Abstract
We investigated the relationship between oct4 gene expression patterns and CpG sites methylation profiles during ES cell differentiation into neurons, and identified relevant binding factor. The oct4 gene expression level gradually declined as ES cell differentiation progressed, and the CpG sites in the oct4 proximal enhancer (PE) and promoter regions were methylated in concert with ES cell differentiation. An electro-mobility shift assay (EMSA) showed that putative proteins bind to CpG sites in the oct4 PE/promoter. We purified CpG binding proteins with DNA-binding purification method, and NonO was identified by liquid chromatography-mass spectrometry. EMSA with specific competitors revealed that NonO specifically binds to the conserved CCGGTGAC sequence in the oct4 promoter. Methylation at a specific cytosine residue (CC* GGTGAC) reduced the binding affinity of NonO for the recognition sequence. Chromatin immunoprecipitation analysis confirmed that NonO binds to the unmethylated oct4 promoter. There were no changes in the NonO mRNA and protein levels between ES cells and differentiated cells. The transcriptional role of NonO in oct4 gene expression was evaluated by luciferase assays and knockdown experiments. The luciferase activity significantly increased threefold when the NonO expression vector was co-transfected with the NonO recognition sequence, indicating that NonO has a transcription activator effect on oct4 gene expression. In accordance with this effect, when NonO expression was inhibited by siRNA treatment, oct4 expression was also significantly reduced. In summary, we purified NonO, a novel protein that binds to the CpG island of oct4 promoter, and positively regulates oct4 gene expression in ES cells.
Keywords: CpG sites, ES cells, methylation, NonO, Oct4
INTRODUCTION
ES cells are pluripotent cells that can self-renew indefinitely or differentiate into various cell types (Magnuson et al., 1982; Thomson et al., 1998). Oct4, sox2 and nanog are known to maintain the stemness of ES cells by silencing the expression of key transcription factors required for differentiation, and by activating the expression of genes important for ES cell maintenance (Boyer et al., 2005; Young, 2011). Among these factors, oct4 has been well documented to maintain the self-renewal and differentiation abilities of ES cells in an expression level-dependent manner (Niwa et al., 2000; Velkey and O’Shea, 2003). A steady state critical amount of oct4 is required to sustain the stemness of ES cells, and the transient up-and down-regulation of oct4 induces divergent differentiation programs. In this context, the evaluation of the up-stream regulatory mechanism of oct4 expression is as important as assessing downstream target gene regulation for the maintenance of stemness and the differentiation of ES cells. A great number of transcription factors that activate or inhibit oct4 gene expression during ES cell maintenance and differentiation have been identified. Transcription factors such as zfp206, sall4, esrrb,-myb, SF1, dax1 and lrh bind to the oct4 distal enhancer (DE)/PE/promoter regions and activate oct4 gene expression (Gu et al., 2005a; Kelly et al., 2010; Tarasov et al., 2008; Yang et al., 2007; Yu et al., 2009; Zhang et al., 2006; 2008). Other transcription factors, such as HIF, GCNF and Tcf3 bind to the oct4 DE/PE/promoter regions and repress oct4 gene expression (Gu et al., 2005b; Moreno-Manzano et al., 2010; Tam et al., 2008). Pml-nuclear body (NB) has dual effects; it activates oct4 expression with transcription factors TR2, SF1 and SP1 in ES cells but is involved in the silencing of oct4 during retinoic acid-induced ES cell differentiation (Chuang et al., 2011). The transcriptional activity of oct4 is closely related to CpG island methylation. Oct4 expression is known to decrease and its CpG islands in the DE/PE/promoter regions are known to be increasingly methylated as differentiation progresses (Feldman et al., 2006; Gidekel and Bergman, 2002; Hattori et al., 2004; Marikawa et al., 2005). As mentioned above, numerous factors that regulate oct4 gene expression have been identified; however, knowledge of the regulatory mechanisms in relation to CpG island methylation and CpG island binding factors is very limited (Gu et al., 2011). Therefore, we tried to find a novel factor that binds to the CpG island and regulates oct4 gene expression.
NonO was originally identified as a non-POU domain-containing octamer binding protein (Yang et al., 1993). NonO is a multi-functional nuclear protein that functions through its two tandem RNA recognition motifs (RRMs) and a helix-turn-helix (HTH) DNA-binding domain (Dong et al., 1993; Shav-Tal and Zipori, 2002; Yang et al., 1993). NonO associates with the 5′ splice site and mediates contacts between RNA polymerase II and snRNPs during the coupled transcription/splicing process (Kameoka et al., 2004; Rosonina et al., 2005). NonO is also reported to be involved in the nuclear retention of edited RNA, inhibiting the translation of mutated proteins by binding to the A-to-I edited RNA and forming a paraspeckle (Chen and Carmichael, 2009; Zhang and Carmichael, 2001). In addition to RNA processing functions, NonO has functions related to DNA replication and repair processes. NonO associates with topoisomerase I and stimulates the jumping of topoisomerase I between separate DNA helices (Straub et al., 1998; 2000). NonO forms a complex with polypyrimidine tract binding protein-associated splicing factor (PSF) and functions as a DNA double-strand break rejoining factor by binding to the DNA substrate in the non-homologous end joining pathway (Bladen et al., 2005; Salton et al., 2010). Finally, NonO is known to function as transcription repressor or activator under many different cellular conditions (Shav-Tal and Zipori, 2002).
In this study, we examined the methylation profile of oct4 promoter CpG islands during ES cell differentiation. We tried to identify novel factors that bind to the oct4 promoter CpG islands. With a CpG island-specific DNA probe, we purified NonO and investigated the transcriptional role of NonO in oct4 gene expression using NonO gain-of-function and loss-of-function experiments.
MATERIALS AND METHODS
Maintenance and differentiation of ES cells
R1 ES cell maintenance and differentiation into neuronal cells were performed as previously described without any modifications (Hwang et al., 2008).
