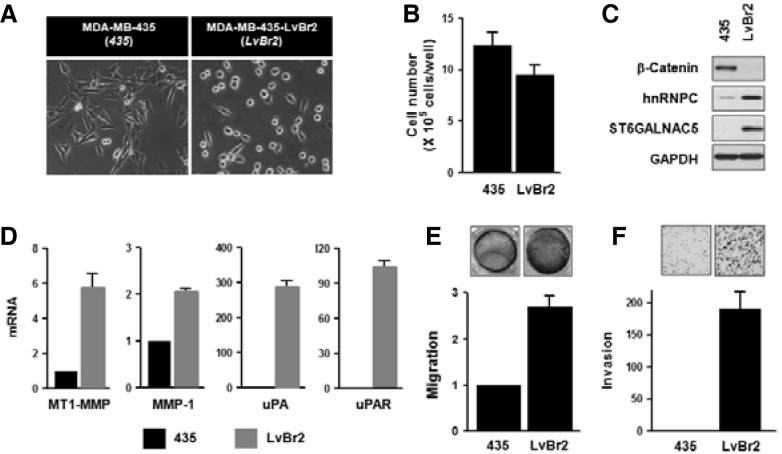
MDA-MB-435 cells were reported to form brain metastasis by injection into the left ventricle (LV) of heart or the internal carotid artery (ICA) (Nam et al., 2008; Price et al., 1990). To investigate the role of miRNA in brain metastasis, MDA-MB-435-LvBr cells were isolated from brain metastasis cells after serial LV injection of parent MDA-MB-435 cells (LvBr1 cells were obtained from the first cycle and LvBr2 cells from the second cycle). Since LvBr1 and LvBr2 cells appeared to display very similar molecular characteristics (Supplementary Fig. 2), we only represent here LvBr2 results. LvBr2 cells preferred to metastasize to the brain and showed morphology very distinct from that of MDA-MB-435 cells (Fig. 1A). While MDA-MB-435 cells had a spindle-like and elongated morphology, LvBr2 displayed a round and aggregated shape. LvBr2 cells proliferated slightly more slowly than MDA-MB-435 cells (Fig. 1B). Western blot analysis revealed that β-catenin expression levels were lower, while hnRNPC expression levels were higher in LvBr2 cells (Fig. 1C). Brain metastasis was validated by analysis of the marker ST6GALNAC5 (alpha2,6-sialyltransferase) (Bos et al., 2009). Additionally, we checked the expression of various matrix proteinases, since metastasis is triggered by degradation of extracellular matrix (ECM); membrane type 1-matrix metalloproteinase (MT1-MMP), matrix metalloproteinase-1 (MMP-1), urokinase-type plasminogen activator (uPA), and uPA receptor (uPAR) were all more highly expressed in LvBr2 cells than in MDA-MB-435 cells (Fig. 1D). In keeping with the elevated expression of matrix proteinases, LvBr2 cells were more migratory and invasive than MDA-MB-435 cells (Figs. 1E and 1F). From above results, we found that brain-trophic LvBr2 cells showed different morphology and more metastatic activity as compared to MDA-MB-435 cells.
Fig. 1.
LvBr2 cells show more metastatic potential than MDA-MB-435 cells. (A) Comparison of cell morphology between MDA-MB-435 cells and the derived brain metastatic cells, LvBr2. (B) MDA-MB-435 and LvBr2 cells were seeded into 6-well plate at a density of 2 × 105 cells per well; 48 h later, cell numbers were determined by direct cell counting. Data are the means and S.D. from three different experiments. (C) The expression of β-catenin, hnRNPC, ST6GALNAC5, and GAPDH was assessed by Western blot analysis of whole-cell lysates from MDA-MB-435 and LvBr2 cells. ST6GALNAC5 expression was used as a marker of brain metastasis. (D) Total RNA was isolated from MDA-MB-435 and LvBr2 cells and the levels of MT1-MMP, MMP-1, uPA, and uPAR mRNA was determined by RT-qPCR analysis. Data are the means and S.D. from three different experiments. (E, F) Equal numbers of MDA-MB-435 and LvBr2 cells were added into Transwells (E) or Matrigel Transwells (F). For migration assay (E), non-migrating cells were completely removed, and migrating cells were stained and photographed. The migratory activity was determined by measuring absorbance at 560 nm. Data are the means and S.D. from three different experiments. For invasion assay (F), invading cells were fixed with 100% methanol, stained with H/E, and counted under a microscope. Data are the means and S.D. from three different experiments.
