Abstract
The transcription factor ATAF2, one of the plant specific NAC family genes, is known as repressor of pathogenesis-related genes and responsive to the diverse defense-related hormones, pathogen infection, and wounding stress. Furthermore, it is important to consider that tryptophan-dependant IAA biosynthesis pathway can be activated by wounding and pathogen. We found that ATAF2pro::GUS reporter was induced upon indole-3-acetonitrile (IAN) treatments. And ataf2 mutant showed reduced sensitivity to IAN whereas 35S::ATAF2 plants showed hyper-sensitivity to IAN. IAN biosynthesis required nitrilase involved in the conversion of IAN to an auxin, indole-3-acetic acid (IAA). We found that the NIT2 gene was repressed in ataf2 knockout plants. Expression of both ATAF2 and NIT2 genes was induced by IAN treatment. Transgenic plants overexpressing ATAF2 showed up-regulated NIT2 expression. ATAF2 activated promoter of the NIT2 gene in Arabidopsis protoplasts. Electrophoretic mobility shift assay revealed that NIT2 promoter region from position −117 to −82 contains an ATAF2 binding site where an imperfect palindrome sequence was critical to the protein-DNA interaction. These findings indicate that ATAF2 regulates NIT2 gene expression via NIT2 promoter binding.
Keywords: Arabidopsis, ATAF2, Auxin, NAC transcription factor, Nitrilase
INTRODUCTION
Plants are under constant threat from environmental stress and attack from pathogens, vertebrates, and invertebrates. To cope with this, plants have evolved elaborate mechanisms in perceiving the attack and responding to the signaling to unite the reception of information to the induction of appropriate defense gene expression (Hegedus et al., 2003). Upon stress, gene expression is tightly regulated in a coordinated and timely manner, and the regulation of gene expression is largely governed by specific transcription factors (Wray et al., 2003).
Plant-specific NAC transcription factor containing the NAC domain, namely NAM (no apical meristem), ATAF1,2 and CUC2 (cup-shaped cotyledon) is one of the largest plant-specific families (Ernst et al., 2004; Riechmann et al., 2000). The members of the NAC protein family are identifiable by the presence of a highly conserved N-terminal NAC domain accompanied by diverse C-terminal domains (Ernst et al., 2004). The DNA binding ability of this protein family has been assigned to the NAC domain (Duval et al., 2002; Ernst et al., 2004; Xie et al., 2000). Only a portion of the NAC genes has been studied to date and the family has been implicated in diverse processes (Fang et al., 2008). Some NAC proteins have a role in biotic and abiotic stress as well as in plant development (Fang et al., 2008). For example, transgenic plants overexpressing one of three different Arabidopsis NAC proteins (ANAC019, ANAC055, and ANAC072) display significantly increased drought tolerance (Tran et al., 2004). The TM2/Arabidopsis NAC domain-containing protein 69 is involved with auxin and salt signaling in seed germination (Park et al., 2011). Thus, NAC proteins are emerging as important proteins in plant development and stress response.
Although NAC family members are very numerous and are known to be transcription factors, little is known about the direct target genes and the DNA-binding specificity of NAC proteins (Olsen et al., 2005). Arabidopsis ATAF1 and ATAF2 were isolated based on their ability to activate the Cauliflower mosaic virus (CaMV) 35S promoter in yeast (Souer et al., 1996). However, only some DNA binding sites in a direct target gene of NAC proteins has been clearly identified to date. For example, ANAC019, ANAC055, and ANAC072 bind to the 63 bp promoter region of the ERD1 gene and the NAC recognition sequence (NACRS), ANNNNNTCNNNNNNNACACGCATGT, containing CACG as the core DNA-binding site was determined using the yeast one-hybrid system (Tran et al., 2004). Furthermore, vascular-related NAC-domain 7 (VND7) and secondary wall-associated NAC domain protein 1 (SND1) directly bind to secondary wall NAC binding element (SNBE) in the target promoters (Zhong et al., 2010).
Previously, ATAF2 was induced by salt stress and pathogen attack and involved in the regulation of biotic and abiotic stress and development (Delessert et al., 2005; Seki et al., 2002). However, ATAF2 has no known biological function in auxin pathway. We report here that ATAF2 directly binds to the promoter of nitrilase 2 (NIT2) that encodes a nitrilase, which converts IAN to auxin, and activates NIT2 transcription in vivo. Furthermore, we analyzed the ATAF2 binding sequence within the NIT2 promoter and demonstrated that the sequence contains an imperfect palindrome, CAAATNNNATTTG.
MATERIALS AND METHODS
Plant materials
Arabidopsis thaliana (ecotype Col-0) were grown in soil under a 16 h-light/8 h-dark photoperiod at 23°C. For chemical treatments, plants were grown aseptically on plant nutrient medium (Haughn and Somerville, 1986) supplemented with 0.5% (w/v) sucrose (PNS) and 0.6% agar or PNS supplemented with 10–50 μM IAN or 0.5–3 μM IAA.
Construction of transgenic plants
The coding region of ATAF2 was obtained by PCR amplification. The resulting PCR fragment of ATAF2 was cloned into XbaI/SacI site in pC2300M2 (CAMBIA). The construct was introduced into Col-0 and ataf2 mutant plants using Agrobacterium-mediated transformation as described previously (Clough and Bent, 1998). The ataf2 mutant was obtained from the Salk mutant collection (SALK_015750).