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde for 20 min and processed for immunocytochemistry as previously described (Hwang et al., 2008). The following dilutions of antibodies were used: rabbit anti-oct4 (Santa Cruz, 1:500), mouse anti-nestin (Chemicon, 1:500), and mouse anti-NonO (Upstate, 1:500).
Real-time RT-PCR
Total RNA was isolated by a ToTALLY RNA™ kit (Ambion, USA) according to the manufacturer’s protocol, and Real-Time RT-PCR was performed as previously described (Hwang et al., 2008). The following primer sequences were used: Oct4-F, 5′-GAGGACTCCCAGGACATG-3′; Oct4-R, 5′-CCTGGGAAAGG TGTCCTGTA-3′; Nestin-F, 5′-AACAGAGGTGGGAGGATGTG-3′; Nestin-R, 5′-CTGGCAGCCTCTAATCCAAG-3′; NonO-F, 5′-ATTCGCTTGGAAACACGAAC-3′; NonO-R, 5′-GAAGGAGCC TTCACTGCATC-3′.
Bisulfite sequencing
The bisulfite sequencing of genomic DNA isolated from ES cells; from differentiated cells at day 3, day 6, day 9; and from a mouse tail was performed as described previously (Kang et al., 2001). The PCR primers used to amplify bisulfite-converted DNA were as follows: Oct4(BSP)F1, 5′-AAGTTGATGAAGTTG AGGTAGG-3′; Oct4(BSP)R1, 5′-CTCTCCTCAAAAACAAAAC CTC-3′; Oct4(BSP)F2, 5′-GGGATTAGGATTGTTTAGTTAAGG-3′; Oct4(BSP)R2, 5′-ACCTATTAACACTACACCCTCTC-3′; Oct4 (BSP)F3, 5′-GGTTTTTTAGAGGATGGTTGAGTG-3′; Oct4(BSP) R3, 5′-TCCAACCCTACTAACCCATCACC-3′.
Electromobility shift assay (EMSA)
EMSA was performed as previously described (Son et al., 2005). Nuclear extracts from ES cells at day 0 and from differentiated cells at day 6 were used in the EMSA. The sequences of the probes and competitors are listed in Table 1. Four picomoles of annealed oligonucleotides was labeled with 20 μCi [α-32P]dCTP (Amersham, Sweden) using the Klenow fragment (Promega, Madison, WI). Labeled probe at a concentration of 40 fM was incubated with 10 μg of nuclear extract in 1× EMSA buffer (20 mM Hepes, pH 7.6, 0.1 Mm KCl, 10% glycerol, 0.1% Nonidet P-40, and 1 mM DTT) containing 2.5 μg poly(dIdC) in a 20 μl reaction mixture. The binding reaction was performed on ice for 20 min and then at room temperature for further 30 min. Protein-DNA complexes were resolved on a 5% native polyacrylamide (29:1) gel. For competition experiments, a 100-fold molar excess of competitor DNA was added along with the P32-labeled probes.
Table 1.
EMSA probes containing CpG sites in the oct4 promoter
| EMSA probe symbols | Position | Sequences for EMSA probes |
|---|---|---|
| A | −1226∼−1209 | 5′-CCTCCTAATCCCGTCTCC-3′ |
| B | −1184∼−1162 | 5′-AATGGGGGAGGGGTGGGTGACGA-3′ |
| C | −771∼−747 | 5′-ATCTTGAGGAAAGAGGCCCCGGCCT-3′ |
| D | −369∼−345 | 5′-AAGGACAGGCCGAGAGGGTGCAGTG-3′ |
| E | −324∼−303 | 5′-ATGGGGCATCCGAGCAACTGGT-3′ |
| F | −297∼−273 | 5 ′-GAGGTGTCCGGTGACCCAAGGCAGG-3′ |
| G | −213∼−195 | 5′-CCTGTCCAGACGTCCCCAA-3′ |
| H | −174∼−159 | 5′-GGCAGATAGCGCTCGC-3′ |
| I | −41∼−14 | 5 ′-TCTTTCCACCAGGCCCCCGGCTCGGGGT-3′ |
| mt1-F | −297∼−273 | 5′-GAGGTGTCCGAATTTCCAAGGCAGG-3′ |
| mt2-F | −297∼−273 | 5′-GAGGTGTCCGGTGTTCCAAGGCAGG-3′ |
| met-F | −297∼−273 | 5′-GAGGTGTCC*GGTGACCCAAGGCAGG-3′ |
Purification of DNA-binding proteins
The proteins that bind to the oct4 promoter CpG island were purified with the DNA-binding Protein Purification Kit (Roche Applied Science) according to the manufacturer’s protocol. Briefly, dsDNA containing EMSA probe F (Table 1) was ligated in tandem and attached to magnetic beads. Day 0 and day 6 cell nuclear extracts were incubated the reaction mixture at 4°C for 1 h. The bound protein was extracted with extraction buffer, separated with SDS/PAGE, and silver-stained. Individual bands were excised, digested with trypsin, and analyzed by 2D-LC/MS/MS (Thermo Electron, US/LRQ, LC/MS/MS).
ChIP-PCR
ES cells at day 0 and differentiated cells at day 6 were cross-linked with 1% formaldehyde (Sigma). The soluble chromatin was extracted and sonicated following a protocol provided by Upstate Biotechnology. The sonicated lysates were immuno-precipitated with 2 μg of anti-NonO antibody. The cell lysate not incubated with the antibody was used as a positive input control. Immune complexes were collected by incubation with protein A Sepharose beads and sheared salmon sperm DNA. The bound DNA was eluted with SDS-proteinase K solution and extracted with phenol/CHCl3. The primer set used for ChIP-PCR was as follows: NonO-ChIP forward primer, 5′-TCTCCAGAGGATGG CTGAGT-3′, and NonO-ChIP reverse primer, 5′-AGCGCTATC TGCCTGTGTCT-3′ (amplified a 234 bp-fragment).