Decreased level of miR-146a in LvBr2 is closely associated with metastatic activity
To screen for miRNAs which display differential expression in brain metastasis, microRNA microarray was performed as described in “Materials and Methods”. This analysis revealed that in LvBr1 and LvBr2 cells, 34 miRNAs were upregulated and 63 miRNAs were downregulated (Supplementary Fig. 3). Among them, miR-146a showed the most significant difference between MDA-MB-435 and LvBr cells. Whereas miR-146a was highly expressed in MDA-MB-435 cells, it was virtually undetectable in LvBr2 cells (Fig. 2A and Supplementary Fig. 4). To investigate the possible role of miR-146a in the metastatic potential of LvBr2, migratory and invasive activities were examined after miR-146 overexpression. As shown in Fig. 2B and 2C, transfection of LvBr2 cells with pre-miR-146a caused a reduction in migration and invasiveness, suggesting that miR-146a is closely associated with the metastatic potential of LvBr2 cells.
Fig. 2.
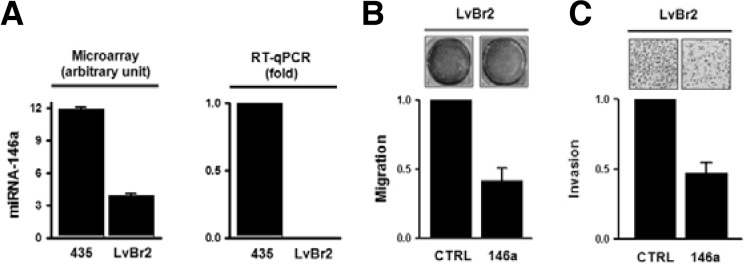
MiR-146a is highly expressed in LvBr2 cells and suppresses their metastatic potential. (A) The level of miR-146a in MDA-MB-435 and LvBr2 cells was validated using TaqMan microRNA assay (right). (B, C) LvBr2 cells were transfected with control siRNA (CTRL) or pre-miR-146a (146a); 48 h later, cells were harvested and equal numbers of MDA-MB-435 and LvBr2 cells were added into Transwells (B) or Matrigel Transwells (C). For the migration assay (B), non-migrating cells were completely removed, and migrating cells were stained and photographed. The migratory activity was determined by measuring absorbance at 560 nm. Data are the means and S.D. from three different experiments. For the invasion assay (C), invading cells were fixed with 100% methanol, stained with H/E, and counted under a microscope. Data are the means and S.D. from three different experiments.
Overexpression of miR-146a suppressed the metastatic activity of LvBr2 cells linked to increased expression of β-catenin
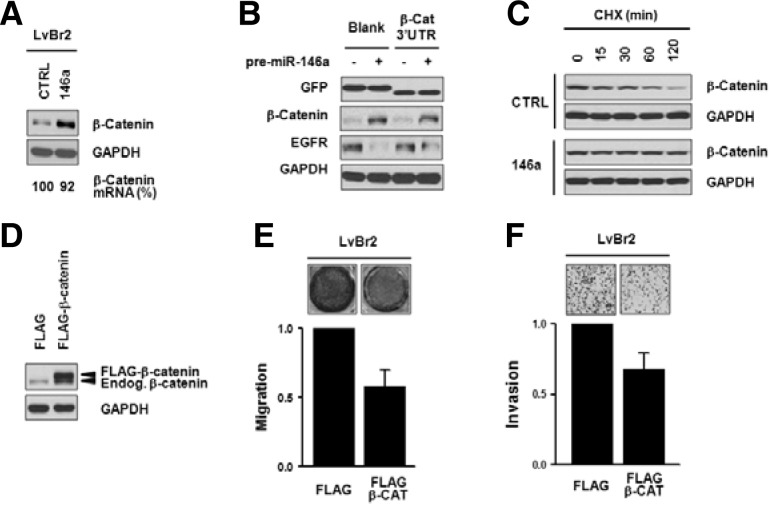
To elucidate the molecular mechanisms whereby miR-146a suppresses migratory and invasive activity, we examined the effect of miR-146a on β-catenin expression. As shown in Fig. 1C, LvBr2 cells showed undetectable levels of β-catenin expression, while MDA-MB-435 cells highly expressed β-catenin. Transfection of LvBr2 cells with pre-miR-146a increased β-catenin expression levels without significantly altering β-catenin mRNA levels (Fig. 3A). Increased expression of β-catenin by miR-146a was also found in Br4 cells which were isolated through two cycles of injection of LvBr2 cells into the internal carotid artery (Supplementary Fig. 2D) (Nam et al., 2008).