To generate ATAF2promoter::GUS transgenic plants, a 2.3 kb fragment upstream from the start codon of ATAF2 was PCR amplified from genomic DNA of Arabidopsis. The 2.3 kb fragment was ligated into BamH1/SmaI site of the promoter-less GUS expression vector pBI101 (Clontech).
To generate NIT2-RNAi transgenic plants, fragment of NIT2 was PCR amplified from cDNA of Arabidopsis, cloned, and inserted into the pHANNIBAL vector in inverted repeat manner. And then, a NotI fragment containing CaMV 35S promoter with which repeated sequence fused was subjected to cloning into the pART27 binary vector. The resulting vector was subjected into Agrobacterium-mediated transformation as described previously (Clough and Bent, 1998).
Construction of deleted promoter regions fused to β-glucuronidase or luciferase reporter genes
For the construction of various chimeric genes with the NIT2 promoter fused to the GUS or LUC reporter genes, initially the 2.4 kb upstream sequence of NIT2 was obtained with PCR using the NIT2F2.4K and NIT2R2.4K primers (Supplementary Table S1). With the 2.4 kb PCR fragment as a template, a series of 5′ deletion constructs of the NIT2 promoter were generated using the same reverse PCR primer, NIT2PROR and specific forward primer set: P1 primer for the 1,467 bp P1 construct (−1,505 to −38); P2 primer for the 665 bp P2 construct (−703 to −38); P3 primer for the 234 bp P3 construct (−272 to −38); P4 primer for the 230 bp P4 construct (−268 to −38); P5 primer for the 199 bp P5 construct (−237 to −38); P6 primer for the 148 bp P6 construct (−186 to −38); P7 primer for the 117 bp P7 construct (−155 to −38); P8 primer for the 90 bp P8 construct (−128 to −38); P9 primer for the 80 bp P9 construct (−118 to −38). The resulting constructs were ligated into a BamHI/NcoI site of the promoter-less pC2300X vector. For the two 3′ deletion constructs, PCR was performed with P2 primer and P10 primer for the 584 bp P10 construct (−665 to −81) or with P2 primer and P11 primer for the 548 bp P11 construct (−665 to −117). The constructs were ligated into the BamHI site of pBKM2 vector in which 35S minimal promoter was fused with the LUC coding region.
RNA blot analysis and qRT-PCR
For expression pattern analyses, three-week-old plants were harvested from PNS agar plates, and then grown hydroponically in a solution containing 300 mM NaCl, 100 μM ABA, 200 μM IAN, 1 mM SNP, and 1 mM salicylic acid (SA). For dehydration stress, plants were transferred on Whatman 3MM paper at 23°C with approximately 60% humidity condition. The total RNA was extracted with TRI REAGENT as described in the manufacturer’s protocol (Molecular Research Center). RNA blot was performed according to a published protocol (Church and Gilbert, 1984).
qRT-PCR was performed with SYBR Green (KapaBiosystems) according to the manufacturer’s instructions and as previously described (Huh et al., 2012). Three independent biological replicate cDNA samples were tested for each experiment. Relative mRNA levels for each transcript were determined by the comparative cycle-threshold (CT) method (Appliedbiosystems). Expression data was normalized with an endogenous Actin7 gene (internal reference). The following primer sets were used: ATAF2F 5′-TCTTCTACGCAGGGAAAGCTCC-3′ and ATAF2R 5′-AGCCATTGTCGTGGTCCTCG-3′, NIT1F 5′-CGTT GCCCCATCCACCAC-3′ and NIT1R 5′-TATGAACGCCAACC GCTAAACC-3′, NIT2F 5′-TCG TTA ATA ATG GAA GAG A-3′ and NIT2R 5′-AAA AAG AAA AGA ACA CAA CTG-3′, NIT3F 5′-CCCCGTTTACGACACTCCTA-3′ and NIT3R 5′-CATCCACCTTCTACCGCAA-3′, Actin7F 5′-AATGGTGAAGGCTGGTTTTG-3′ and Actin7R 5′-TGCCTCTGTGAGTAGAACTG-3′.
Transient expression assay using Arabidopsis protoplasts
For transactivation analysis in Arabidopsis protoplasts, 2 × 104 protoplasts were transfected with 21 μg plasmid DNA with different combinations of reporter, effector, and internal control. This procedure was performed as described previously (Jin et al., 2001).
Bacterial expression of glutathione S-transferase (GST)-ATAF2 fusion protein
ATAF2 fragment was prepared by PCR amplification and cloned into the XbaI and SacI sites of the pGEX-3X vector (Amersham). The resulting in-frame fusion plasmid was transformed into Escherichia coli strain BL21 (DE3). The GST fusion protein was recovered by affinity chromatography on glutathione-sepharose beads (Pharmacia).