Luciferase assay
The NonO coding sequence was amplified by PCR and cloned into the PCNA vector (Invitrogen) for NonO over-expression (PCNA-NonO). NonO recognition sequence F was cloned into the pGL3-Basic vector (Promega) for use in a luciferase assay (pGL3-F). One microgram of PCNA-NonO and/or pGL3-F was transfected into 293T cells with Lipofectamine 2000 (Invitrogen), and the luciferase activity was quantified with a Multilabel Counter (PerkinElmer) 24 h after transfection.
siRNA treatment
The siRNA targeting mouse NonO and a non-targeting control siRNA were constructed based on a previous report with minor modifications (Song et al., 2008). The siRNAs were transfected into ES cells, and the NonO and Oct4 levels were examined 24 h after transfection by qRT-PCR and immunocytochemistry. The sequences of the siRNAs were as follows: NonO-siRNA, 5′-GUCGAACGAACUGCUGGAA(dTdT)-3′, and control-siRNA, 5′-CCUACGCCACCAAUUUCGU(dTdT)-3′.
RESULTS
Expression pattern of oct4 and methylation profile of oct4 PE/promoter CpG sites during ES cell differentiation
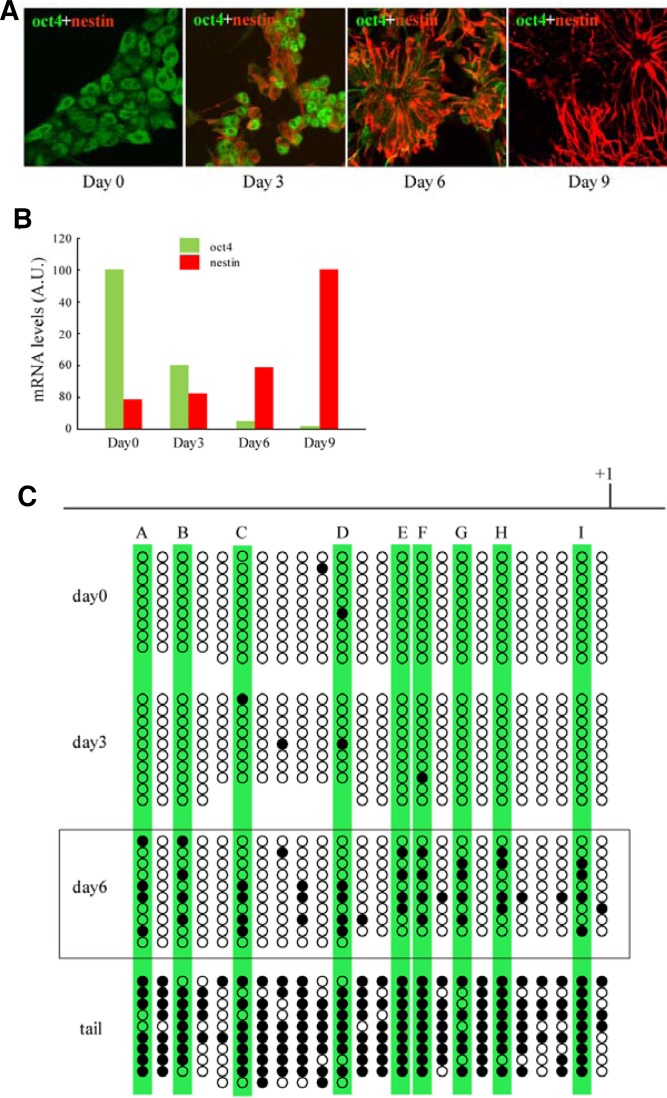
In the first step, we examined the regulatory mechanism of oct4 gene expression during ES cell differentiation. We differentiated ES cells into neurons and monitored the genetic and epigenetic changes related to the oct4 gene. Mouse ES cells were differentiated into neurons with the monolayer culture method (Ying et al., 2003), and the oct4 and nestin expression patterns were examined by immunocytochemistry and quantitative RT-PCR. The immunocytochemistry results showed that oct4 was expressed in undifferentiated ES cells at day 0 and that this level of expression decreased gradually and disappeared at day 9 of differentiation, whereas nestin, a type VI intermediate filament that is specific for neural cells, appeared on day 3 and gradually increased to day 9 (Fig. 1A). The quantitative RT-PCR results were consistent with the expression profile of immunocytochemistry (Fig. 1B).
Fig. 1.
Expression pattern of oct4 and DNA methylation profile of CpG sites in the oct4 promoter. Mouse ES cells were differentiated into neurons in adherent monolayer cultures, and the oct4 and nestin expression patterns were detected by immunocytochemistry (A) and quantitative RT-PCR (B). Green and red colors represent oct4 and nestin, respectively. (C). The DNA methylation profile of 24 CpG sites located in the oct4 promoter from-1200 to the ATG start codon was determined by bisulfite sequencing at 0, 3, and 6 days after the start of ES cell differentiation. Mouse tail DNA was used as a positive control. The open circles represent unmethylated CpG sites, and the black closed circles represent methylated CpG sites. Capital letters A to I represent CpG sites that are methylated at day 6.
We next examined the methylation profile of the oct4 PE/promoter CpG sites during ES cell differentiation with bisulfite sequencing. We monitored 1.2 kb upstream of the oct4 promoter CpG sites and found that the methylation profile exhibited an inverse correlation with the oct4 gene expression pattern. In day 0 undifferentiated ES cells, most CpG sites were unmethylated, but they were gradually methylated as differentiation progressed, as demounted by the results for day 3 and day 6 of differentiation (Fig. 1C). Fully differentiated adult mouse-tail DNA exhibited complete methylation. Of interest, we found 9 CpG sites that were vulnerable to methylation even in the early stage of differentiation (marked A to I in Fig. 1C). We assumed that the affinity of DNA-binding proteins for these CpG sites is altered depending on the methylation status, and this altered DNA binding regulates oct4 gene expression during ES cell differentiation. Therefore, we performed experiments to identify DNA-binding proteins that recognize these methylation-sensitive CpG sites.