Fig. 3.
Overexpression of miR-146a suppresses the metastatic potential of LvBr2 cells associated with upregulation of β-catenin. (A) LvBr2 cells were transfected with control siRNA (CTRL) or pre-miR-146a (146a); 48 h later, whole-cell lysates and total RNA were prepared. The level of β-catenin protein and mRNA was determined by Western blot and RT-qPCR analyses, respectively. (B) To investigate whether increased expression of β-catenin by miR-146 was mediated through binding to 3′UTR of β-catenin mRNA, EGFP vector containing the β-catenin 3′UTR was constructed. Twenty-four h after transfecting LvBr2 cells with control siRNA (CTRL) or pre-miR-146a (146a), cells were resuspended into 6-well plates and transfected with the GFP vectors. GFP expression was assessed by Western blot analysis and EGFR expression was checked to verify miR-146a transfection. (C) To check the effect of miR-146a overexpression on β-catenin protein stability, LvBr2 cells were transfected with control siRNA (CTRL) or pre-miR-146a (146a). Cells were resuspended into 6-well plates and treated with cycloheximide; at the times indicated, cells were harvested and whole-cell lysates were prepared. The level of β-catenin expression was determined by Western blot analysis. (D-F) To elucidate the role of β-catenin in suppression of metastatic potential by miR-146a, β-catenin was overexpressed in LvBr2 cells (D). Equal numbers of transfected cells were added into Transwells (E) or Matrigel Transwells (F). For migration assay (E), non-migrating cells were completely removed, and migrating cells were stained and photographed. The migratory activity was determined by measuring absorbance at 560 nm. Data are the means and S.D. from three different experiments. For invasion assay (F), invading cells were fixed, stained with H/E, and counted using a microscope. Data are the means and S.D. from three different experiments.
To evaluate whether miR-146a influenced β-catenin expression through the β-catenin 3′UTR, we used a reporter vector that expressed a heterologous GFP mRNA carrying the β-catenin 3′UTR. As shown in Fig. 3B, GFP expression was not influenced by miR-146a overexpression, indicating that miR-146a was unlikely to function through its interaction with the β-catenin 3′UTR. In order to verify overexpression of miR-146a, we checked the level of EGFR expression which is known as miR-146a target. β-catenin expression levels are regulated via its phosphorylatioin-dependent degradation by ubiquitin/proteasome system. We studied the stability of β-catenin protein in miR-146a-overexpressing LvBr2 cells by treating cells with cycloheximide (10 μg/ml) to stop protein synthesis and, at the indicated times, β-catenin expression levels were determined by Western blot analysis. As shown, β-catenin was more stable in miR-146a-transfected LvBr2 cells than in control-transfected cells (Fig. 3C), while GADPH levels were unchanged between these two groups.
To investigate whether the miR-146a-mediated increase of β-catenin is required for its anti-metastatic action, we increased β-catenin abundance in LvBRr2 cells by transfection with a vector expressing β-catenin-FLAG (Fig. 3D). Importantly, LvBr2 cells overexpressing β-catenin showed diminished migratory and invasive activity as compared to FLAG-transfected LvBr2 cells (Figs. 3E and 3F). These results indicated that miR-146a increased β-catenin expression by increasing its stability, which suppressed metastatic features of LvBr2 cells.