Electrophoretic mobility shift assay (EMSA)
DNA binding reactions containing specified concentrations of protein (50–150 ng), 5 μl of 4× reaction buffer [200 mM KCl, 100 mM HEPES/KOH at pH 7.9, 2 mM EDTA, 2 mM dithio-threitol (DTT), 20% glycerol, 20 μg/μl bovine serum albumin (BSA)], 1 μg poly dIdC (Pharmacia), and 1 μl 1% Nonidet P-40 in a final volume of 18 μl. The labeled DNA probe (25 ng) was then added to the reaction mixture. After incubation for 30 min at room temperature, 10 μl of the mixture was loaded on a 6% non-denatured polyacrylamide gel. Electrophoresis was performed in 0.5 × TBE (54 mM Tris-borate, pH 8.3, 1 mM EDTA) for 2.5 h. Gels were dried and exposed for autoradiography. The DNA probes used in the analyses were: F1S, 5′-AAAT AAGAAGGCAAATATAATTTGATAACAACTATT-3′; F1AS, 5′-AATAGTTGTTATCAAATTATATTTGCCTTCTTATTT-3′; F2S, 5′-ATATTTGGGTGAATATTTTTCCAAATTAACTGTAGC-3′; F2AS, 5′-GCTACAGTTAATTTGGAAAAATATTCACCCAAATA T-3′; F3S, 5′-ATGGAGTAATAATATATTTTGTTTAATTGTCA GTCT-3′; F3AS, 5′-AGACTGACAATTAAACAAAATATATTAT TACTCCAT-3′.
RESULTS
ATAF2pro::GUS reporter is activated by IAN treatment
Previously, it was shown that gene expression of ATAF2 was increased upon salicylic acid (SA) and methyl jasmonic acid (MeJA) treatments but was not changed upon abscisic acid (ABA) treatment (Delessert et al., 2005). We performed microarray experiment using ataf2 mutant. Interestingly, expression of some of auxin-related genes was repressed in ataf2 mutant (data not shown). However, biological functions of ATAF2 related to plant auxin hormones are still not clear. To identify biological functions of ATAF2 in auxin hormone response, we first generated ATAF2pro::GUS reporter system which allowed to reveal gene expression patterns by histochemical staining of plant tissue upon plant hormone treatments. Weak expression of the ATAF2pro::GUS fusion gene was observed in several regions of rosette leaf while strong expression was revealed in the hydathodes under normal growth conditions (Fig. 1A). By contrast, ATAF2pro::GUS reporter was activated by IAN treatment (Fig. 1B). By IAN treatment, intense staining was shown in rosette leaves and roots (Fig. 1B). ATAF2 function of auxin pathway has not been clearly identified although ATAF2 is involved in the plant defense via repressing the expression of pathogenesis-related genes and is known as target of replicase protein of Tobacco mosaic virus (TMV) (Delessert et al., 2005; Wang et al., 2009). To confirm the expression result, we also performed RNA blot analysis upon IAN treatment. And ATAF2 was strongly induced within 2 h in response to IAN treatment (Fig. 1C). These results indicated that ATAF2 might be involved in auxin signaling.
Fig. 1.
Histochemical assay of the GUS reporter gene under the control of ATAF2 promoter in transgenic plants upon IAN treatment. (A) Histochemical assay was done with 18 days old ATAF2pro::GUS transgenic plants treated with water for 5 h as control. Enlarged images show leaf and root tissue. Arrows indicate the staining of the hydathodes. (B) ATAF2pro::GUS transgenic plants were treated with 200 μM IAN for 5 h and histochemical assay was performed. (C) Expression pattern of ATAF2 under IAN treatment. Three-week-old Arabidopsis seedlings grown vertically on PNS medium were used and 200 μM IAN treatment was conducted for indicated time periods. RNA blot analysis was performed with ATAF2 gene-specific probe. Ribosomal RNA (rRNA) was used as a loading control.
ATAF2 is required for full induction of NIT2 in Arabidopsis
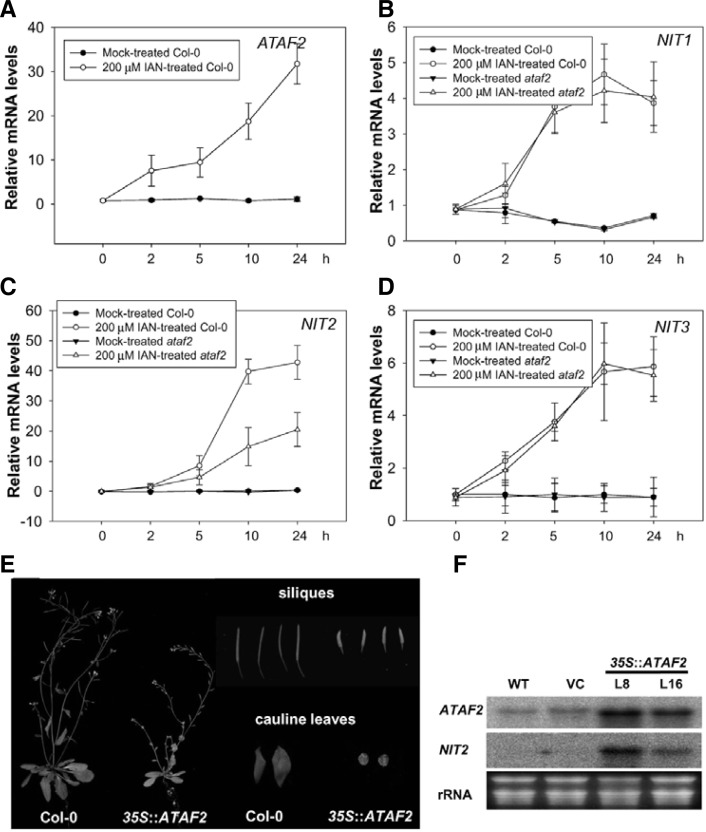