Purification of NonO, a protein that binds to the CpG island of oct4 promoter
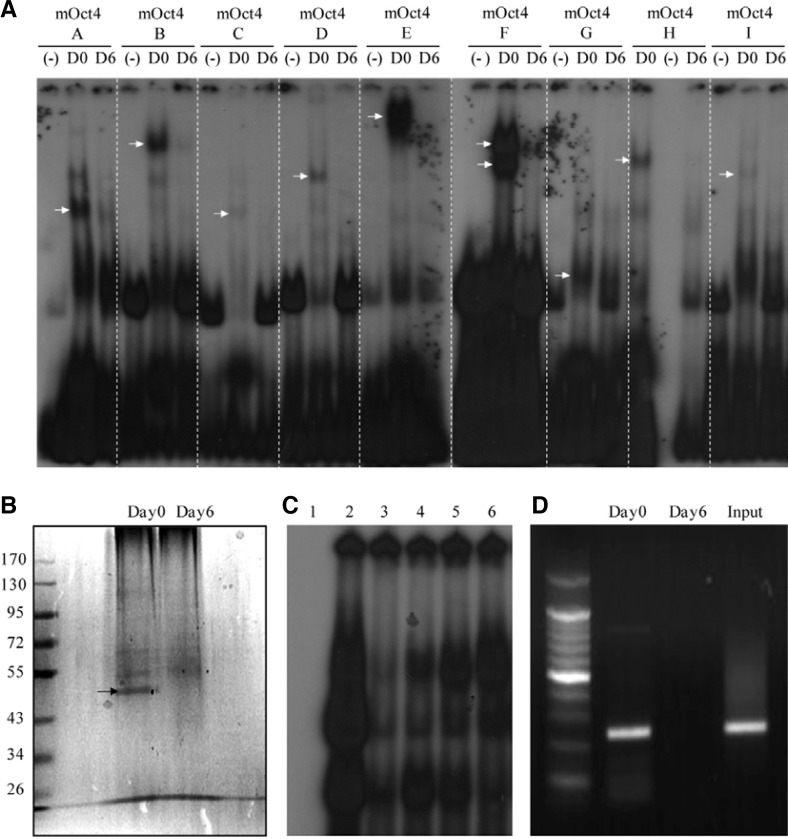
To find oct4 promoter binding proteins, we designed dsDNA probes containing CpG sites (probes A to I, Table 1) and performed Electro-mobility shift assays (EMSAs) with nuclear extracts from undifferentiated ES cells (day 0) and differentiated cells (day 6). There were many shifted bands in the day 0 nuclear extracts (Fig. 2A, arrows). However, there were no shift bands in the day 6 nuclear extracts, as confirmed by the absence of shifted band in control lanes (no nuclear extracts). These results indicate that putative DNA-binding proteins bind to the CpG sites of the oct4 PE/promoter in undifferentiated ES cells. Therefore, we purified the DNA-binding proteins using a DNA-binding purification kit (Roche). Each dsDNA probe (A-I) was elongated and ligated to magnetic beads, which were then mixed with nuclear extracts. The reaction mixtures were separated by SDS-PAGE and silver-stained. Among these probes, dsDNA probe F detected a specific DNA-binding protein in the day 0 nuclear extract (Figs. 2A and 2B, arrow). This protein was determined to be NonO by Liquid Chromatography-Mass Spectrophotometry (LC-MS) analysis. Subsequent sequence analysis revealed that Probe F contains a previously reported NonO recognition sequence (GTGAC) that is located 283–287 bp upstream of the oct4 promoter and is followed by a CpG site (Basu et al., 1997). The DNA binding specificity of NonO was examined with unlabeled probe (cold-F) and the mutated probes mt1-F and mt2-F. The unlabeled cold-F efficiently competed with the labeled probe F for NonO binding (Fig. 2C, lane 3). The mt1-F probe, which contains a 5 bp mutation in the recognition sequence (AATTT), and the mt2-F probe, which has a 2-bp mutation within the recognition sequence (GTGTT), failed to compete with probe F for NonO binding (Fig. 2C, lanes 4 and 5). These results indicate that NonO specifically binds to the GTGAC motif located between-287 and-283 in the oct4 promoter.
Fig. 2.
Purification and characterization of NonO, a novel oct4 promoter binding protein. (A) EMSA was performed using DNA probes containing CpG sites (A to I), and day 0 or day 6 cell nuclear extracts (NE). (−), no NE; D0, day 0 cell NE; D6, day 6 cell NE. The arrows at D0 indicate shifted bands. The shifted band for probe F (arrows) was selected for purification. (B) The protein biding to probe F was purified with the DNA-binding Protein Purification Kit (Roche), separated by SDS-PAGE, and silver-stained. The specific band for day 0 (arrow) was analyzed by liquid chromatography-mass spectrophotometry (LC-MS) and was determined to be NonO. (C) EMSA with various competitors; lane 1, no NE; lane 2, day 0 NE; lane 3, day 0 NE + cold (100X); lane 4, day 0 NE + mt1 (100X); lane 5, day 0 NE + mt2 (100X); lane 6, day 0 NE + methylated (100X). (D) ChIP PCR analysis was performed using an anti-NonO antibody with day 0 and day 6 cells. Genomic DNA left over before the ChIP analysis was used as a positive control.
We then tested whether methylation at the 5′ of the end recognition sequence interferes with NonO binding. Interestingly, met-F, which contains a methylated cytosine at the 5′ end of the recognition sequence (CC*GGTGAC), failed to compete with probe F for NonO binding, a result that supports the hypothesis that methylation at the CpG site affects the binding of NonO to its binding site (Fig. 2C, lane 6). To confirm this hypothesis, we performed ChIP-PCR analysis using a NonO antibody, and we detected the specific binding of NonO only in undifferentiated day 0 ES cells (Fig. 2D). Given that undifferentiated ES cells contain unmethylated CCGGTGAC, whereas differentiated day 6 cells contain methylated CC*GGTGAC (Fig. 1A), the results from the CHIP-PCR analysis support the conclusion that NonO binds to its recognition site in a methylation-dependent manner.