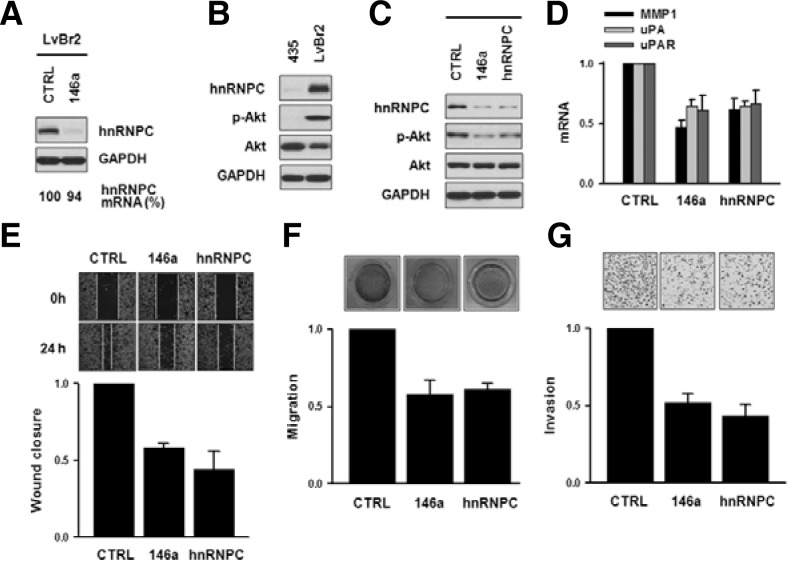
Suppression of hnRNPC is necessary to inhibit the metastatic potential by miR-146a in LvBr2 cells
As shown in Fig. 1C, hnRNPC expression levels in LvBr2 cells were higher than in parent MDA-MB-435 cells. LvBr1 cells showed similar expression as LvBr2 (Supplementary Figs. 2A and 2B). Forty-eight h after transfection of LvBr2 cells with premiR-146a, Western blot analysis revealed that hnRNPC levels were reduced without changes in mRNA levels (Fig. 4A). Together with the level of hnRNPC expression, we found significant differences in the activation of Akt between MBA-MB-435 and LvBr2 cells (Fig. 4B). Diminished expression of hnRNPC by miR-146 or hnRNPC-specific siRNA inactivated Akt pathway in LvBr2 cells, suggesting that the decreased expression of hnRNPC was necessary for inhibiting metastasis by miR-146a (Fig. 4C). As with suppression of Akt, expression of matrix-degrading enzymes including MMP and uPA was decreased by overexpression of miR-146a in LvBr2 cells (Fig. 4D); similarly, silencing of hnRNPC in both LvBr1 and LvBr2 cells diminished the expression levels of various proteolytic enzymes (Supplementary Fig. 5). To investigate the effect of miR-146a-elicited hnRNPC suppression on the metastatic activity of LvBr2 cells, the migratory activity of LvBr2 cells was assessed by the wound healing and Transwell migration assays after transfection with pre-miR-146a or hnRNPC siRNA (Figs. 4E and 4F, respectively). Both the overexpression of miR-146a and the silencing of hnRNPC significantly inhibited the migratory activity in LvBr2 cells. The invasive activity of transfected LvBr2 cells was investigated by using a Matrigel Invasion Chamber. As shown in Fig. 4G, the number of invading LvBr2 cells decreased in miR-146aoverexpressing and hnRNPC-silenced cells. The above results indicate that miR-146a inhibited metastatic traits of LvBr2 cells, including their ability to migrate and invade by suppressing hnRNPC expression. Taken together, our results suggest that the anti-metastatic activity of miR-146a includes two regulatory events: an increase in β-catenin and the suppression of hnRNPC.
Fig. 4.
Suppression of hnRNPC is necessary to inhibit the metastatic potential engendered by miR-146a in LvBr2 cells. (A) LvBr2 cells were transfected with control siRNA (CTRL) or pre-miR-146a (146a) and 48 h later, whole-cell lysates and total RNA were prepared to detect hnRNPC protein levels and mRNA levels by Western blot and RT-qPCR analysis, respectively. (B) Activation of Akt was determined by Western blot analysis of whole-cell lysates of MDA-MB-435 and LvBr2 cells. (C) LvBr2 cells were transfected with control siRNA (CTRL), pre-miR-146a (146a), or hnRNPC siRNA (hnRNPC); 48 h later, whole-cell lysates were prepared and the activation of Akt was determined by Western blot analysis. (D) LvBr2 cells were transfected as above and total RNA was prepared. The mRNA level of proteolytic enzymes including MMP-1, uPA, and uPAR was determined by RT-qPCR. (E-G) To investigate the anti-metastatic effect of miR-146a, the migratory and invasive activities were determined by wound healing (E), migration (F), and invasion (G) assays. LvBr2 cells were transfected as above, transferred into 12-well plates and 24 h later, scratch was made using a plastic pipet tip. The migratory activity was determined by measuring the distance of wound closure (E). Equal numbers of transfected cells were added into Transwells (F) or Matrigel Transwells (G). For migration assay (F), non-migrating cells were completely removed, and migrating cells were stained and photographed. The migratory activity was determined by measuring absorbance at 560 nm. Data are the means and S.D. from three different experiments. Invading cells were fixed with 100% methanol, stained with H/E, and counted under microscope (G). Data are the means and S.D. from three different experiments.