Arabidopsis nitrilases (NIT1, NIT2, and NIT3) convert IAN to IAA and indole-3-acetamide (IAM) (Pollmann et al., 2002). IAA is an auxin and some microarrays analyses have demonstrated up-regulation of NIT1 and NIT2 in response to IAN and NaCl treatment (Grsic et al., 1998; Jiang et al., 2007; Woodward and Bartel, 2005). Furthermore, NAC transcription factor also controls salinity stress via the auxin pathway (He et al., 2005; Park et al., 2011). Therefore, we wanted to understand whether ATAF2 might be related to the auxin pathway by regulating NIT genes at the transcriptional level under environmental stress conditions. We first analyzed gene expression of NIT1, NIT2, and NIT3 in ataf2 mutant upon IAN treatment. NIT1 and NIT3 transcript levels were not affected in ataf2 mutant upon IAN treatment (Figs. 2B and 2D). However, NIT2 gene expression in the ataf2 mutant was repressed (by about 2-fold) compared to Col-0 plants upon IAN treatment (Fig. 2C), suggesting that ATAF2 specifically affects one of three NIT genes (Fig. 2C). ATAF2 transcripts were induced within 2 h after IAN treatment (Fig. 2A). NIT2 transcripts were also elevated within 2–5 h after IAN treatment (Fig. 2C), suggesting that expressions of both genes are related each other (Fig. 2C). We also performed RNA blot analysis. Transcript levels of ATAF2 and NIT2 almost did not change under this condition (Supplementary Fig. S1F). The transcripts of ATAF2 started to accumulate within 1 h after NaCl, SNP, SA, and dehydration treatment and most of time gradually increased further (Supplementary Figs. S1A, S1C, S1D, and S1E). NIT2 gene expression upon diverse stress conditions was increased within 5 h, later than that of ATAF2 (Supplementary Figs. S1A-S1E). For the ABA treatment, the transcript level of NIT2 was increased but ATAF2 gene did not respond (Supplementary Fig. S1B). These results suggest that the ATAF2 induction by osmotic stress might occur through the ABA-independent pathway while the ABA-dependent pathway could involve the NIT2 induction. And ATAF2-NIT2 regulation might be closely related to diverse biotic and abiotic stress response.
Fig. 2.
ATAF2 requirement for full expression of NIT2 but not for NIT1 and NIT3 upon IAN treatment. (A) Induction of ATAF2 upon IAN treatment was confirmed by qRT-PCR. (B-D) Expression pattern of NIT1, NIT2, and NIT3 in ataf2 knock-out mutant and Col-0 plants upon IAN treatment. Samples were harvested at the indicated time points following mock and 200 μM IAN treatments. Three independent biological replicate cDNA samples were tested for each experiment. Expression data was normalized with an endogenous Actin7 gene (internal reference). (E) Phenotypic abnormalities in 35S::ATAF2 plants. Six-week-old 35S::ATAF2 transgenic plants showed reduced size and height and lighter green leaf colors when compared with Col-0 plants. 35S::ATAF2 transgenic plants also exhibit stunted siliques and round-shaped cauline leaves. (F) RNA blot analysis of ATAF2 and NIT2 gene in wild type and 35S::ATAF2 transgenic plants. Twelve-day-old seedlings of wild type (WT), vector control (VT), 35S::ATAF2 transgenic (L8 and L16) plants were hydroponically cultured and harvested. The blot was hybridized with ATAF2 and NIT2 gene-specific probes. Ribosomal RNA (rRNA) was used as a loading control.
We also generated ATAF2-overexpressing plants under the control of the CaMV 35S promoter and analyzed the NIT2 gene expression in the 35S::ATAF2 transgenic plants. ATAF2-overexpressing transgenic plants exhibited abnormal developmental phenotypes such as dwarfism, and leaf-yellowing (Fig. 2E). This developmental change does not appear to be associated with ATAF2 overexpression in the C24 background with respect to the dwarfism, but leaf-yellowing phenotype was similar (Delessert et al., 2005). We do not know why some of the ATAF2 overexpression phenotypes are different depending on the background. The NIT2 transcripts were highly expressed in two independent 35S::ATAF2 transgenic plants, compared with wild type and vector control plants (Fig. 2F). This result suggested that ATAF2 might directly regulate NIT2 gene expression and affect developmental processes.