Functional role of NonO in the regulation of oct4 gene expression
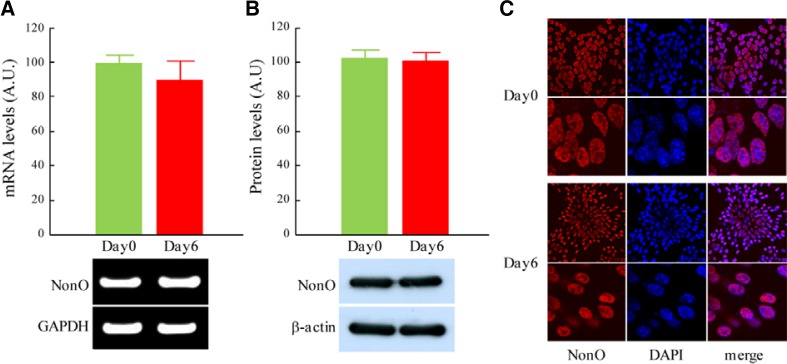
We examined the NonO mRNA and protein levels in ES cells and differentiated cells. Quantitative RT-PCR analysis showed no differences in the expression of NonO between day 0 ES cells and day 6 differentiated cells (Fig. 3A). Western blot analysis confirmed that there were no quantitative differences in the NonO protein levels (Fig. 3B). Immunocytochemistry with an anti-NonO antibody revealed that NonO was specifically expressed in the nucleus of both day 0 ES cells and differentiated day 6 cells (Fig. 3C).
Fig. 3.
Expression of NonO showed no significant differences between ES cells and differentiated cells. Mouse ES cells were differentiated into neurons by adherent monolayer cultures, and NonO mRNA and protein levels were examined with RT-PCR, Western blot analysis and immunocytochemistry. (A) Relative mRNA levels of NonO in ES cells (Day0) and differentiated cells (Day6) were determined by quantitative RT-PCR analysis, normalized with GAPDH and expressed as mean ± SEM (n = 6). (B) Arbitrary levels of NonO protein at Day0 and Day6 of differentiation were determined by Western blot analysis, normalized with β-actin and expressed as mean ± SEM (n = 6). (C) NonO expression at Day0 and Day6 cells was detected with immunocytochemistry. Red color represents NonO protein and blue color represents DAPI staining of nucleus. Lower panels show higher magnification of NonO staining in the nucleus.
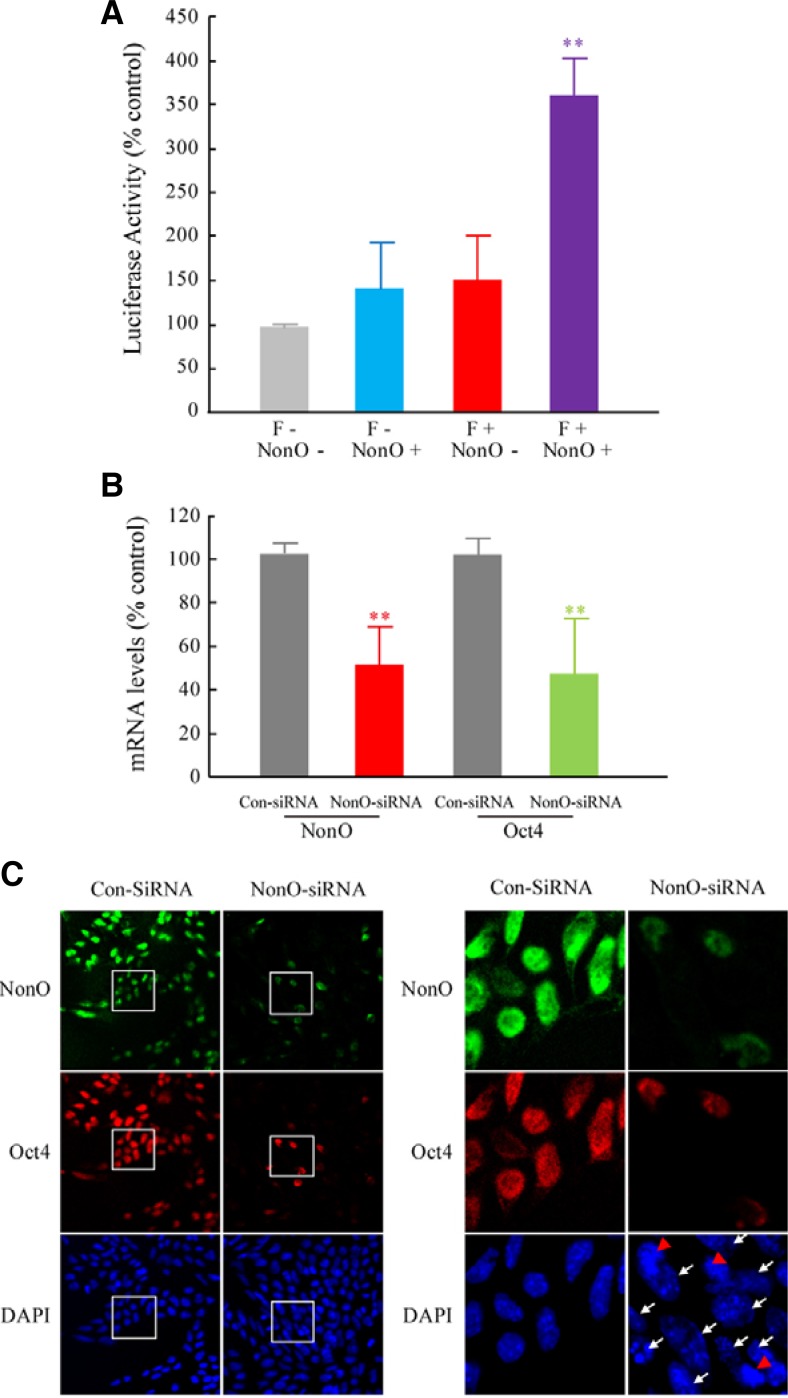
The transcriptional role of NonO on oct4 gene expression was evaluated in vitro using a luciferase assay. A NonO expression vector (PCNA-NonO) and a pGL3 luciferase vector containing recognition sequence F (pGL3-F) were constructed and transfected into the 293T cells, and the luciferase activity was quantified to determine the NonO transcriptional activity. Mock-transfected cells (PCNA with no NonO + pGL3 with no F) were used as a control and showed basal luciferase activity.