DISCUSSION
Brain metastases are the most common tumors of the central nervous system and are mainly originated from lung cancer, breast cancer, and melanoma. Even though about 40-60% of melanoma patients suffer from brain metastases, the molecular basis of the metastatic colonization of melanoma in brain is still obscure. In this report, we sought to identify miRNAs expressed in brain-trophic metastasis developed through a xenograft model. Among the differentially expressed miRNAs, we became interested in miR-146a, as its expression was lost in the metastatic cells (LvBr2). Moreover, overexpression of miR-146a inhibited several metastatic traits of LvBr2 cells, particularly migration and invasion. MiR-146a elicits these effects via two mechanisms: by upregulating β-catenin and by downregulating hnRNPC (Figs. 3 and 4).
The Wnt pathway is critical for signaling in tumorigenesis and metastasis. Deregulation of Wnt signal abnormally stabilizes β-catenin, which results in increased β-catenin levels in both the cytoplasm and the nucleus (Giles et al., 2003). In many types of cancer, increased expression of β-catenin is closely associated with enhanced cancer phenotype and is found in the invasive front of most colon cancers. Moreover, its target genes are used in the prognosis and disease-free survival of cancer patients (Giles et al., 2003). In contrast to earlier reports, β-catenin was associated with good prognosis and increased overall survival (Chien et al., 2009; Gould Rothberg et al., 2009) and its reduced expression was linked to cancer progression including metastasis in melanoma (Maelandsmo et al., 2003). In this regard, Arozarena et al. (2011) recently reported that MITF, a melanoma-specific protein, opposes the function of β-catenin in melanoma. The β-catenin suppresses the invasive activity of melanoma and affects the invasive morphology of melanoma through inhibition of Rho-GTPase (Arozarena et al., 2011). We found that overexpression of miR-146a in LvBr2 cells increased the stability and hence the expression of β-catenin, which inhibit metastatic activity (Figs. 3F and 3G). Furthermore, since β-catenin is known to inhibit NF-κB DNA binding and transactivation by forming a complex with NF-κB (Deng et al., 2002), NF-κB might be involved in the anti-metastatic action of miR-146a; however, additional work is needed to investigate this hypothesis.
HnRNPC is an RNA-binding protein that influences mRNA metabolism including stability and translation (Christian et al., 2008). It binds the uPAR 3′UTR and increases uPAR expression by enhancing uPAR mRNA stability (Bhandary et al., 2009; Shetty, 2005). By comparing the expression of metastasis-associated genes, we found that Akt activation is markedly different between MDA-MB-435 and LvBr2 cells. Silencing of Akt suppresses cell migration and invasion, suggesting that Akt activation is necessary for metastatic potential (Qiao et al., 2008). Overexpression of miR-146a in LvBr2 cells suppressed the activation of Akt via downregulation of hnRNPC (Fig. 4C), which is associated with the decreased expression of proteolytic enzymes such as MMPs and uPA. Collectively, we found that miR-146a suppresses the metastatic activity of LvBr2 cells by upregulating β-catenin and downregulating hnRNPC.
Acknowledgments
We thank Prof. Sunjoo Jeong (Dankook University, Korea) for a reporter vector expressing GFP mRNA carrying the β-catenin 3′UTR. This study was supported by a grant from the Basic Science Research Program, National Research Foundation of Korea by the Ministry of Education, Science, and Technology (2011-009329 to H. H. Kim) and the Korea Health Care technology R&D project, Ministry of Health & Welfare Affairs, Republic of Korea (A092255). Myriam Gorospe was supported by the NIA-IRP, NIH.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Aigner A. MicroRNAs (miRNAs) in cancer invasion and metastasis: therapeutic approaches based on metastasis-related miRNAs. J. Mol. Med. 2011;89:445–457. doi: 10.1007/s00109-010-0716-0. [DOI] [PubMed] [Google Scholar]
- Akslen L.A., Hove L.M., Hartveit F. Metastatic distribution in malignant melanoma. A 30-year autopsy study. Invasion Metastasis. 1987;7:253–263. [PubMed] [Google Scholar]