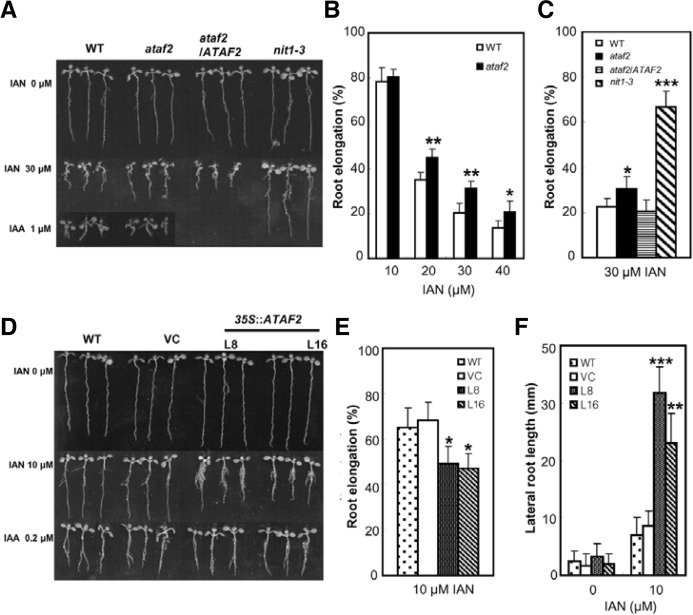
ataf2 plants exhibit reduced IAN sensitivity and 35S::ATAF2 plants show increased sensitivity to IAN
A previous study demonstrated the IAN insensitivity of nit1 mutant plants and increased sensitivity to IAN of NIT2-overexpressing transgenic plants (Normanly et al., 1997). We showed that ATAF2 regulated NIT2 expression from the study of ataf2 mutant and 35S::ATAF2 transgenic plants (Figs. 2C and 2F). Based on these results, we tested whether sensitivity to IAN was altered in ataf2 mutant and ataf2 complementation lines. When germinated on the medium with 20–40 μM IAN, ataf2 mutant plants showed reduced sensitivity to IAN (Figs. 3A and 3B). After 8 days growth in the presence of 30 μM IAN, the ataf2 mutant plants exhibited about 30% increased primary root elongation compared with wild type plants, although the difference in root elongation was not as dramatic as the nit1-3 mutant (Fig. 3C) (Normanly et al., 1997). In contrast, when grown in the presence of 1 μM IAA, ataf2 root elongation was not different from that of wild type plants (Fig. 3A). This phenomenon also reported (Normanly et al., 1997); that is, nitrilase converts indole-3-acetonitrile (IAN) to indole-3-acetic acid (IAA) and IAA treatment did not make any difference in root growth in WT and auxin-related mutant plants. Furthermore, ataf2 complementation transgenic plants showed comparable root elongation with wild type plants on IAN (Fig. 3C), indicating that sensitivity to IAN in the ataf2 mutant plants resulted from disruption of the ataf2 gene.
Fig. 3.
Indole-3-acetonitrile (IAN) sensitivity test of ataf2 knockout and 35S::ATAT2 plants. (A) Growth of seedlings on plant nutrient medium with sucrose (PNS) or PNS containing 30 μM IAN or 1 μM IAA. (B) IAN dose response of primary root growth. Shown is the percentage of elongation for IAN treatment at indicated concentration compared with that of the PNS control. Ten seedlings of each ge-notype were measured at individual IAN concentration. (C) Root elongation of ataf2, ataf2/ATAF2 complementation transgenic line, nit1-3, and wild type plants on PNS with 30 μM IAN. Twelve seedlings of each genotype were used. (D) IAN sensitivity of 35S::ATAT2 transgenic plants. Eight-day-old seedlings were grown on PNS or PNS containing 10 μM IAN or 0.2 μM IAA. (E) IAN sensitivity of primary root elongation in WT, VC, and two independent ATAF2-overexpressing transgenic line seedlings upon IAN treatment. The percent root growth compared to non-treated control was measured. Twelve seedlings of each genotype were used for each condition. (F) Lateral root length of WT, VC, and two ATAF2-overexpressing transgenic line plants on PNS with and without 10 μM IAN. At least 12 seedlings of each genotype were measured for each condition (Student’s t-tests; *P < 0.05, **P < 0.001, ***P < 0.0001).
We obtained consistent gain-of-function results from ATAF2-overexressing plants, which displayed increased sensitivity to IAN (Fig. 3D). The primary root elongation in 35S::ATAF2 transgenic plants was inhibited about 30% compared with wild type plants when both were grown on 10 μM IAN (Fig. 3E). However, the 35S::ATAF2 plants showed a 3–4 fold increase in total lateral root length compared with that of the wild type or a vector control transgenic plants (Fig. 3F).
ATAF2 transactivates the NIT2 gene promoter
From the gene expression and phenotypic analysis in the plants with gain and loss of ATAF2 function, we speculated that ATAF2 might activate the NIT2 gene at the transcriptional level. Thus, we performed the transient transactivation assay with NIT2 promoter using Arabidopsis protoplasts. Reporter constructs containing the region spanning positions from −1459 to +48 of the NIT2 promoter (P1) and ATAF2 expression vector or empty vector were co-transfected into Arabidopsis protoplasts. And indeed ATAF2 induced the expression of the NIT2 promoter-driven GUS reporter gene (Fig. 4A).
Fig. 4.
Activation of the NIT2 promoter::GUS (or LUC) fusion gene by ATAF2 protein in Arabidopsis protoplasts. (A) and (B) 35S::LUC (or GUS) plasmid was co-transfected as a control for cell viability and transfection efficiency analysis. Arabidopsis protoplasts were transfected with effecter plasmids and various reporter plasmids with different length sets of NIT2 promoter.
To define the region of the NIT2 promoter that responds to ATAF2 transactivation, a series of 5′ deletion mutants of the NIT2 promoter construct were generated (from P2 to P9). Deletion of positions from −665 to −117 of the NIT2 promoter region (P2 to P7) had no significant impact on the response to ATAF2 (Fig. 4A). Further deletions (P8 and P9) from position −117 to −82 resulted in a complete loss of ATAF2 responsiveness.
To further confirm the location of the ATAF2-responsive cis-elements, two 3′ deletion mutants (P10 and P11) fused with 35S minimal promoter and luciferase (LUC) reporter were constructed. Only the P10 construct that contained a sequence from position −117 to −82 was responsive to ATAF2 (Fig. 4B). Taken together, these results demonstrate the existence of a discrete ATAF2 responsive element that localizes to the 36 base pair region from position −117 to −82 in the NIT2 promoter.