There were no significant differences between the PCNA-NonO-transfected cells and the pGL3-F-transfected cells. However, the luciferase activity was significantly increased by 3.5-fold in the PCNA-NonO and pGL3-F co-transfected cells (Fig. 4A). These results suggest that NonO acts as a transcriptional activator of oct4 gene expression. The transcriptional role of NonO in oct4 gene expression was further evaluated with a knockdown experiment. The siRNA targeting NonO and a non-targeting control siRNA were constructed based on a previous report with minor modifications (Song et al., 2008). These siRNAs were transfected into ES cells, and the expression levels of oct4 were determined by qRT-PCR and immunocytochemistry. As previously shown in Figs. 1B and 3, both oct4 and NonO were expressed at high levels in ES cells. Quantitative RT-PCR showed that NonO expression was inhibited by 50% by siRNA treatment (Fig. 4B). Surprisingly, the oct4 expression was reduced by 50%, a significant change, in parallel with NonO inhibition (Fig. 4B). The immunocytochemistry results confirmed the above results at the single-cell level. Oct4 was co-expressed with NonO in ES cells (Fig. 4C, control and red arrows). However, the oct4 expression was dramatically reduced when NonO expression was inhibited by siRNA treatment (Fig. 4C, white arrows). These results, along with the luciferase activity data, strongly support the conclusion that NonO positively regulates oct4 gene expression in ES cells.
Fig. 4.
NonO functions as a transcriptional activator of oct4 gene expression. (A) Transcriptional effect of NonO on the oct4 promoter was determined with a luciferase assay. A NonO expression vector (PCNA-NonO) and a pGL3-F luciferase vector containing the NonO recognition sequence were constructed and transfected into 293T cells in various combinations (NonO−:F−, NonO+: F−, NonO−:F+ and NonO+:F+). Student’s t-test was used for the statistical analysis. **p < 0.01 vs. F−:NonO− (n = 6). (B, C) Positive regulation of oct4 gene expression by NonO. An siRNA against NonO and a control siRNA were constructed and transfected into ES cells, and oct4 expression was determined 24 h after siRNA treatment. (B) Quantitative RT-PCR analysis of the NonO and oct4 mRNA levels in the control and NonO-siRNA treated cells. Student’s t-test was used for the statistical analysis. **p < 0.01 vs. ConSiRNA (n = 6). (C) Immunocytochemical analysis of NonO and oct4 (green color: NonO, red color: oct4, blue color: DAPI). Red arrows show the co-expression of NonO and oct4 in the same cells, and white arrows show the inhibition of oct4 in the NonO-siRNA treated cells.
DISCUSSION
In this study, we investigated the relationships among oct4 promoter CpG site methylation, NonO binding to the CpG site and oct4 gene expression during the course of ES cell differentiation. First, we analyzed 24 CpG sites in the PE/promoter regions of the oct4 gene and found 9 CpG sites (5 in the PE and 4 in the promoter) which were vulnerable to methylation even at day 6 of differentiation (Fig. 1C). Taking the developmental stage and tissue-specific methylation profiles of each CpG site of the oct4 PE/promoter regions into account (Gidekel and Bergman, 2002; Hattori et al., 2004; Marikawa et al., 2005), there is a possibility that temporal differences in the methylation of each CpG site results in the recruitment of different of transcription factors and regulate quantitative oct4 expression during ES cell differentiation. Many factors that regulate oct4 gene expression have been identified; however, the sequential and quantitative regulation of oct4 gene expression during ES cell differentiation is largely unknown. The EMSA results in Fig. 2A imply the existence of novel factors that bind to methylation-vulnerable CpG sites and regulate the temporal expression of oct4 during ES cell differentiation. Therefore, we tried to identify novel factors with probes containing CpG sites, and we purified NonO, a novel oct4 promoter binding protein. NonO binds to probe F, which contains a CpG site located 288–289 bp upstream of the oct4 promoter. EMSA with mutated and methylated competitors showed that NonO binds to its recognition sequence ‘CCGGTGAC’ only when the CpG is not methylated in ES cells. The results from the CHIP analysis with DNA from ES cells and differentiated cells (Fig. 2C) support these EMSA results.
According to the luciferase assays and the NonO-siRNA treatment studies (Fig. 4), NonO functions as a transcriptional activator of oct4 gene expression. NonO has been reported to regulate transcription in many different ways. NonO directly binds to the CYP17 promoter with the Sin3A and HDAC transcriptional repressor complex and inhibits the expression of CYP17, which participates in steroid hormone biosynthesis (Sewer et al., 2002). In addition to direct binding to target DNA, NonO binds to transcription factors and modulates their transcriptional activity. For example, NonO binds to the DNA-binding domain of steroid hormone receptors and suppresses transcriptional activity in the absence of hormone ligand. (Dong et al., 2009). In contrast to the many reported repressive effects, NonO has a transcriptional coactivator function in androgen receptor-mediated transcription (Ishitani et al., 2003; Kuwahara et al., 2006). NonO also activates transcription by enhancing the association of many transcription factors, such as E47, OTF-1 and OTF-2, with their target DNAs (Yang et al., 1997). Our manuscript is the first report that NonO binds to the CpG island of the oct4 promoter region and that this binding is modulated by DNA methylation. Another novel finding from our study is that NonO regulates oct4 gene expression. It is uncertain whether NonO regulates oct4 expression by modulating epigenetic processes. There are some examples of transcription factors that regulate oct4 expression by modulating epigenetic processes. The Lsh/Dnmt3 complex induces DNA methylation, and the cdx2/brg1 complex induces chromosome remodeling and suppresses oct4 expression during ES cell differentiation and trophectoderm formation, respectively (Wang et al., 2010; Xi et al., 2009). In contrast, CARM1 enhances oct4 gene expression by binding to the PE of oct4 and modulating histone H3 R17/26 methylation (Wu et al., 2009). Considering these facts and a previous report that NonO can bind to Sin3A and HDAC, it is possible that NonO modulates the epigenetic remodeling of the oct4 promoter and regulates oct4 expression during ES cell differentiation. The explanation for the epigenetic regulation of oct4 gene expression by NonO remains to be determined in further experiments. Most studies concerning the upstream regulation of oct4 gene expression have been performed using ChIP analyses, and therefore, there is no detailed information available about the binding sites of these transcription factors. Our study helps to explain the regulation of oct4 expression in terms of CpG island methylation profiles and CpG binding partners in ES cells.