- Arozarena I., Bischof H., Gilby D., Belloni B., Dummer R., Wellbrock C. In melanoma, beta-catenin is a suppressor of invasion. Oncogene. 2011;30:4531–4543. doi: 10.1038/onc.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandary Y.P., Velusamy T., Shetty P., Shetty R.S., Idell S., Cines D.B., Jain D., Bdeir K., Abraham E., Tsuruta Y., et al. Post-transcriptional regulation of urokinase-type plasminogen activator receptor expression in lipopolysaccharide-induced acute lung injury. Am. J. Respir. Crit. Care Med. 2009;179:288–298. doi: 10.1164/rccm.200712-1787OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik D., Scott G.K., Schokrpur S., Patil C.K., Campisi J., Benz C.C. Expression of microRNA-146 suppres-ses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27:5643–5647. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos P.D., Zhang X.H., Nadal C., Shu W., Gomis R.R., Nguyen D.X., Minn A.J., van de Vijver M.J., Gerald W.L., Foekens J.A., et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers A.F., Groom A.C., MacDonald I.C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- Chien A.J., Moore E.C., Lonsdorf A.S., Kulikauskas R.M., Rothberg B.G., Berger A.J., Major M.B., Hwang S.T., Rimm D.L., Moon R.T. Activated Wnt/beta-catenin signaling in melanoma is associated with decreased proliferation in patient tumors and a murine melanoma model. Proc. Natl. Acad. Sci USA. 2009;106:1193–1198. doi: 10.1073/pnas.0811902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian K.J., Lang M.A., Raffalli-Mathieu F. Interaction of heterogeneous nuclear ribonucleoprotein C1/C2 with a novel cis-regulatory element within p53 mRNA as a response to cytostatic drug treatment. Mol. Pharmacol. 2008;73:1558–1567. doi: 10.1124/mol.107.042507. [DOI] [PubMed] [Google Scholar]
- Deng J., Miller S.A., Wang H.Y., Xia W., Wen Y., Zhou B.P., Li Y., Lin S.Y., Hung M.C. beta-catenin interacts with and inhibits NF-kappa B in human colon and breast cancer. Cancer Cell. 2002;2:323–334. doi: 10.1016/s1535-6108(02)00154-x. [DOI] [PubMed] [Google Scholar]
- Giles R.H., van Es J.H., Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- Gould Rothberg B.E., Berger A.J., Molinaro A.M., Subtil A., Krauthammer M.O., Camp R.L., Bradley W.R., Ariyan S., Kluger H.M., Rimm D.L. Melanoma prognostic model using tissue microarrays and genetic algorithms. J. Clin. Oncol. 2009;27:5772–5780. doi: 10.1200/JCO.2009.22.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst D.R., Edmonds M.D., Welch D.R. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495–7498. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Vandenboom T.G., 2nd, Wang Z., Kong D., Ali S., Philip P.A., Sarkar F.H. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70:1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maelandsmo G.M., Holm R., Nesland J.M., Fodstad O., Florenes V.A. Reduced beta-catenin expression in the cytoplasm of advanced-stage superficial spreading malignant melanoma. Clin. Cancer Res. 2003;9:3383–3388. [PubMed] [Google Scholar]
- Nam D.H., Jeon H.M., Kim S., Kim M.H., Lee Y.J., Lee M.S., Kim H., Joo K.M., Lee D.S., Price J.E., et al. Activation of notch signaling in a xenograft model of brain metastasis. Clin. Cancer Res. 2008;14:4059–4066. doi: 10.1158/1078-0432.CCR-07-4039. [DOI] [PubMed] [Google Scholar]
- Nguyen D.X., Bos P.D., Massague J. Metastasis: from dissemination to organ-specific colonization. Nat. Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- Nicoloso M.S., Spizzo R., Shimizu M., Rossi S., Calin G.A. MicroRNAs--the micro steering wheel of tumour metastases. Nat. Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- Price J.E., Polyzos A., Zhang R.D., Daniels L.M. Tumorigenicity and metastasis of human breast carcinoma cell lines in nude mice. Cancer Res. 1990;50:717–721. [PubMed] [Google Scholar]
- Qiao M., Sheng S., Pardee A.B. Metastasis and AKT activation. Cell Cycle. 2008;7:2991–2996. doi: 10.4161/cc.7.19.6784. [DOI] [PubMed] [Google Scholar]
- Shetty S. Regulation of urokinase receptor mRNA stability by hnRNP C in lung epithelial cells. Mol. Cell. Biochem. 2005;272:107–118. doi: 10.1007/s11010-005-7644-2. [DOI] [PubMed] [Google Scholar]
- Xiao B., Zhu E.D., Li N., Lu D.S., Li W., Li B.S., Zhao Y.L., Mao X.H., Guo G., Yu P.W., et al. Increased miR-146a in gastric cancer directly targets SMAD4 and is involved in modulating cell proliferation and apoptosis. Oncol. Rep. 2012;27:559–566. doi: 10.3892/or.2011.1514. [DOI] [PubMed] [Google Scholar]