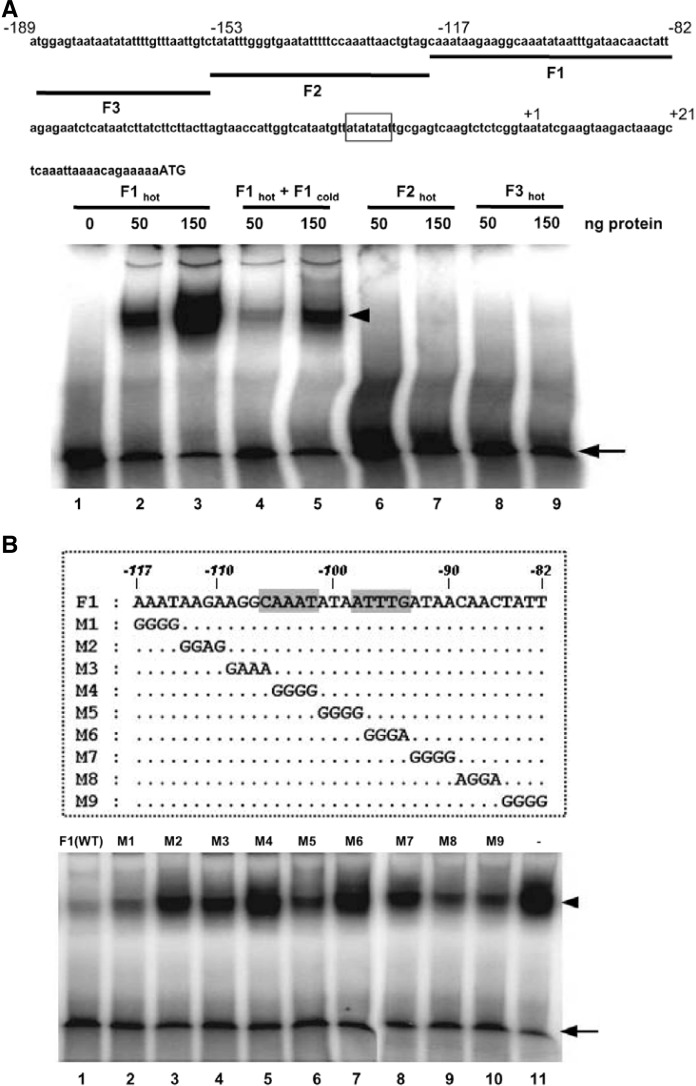
ATAF2 binds to the −117 to −82 region of the NIT2 promoter in vitro
To test whether ATAF2 could bind to the ATAF2-responsive cis-elements, EMSA was performed. For a labeled probe in EMSA, three tandemly arrayed 36 nucleotide sequences of NIT2 promoter were chosen to test the specificity of binding (Fig. 5A). Among the three fragments, only the 36-bp segment from −117 to −82 that contains the ATAF2-responsive element was bound by the GST-ATAF2 protein, as also confirmed by a competition assay using unlabeled probe (Fig. 5A). Interestingly, that ATAF2-responsive element was shown to contain an imperfect palindrome sequence (Fig. 5B), as previously shown in a few NAC proteins (Olsen et al., 2005; Zhong et al., 2010). Thus, mutational analysis of the ATAF2-responsive element was performed. Two oligonucleotides (M4 and M6) containing mutations in the palindrome sequence were unable to compete for the binding at a 100-fold molar excess (Fig. 5B, compare lanes 1 and 11 to lanes 5 and 7). Three oligonucleotides carrying mutations M2, M3, and M7 were also unable to compete for the complex formation, but with a slightly increased competition compared to M4 and M6 (Fig. 5B, compare lanes 1 and 11 to lanes 3, 4, and 8). In contrast, mutations M1, M5, M8, or M9 only marginally influenced the ability of the oligonucleotides to compete for ATAF2 binding to the sequence from position −117 to −82 (Fig. 5B, compare lanes 1 and 11 to lanes 2, 6, 9, and 10). Collectively, these results suggest that the region from positions −117 to −82 contains an important motif for ATAF2 to bind to the NIT2 promoter.
Fig. 5.
Binding of recombinant GST-ATAF2 fusion protein to the −117 to −82 fragment of NIT2 promoter in eletrophoretic mobility shift assay (EMSA). (A) For labeled probes preparation in EMSA, three tandemly arrayed 36 nucleotide sequences of NIT2 promoter (underlined, F1–F3) were selected to examine the specificity of binding. Lane 1, 500 ng of GST; lanes 2, 4, 6, 8, 50 ng of GST-ATAF2; lanes 3, 5, 7, 9, 150 ng of GST-ATAF2; lanes 2 and 3 contained 32P-labeled F1 fragment as probe; lanes 4 and 5 contained the radio-labeled F1 fragment as probe and 100-fold unlabeled F1 fragment as competitor; lanes 6–7 and 8–9 contained 32P-labeled F2 and F3 fragments as probes, respectively. (B) EMSA of the −117 to −82 fragment of NIT2 promoter incubated with the oligonucleotide carrying mutations as competitors. Shown are the sequences of the mutant oligonucleotides (M1–M9) corresponding to the −117 to −82 (F1) of NIT2 promoter which were used as competitors in EMSA. EMSAs were performed using 32P-labeled F1 fragment as probe. Lane 1 contained cold-labeled F1 fragment as competitor; lanes 2–10 contained base-substituted mutant competitors shown in the box; lane 11, no competitor.