Acknowledgments
This research was supported by the Korea Research Foundation, funded by the Korean Government (Ministry of Education, Science and Technology) (Grant KRF-20120007428).
REFERENCES
- Basu A., Dong B., Krainer A.R., Howe C.C. The intracisternal A-particle proximal enhancer-binding protein activates transcription and is identical to the RNA-and DNA-binding protein p54nrb/NonO. Mol. Cell. Biol. 1997;17:677–686. doi: 10.1128/mcb.17.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bladen C.L., Udayakumar D., Takeda Y., Dynan W.S. Identification of the polypyrimidine tract binding protein-associated splicing factor.p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J. Biol. Chem. 2005;280:5205–5210. doi: 10.1074/jbc.M412758200. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.L., Carmichael G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang Y.S., Huang W.H., Park S.W., Persaud S.D., Hung C.H., Ho P.C., Wei L.N. Promyelocytic leukemia protein in retinoic acid-induced chromatin remodeling of Oct4 gene promoter. Stem Cells. 2011;29:660–669. doi: 10.1002/stem.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B., Horowitz D.S., Kobayashi R., Krainer A.R. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res. 1993;21:4085–4092. doi: 10.1093/nar/21.17.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Yu C., Shynlova O., Challis J.R., Rennie P.S., Lye S.J. p54nrb is a transcriptional corepressor of the progesterone receptor that modulates transcription of the laborassociated gene, connexin 43 (Gja1) Mol. Endocrinol. 2009;23:1147–1160. doi: 10.1210/me.2008-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N., Gerson A., Fang J., Li E., Zhang Y., Shinkai Y., Cedar H., Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Gidekel S., Bergman Y. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J. Biol. Chem. 2002;277:34521–34530. doi: 10.1074/jbc.M203338200. [DOI] [PubMed] [Google Scholar]
- Gu P., Goodwin B., Chung A.C., Xu X., Wheeler D.A., Price R. R., Galardi C., Peng L., Latour A.M., Koller B.H., et al. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol. Cell. Biol. 2005a;25:3492–3505. doi: 10.1128/MCB.25.9.3492-3505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P., LeMenuet D., Chung A.C., Mancini M., Wheeler D.A., Cooney A.J. Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation. Mol. Cell. Biol. 2005b;25:8507–8519. doi: 10.1128/MCB.25.19.8507-8519.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Gu P., Xu X., Le Menuet D., Chung A.C., Cooney A.J. Differential recruitment of methyl CpG-binding domain factors and DNA methyltransferases by the orphan receptor germ cell nuclear factor initiates the repression and silencing of Oct4. Stem Cells. 2011;29:1041–1051. doi: 10.1002/stem.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori N., Nishino K., Ko Y.G., Ohgane J., Tanaka S., Shiota K. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 2004;279:17063–17069. doi: 10.1074/jbc.M309002200. [DOI] [PubMed] [Google Scholar]
- Hwang M., Gorivodsky M., Kim M., Westphal H., Geum D. The neuronal differentiation potential of Ldb1-null mutant embryonic stem cells is dependent on extrinsic influences. Stem Cells. 2008;26:1490–1495. doi: 10.1634/stemcells.2007-1099. [DOI] [PubMed] [Google Scholar]
- Ishitani K., Yoshida T., Kitagawa H., Ohta H., Nozawa S., Kato S. p54nrb acts as a transcriptional coactivator for activation function 1 of the human androgen receptor. Biochem. Biophys. Res. Commun. 2003;306:660–665. doi: 10.1016/s0006-291x(03)01021-0. [DOI] [PubMed] [Google Scholar]
- Kameoka S., Duque P., Konarska M.M. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 2004;23:1782–1791. doi: 10.1038/sj.emboj.7600187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.K., Koo D.B., Park J.S., Choi Y.H., Chung A.S., Lee K.K., Han Y.M. Aberrant methylation of donor genome in cloned bovine embryos. Nat. Genet. 2001;28:173–177. doi: 10.1038/88903. [DOI] [PubMed] [Google Scholar]
- Kelly V.R., Xu B., Kuick R., Koenig R.J., Hammer G.D. Dax1 up-regulates Oct4 expression in mouse embryonic stem cells via LRH-1 and SRA. Mol. Endocrinol. 2010;24:2281–2291. doi: 10.1210/me.2010-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara S., Ikei A., Taguchi Y., Tabuchi Y., Fujimoto N., Obinata M., Uesugi S., Kurihara Y. PSPC1, NONO, and SFPQ are expressed in mouse Sertoli cells and may function as coregulators of androgen receptor-mediated transcription. Biol. Reprod. 2006;75:352–359. doi: 10.1095/biolreprod.106.051136. [DOI] [PubMed] [Google Scholar]
- Magnuson T., Epstein C.J., Silver L.M., Martin G.R. Pluripotent embryonic stem cell lines can be derived from tw5/tw5 blastocysts. Nature. 1982;298:750–753. doi: 10.1038/298750a0. [DOI] [PubMed] [Google Scholar]
- Marikawa Y., Fujita T.C., Alarcon V.B. Heterogeneous DNA methylation status of the regulatory element of the mouse Oct4 gene in adult somatic cell population. Cloning Stem Cells. 2005;7:8–16. doi: 10.1089/clo.2005.7.8. [DOI] [PubMed] [Google Scholar]
- Moreno-Manzano V., Rodriguez-Jimenez F.J., Acena-Bonilla J.L., Fustero-Lardies S., Erceg S., Dopazo J., Montaner D., Stojkovic M., Sanchez-Puelles J.M. FM19G11, a new hypoxia-inducible factor (HIF) modulator, affects stem cell differentiation status. J. Biol. Chem. 2010;285:1333–1342. doi: 10.1074/jbc.M109.008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or selfrenewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Rosonina E., Ip J.Y., Calarco J.A., Bakowski M.A., Emili A., McCracken S., Tucker P., Ingles C.J., Blencowe B.J. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol. Cell. Biol. 2005;25:6734–6746. doi: 10.1128/MCB.25.15.6734-6746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton M., Lerenthal Y., Wang S.Y., Chen D.J., Shiloh Y. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 2010;9:1568–1576. doi: 10.4161/cc.9.8.11298. [DOI] [PubMed] [Google Scholar]
- Sewer M.B., Nguyen V.Q., Huang C.J., Tucker P.W., Kagawa N., Waterman M.R. Transcriptional activation of human CYP17 in H295R adrenocortical cells depends on complex formation among p54(nrb)/NonO, protein-associated splicing factor, and SF-1, a complex that also participates in repression of transcription. Endocrinology. 2002;143:1280–1290. doi: 10.1210/endo.143.4.8748. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y., Zipori D. PSF and p54(nrb)/NonO-multifunctional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- Son G.H., Geum D., Chung S., Park E., Lee K.H., Choi S., Kim K. A protective role of 27-kDa heat shock protein in glucocorticoid-evoked apoptotic cell death of hippocampal progenitor cells. Biochem. Biophys. Res. Commun. 2005;338:1751–1758. doi: 10.1016/j.bbrc.2005.10.152. [DOI] [PubMed] [Google Scholar]
- Song K.S., Kim K., Chung K.C., Seol J.H., Yoon J.H. Interaction of SOCS3 with NonO attenuates IL-1beta-dependent MUC8 gene expression. Biochem. Biophys. Res. Commun. 2008;377:946–951. doi: 10.1016/j.bbrc.2008.10.084. [DOI] [PubMed] [Google Scholar]
- Straub T., Grue P., Uhse A., Lisby M., Knudsen B.R., Tange T.O., Westergaard O., Boege F. The RNA-splicing factor PSF/p54 controls DNA-topoisomerase I activity by a direct interaction. J. Biol. Chem. 1998;273:26261–26264. doi: 10.1074/jbc.273.41.26261. [DOI] [PubMed] [Google Scholar]
- Straub T., Knudsen B.R., Boege F. PSF/p54(nrb) stimulates “jumping” of DNA topoisomerase I between separate DNA helices. Biochemistry. 2000;39:7552–7558. doi: 10.1021/bi992898e. [DOI] [PubMed] [Google Scholar]
- Tam W.L., Lim C.Y., Han J., Zhang J., Ang Y.S., Ng H.H., Yang H., Lim B. T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells. 2008;26:2019–2031. doi: 10.1634/stemcells.2007-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov K.V., Tarasova Y.S., Tam W.L., Riordon D.R., Elliott S. T., Kania G., Li J., Yamanaka S., Crider D.G., Testa G., et al. B-MYB is essential for normal cell cycle progression and chromosomal stability of embryonic stem cells. PLoS One. 2008;3:e2478. doi: 10.1371/journal.pone.0002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Velkey J.M., O’Shea K.S. Oct4 RNA interference induces trophectoderm differentiation in mouse embryonic stem cells. Genesis. 2003;37:18–24. doi: 10.1002/gene.10218. [DOI] [PubMed] [Google Scholar]
- Wang K., Sengupta S., Magnani L., Wilson C.A., Henry R.W., Knott J.G. Brg1 is required for Cdx2-mediated repression of Oct4 expression in mouse blastocysts. PLoS One. 2010;5:e10622. doi: 10.1371/journal.pone.0010622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Bruce A.W., Jedrusik A., Ellis P.D., Andrews R.M., Langford C.F., Glover D.M., Zernicka-Goetz M. CARM1 is required in embryonic stem cells to maintain pluripotency and resist differentiation. Stem Cells. 2009;27:2637–2645. doi: 10.1002/stem.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi S., Geiman T.M., Briones V., Guang Tao Y., Xu H., Muegge K. Lsh participates in DNA methylation and silencing of stem cell genes. Stem Cells. 2009;27:2691–2702. doi: 10.1002/stem.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.S., Hanke J.H., Carayannopoulos L., Craft C.M., Capra J.D., Tucker P.W. NonO, a non-POU-domain-containing, octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss. Mol. Cell. Biol. 1993;13:5593–5603. doi: 10.1128/mcb.13.9.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.S., Yang M.C., Tucker P.W., Capra J.D. NonO enhances the association of many DNA-binding proteins to their targets. Nucleic Acids Res. 1997;25:2284–2292. doi: 10.1093/nar/25.12.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.M., Do H.J., Kim D.K., Park J.K., Chang W.K., Chung H.M., Choi S.Y., Kim J.H. Transcriptional regulation of human Oct4 by steroidogenic factor-1. J. Cell. Biochem. 2007;101:1198–1209. doi: 10.1002/jcb.21244. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Stavridis M., Griffiths D., Li M., Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Young R.A. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.B., Kunarso G., Hong F.H., Stanton L.W. Zfp206, Oct4, and Sox2 are integrated components of a transcriptional regulatory network in embryonic stem cells. J. Biol. Chem. 2009;284:31327–31335. doi: 10.1074/jbc.M109.016162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Carmichael G.G. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- Zhang J., Tam W.L., Tong G.Q., Wu Q., Chan H.Y., Soh B.S., Lou Y., Yang J., Ma Y., Chai L., et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang J., Wang T., Esteban M.A., Pei D. Esrrb activates Oct4 transcription and sustains self-renewal and pluripotency in embryonic stem cells. J. Biol. Chem. 2008;283:35825–35833. doi: 10.1074/jbc.M803481200. [DOI] [PubMed] [Google Scholar]