DISCUSSION
In this study, we found biological function of ATAF2 was involved in auxin biosynthesis via NIT2 gene regulation. The induction level of NIT2 was repressed in ataf2 mutant upon IAN treatment. Meanwhile, the stable expressions ATAF2 in Arabidopsis Col-0 ecotype plants increase NIT2 transcript level. To confirm of IAN-sensitivity (of root elongation) in the nit2 loss-of-function mutant, we obtained NIT2-RNAi transgenic plants. However, NIT2-RNAi plant repressed both NIT1 and NIT2 gene expressions because NIT1 and NIT2 have highly similar nucleotide sequence (Supplementary Fig. S2A). The NIT2-RNAi transgenic plants exhibited slightly more enhanced IAN-insensitivity compared with nit1-3 mutant (Supplementary Fig. S2B). Thus, ATAF2-NIT2 regulation could be involved in auxin signaling. We also performed a microarray with 27K Arabidopsis gene chip (Green Gene BioTech Inc.) and NIT2 expression was repressed about 0.5 fold in ataf2 mutant (data not shown). ATAF2 was able to transactivate the NIT2 promoter in vivo when protoplasts were used. Moreover, ATAF2 protein specifically bound to the NIT2 promoter region where an imperfect palindrome sequence exists.
ATAF2 is responsive to diverse stresses such as plant defense-related hormone, pathogen infection, wounding, and environmental stress (Delessert et al., 2005; Seki et al., 2002; Wang et al., 2009). We also showed ATAF2 and NIT2 transcripts were induced under diverse stress conditions (Supplementary Fig. S1). These results mean that ATAF2 functions could be involved in biotic and abiotic stress via NIT2 regulation. Interestingly, ATAF2 was known to repress the pathogenesis-related gene expressions although the exact mechanism of ATAF2 function in plant defense is not known. It was recently shown that some defense genes were repressed by ATAF2-TMV replicase interaction upon TMV infection (Wang et al., 2009). And lesion formation, wounding, and pathogen infection also activate tryptophan-dependent IAA biosynthesis pathways (Normanly and Bartel, 1999; Zhao and Last, 1996). Furthermore, the tryptophan biosynthesis pathway is also involved in the defense responses (Ishihara et al., 2008). Thus, we can postulate that ATAF2 might regulate auxin biosynthesis pathway and this regulation could be connected with ATAF2-mediated plant defense system. And the regulation of ATAF2-NIT2 could affect IAA levels. However, it can be expected that NIT2 induction via ATAF2 may not give rise to dramatic changes in IAN sensitivity because Arabidopsis has three nitrilase genes and, one of them, NIT1, accounts for the significant portions (60% at 30 μM IAN) of total nitrilase activity (Normanly et al., 1997). Auxin-related mutant showed dwarfism and ATAF2-overexpressing transgenic plant also exhibited similar phenotype (Fig. 2E). However, this phenotype itself does not exclude the possibility of ATAF2 involvement in other plant hormones such as salicylic acid (SA) or jasmonic acid (JA) because ATAF2 could regulate pathogenesis-related genes (Wang et al., 2009). Furthermore, NAC transcription factors interact each other and exhibit diverse functions (Fang et al., 2008). Thus, there is a possibility of ATAF2 and NIT2 involvement in complex plant signaling pathway.
NAC domains from NAC1, AtNAM, and ANAC bind specifically to as-1 like sequence within the CaMV 35S promoter (Duval et al., 2002; Xie et al., 2000). Three dehydration-inducible NAC proteins (ANAC019, ANAC055, and ANAC072) bind specifically to the CATGTG motif of early response to dehydration 1 (ERD1) promoter both in vitro and in vivo (He et al., 2005). It was reported that the NAC proteins recognize a sequence containing CATGT and harboring CACG as the core DNA binding site (Tran et al., 2004). Furthermore, vascular-related NAC-domain 7 (VND7) and secondary wall-associated NAC domain protein 1 (SND1) directly bind to secondary wall NAC binding element (SNBE) in the target promoters (Yamaguchi et al., 2011; Zhong et al., 2010). However, still little is known about the NAC protein binding site in biological target gene of NAC. The present study reports for the first time that the ATAF2 protein specifically binds to the imperfect palindromic sequence −117 to −82 of the NIT2 promoter. These results demonstrate that the ATAF2 protein functions as a transcriptional activator with NIT2 as an immediate target gene of ATAF2, and infer that a NAC domain might be able to recognize more than one sequence apparently unrelated to each other and/or that the sequence preference of a NAC protein depends on the presence of an additional cis-regulatory element. For example, ATAF2 share 95% sequence identity to ANAC102. Furthermore, anac102 mutant also exhibited less sensitivity to IAN and ANAC102 bound to ATAF2-responsive element (data not shown). These results demonstrate that NIT2 gene could be regulated by ATAF2 and other NAC proteins via their binding to the imperfect palindromic sequence −117 to −82 of the NIT2 promoter.
Supplementary Material
Acknowledgments
This work was supported by the Science Research Center-Engineering Research Center Program (Plant Signaling Network Research Center) of the Ministry of Education, Science and Technology (R11-2003-008-02001-0), the Mid-career Researcher-Program through a National Research Foundation grant funded by the Mest (2009-0085565), and the Wujangchoon Project (PJ007850) from the Rural Development Administration, Republic of Korea.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Church G.M., Gilbert W. Genomic sequencing. Proc. Natl. Acad. Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delessert C., Kazan K., Wilson I.W., Van Der Straeten.D.,Manners J., Dennis E.S., Dolferus R. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 2005;43:745–757. doi: 10.1111/j.1365-313X.2005.02488.x. [DOI] [PubMed] [Google Scholar]
- Duval M., Hsieh T.F., Kim S.Y., Thomas T.L. Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 2002;50:237–248. doi: 10.1023/a:1016028530943. [DOI] [PubMed] [Google Scholar]
- Ernst H.A., Olsen A.N., Larsen S., Lo Leggio L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004;5:297–303. doi: 10.1038/sj.embor.7400093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., You J., Xie K., Xie W., Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet Genomics. 2008;280:547–563. doi: 10.1007/s00438-008-0386-6. [DOI] [PubMed] [Google Scholar]
- Grsic S., Sauerteig S., Neuhaus K., Albrecht M., Rossiter J., Ludwig-Muller J. Physiological analysis of transgenic Arabidopsis thaliana plants expressing one nitrilase isoform in sense or antisense direction. J. Plant Physiol. 1998;153:446–456. [Google Scholar]
- He X.J., Mu R.L., Cao W.H., Zhang Z.G., Zhang J.S., Chen S.Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005;44:903–916. doi: 10.1111/j.1365-313X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- Hegedus D., Yu M., Baldwin D., Gruber M., Sharpe A., Parkin I., Whitwill S., Lydiate D. Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol. Biol. 2003;53:383–397. doi: 10.1023/b:plan.0000006944.61384.11. [DOI] [PubMed] [Google Scholar]
- Huh S.U., Lee I.J., Ham B.K., Paek K.H. Nicotiana tabacum Tsip1-interacting ferredoxin 1 affects biotic and abiotic stress resistance. Mol Cells. 2012;34:43–52. doi: 10.1007/s10059-012-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ishihara A., Hashimoto Y., Tanaka C., Dubouzet J.G., Nakao T., Matsuda F., Nishioka T., Miyagawa H., Wakasa K. The tryptophan pathway is involved in the defense responses of rice against pathogenic infection via serotonin production. Plant J. 2008;54:481–495. doi: 10.1111/j.1365-313X.2008.03441.x. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Yang B., Harris N.S., Deyholos M.K. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 2007;58:3591–3607. doi: 10.1093/jxb/erm207. [DOI] [PubMed] [Google Scholar]
- Jin J.B., Kim Y.A., Kim S.J., Lee S.H., Kim D.H., Cheong G.W., Hwang I. A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell. 2001;13:1511–1526. doi: 10.1105/TPC.000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J., Bartel B. Redundancy as a way of life-IAA metabolism. Curr. Opin. Plant Biol. 1999;2:207–213. doi: 10.1016/s1369-5266(99)80037-5. [DOI] [PubMed] [Google Scholar]
- Normanly J., Grisafi P., Fink G.R., Bartel B. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell. 1997;9:1781–1790. doi: 10.1105/tpc.9.10.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Park J., Kim Y.S., Kim S.G., Jung J.H., Woo J.C., Park C.M. Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol. 2011;156:537–549. doi: 10.1104/pp.111.177071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollmann S., Muller A., Piotrowski M., Weiler E.W. Occurrence and formation of indole-3-acetamide in Arabidopsis thaliana. Planta. 2002;216:155–161. doi: 10.1007/s00425-002-0868-4. [DOI] [PubMed] [Google Scholar]
- Riechmann J.L., Heard J., Martin G., Reuber L., Jiang C., Keddie J., Adam L., Pineda O., Ratcliffe O.J., Samaha R.R., et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Seki M., Narusaka M., Ishida J., Nanjo T., Fujita M., Oono Y., Kamiya A., Nakajima M., Enju A., Sakurai T., et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Souer E., van Houwelingen A., Kloos D., Mol J., Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85:159–170. doi: 10.1016/s0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Goregaoker S.P., Culver J.N. Interaction of the Tobacco mosaic virus replicase protein with a NAC domain transcription factor is associated with the suppression of systemic host defenses. J. Virol. 2009;83:9720–9730. doi: 10.1128/JVI.00941-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A.W., Bartel B. Auxin: regulation, action, and interaction. Ann. Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray G.A., Hahn M.W., Abouheif E., Balhoff J.P., Pizer M., Rockman M.V., Romano L.A. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Xie Q., Frugis G., Colgan D., Chua N.H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14:3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Mitsuda N., Ohtani M., Ohme-Takagi M., Kato K., Demura T. VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 2011;66:579–590. doi: 10.1111/j.1365-313X.2011.04514.x. [DOI] [PubMed] [Google Scholar]
- Zhao J., Last R.L. Coordinate regulation of the tryptophan biosynthetic pathway and indolic phytoalexin accumulation in Arabidopsis. Plant Cell. 1996;8:2235–2244. doi: 10.1105/tpc.8.12.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Lee C., Ye Z.H. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant. 2010;3:1087–1103. doi: 10.1093/mp/ssq062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.