Abstract
Oxidative stress promotes damage to cellular proteins, lipids, membranes and DNA, and plays a key role in the development of cancer. Reactive oxygen species disrupt redox homeostasis and promote tumor formation by initiating aberrant activation of signaling pathways that lead to tumorigenesis. We used shotgun proteomics to identify proteins containing oxidation-sensitive cysteines in tissue specimens from colorectal cancer patients. We then compared the patterns of cysteine oxidation in the membrane fractions between the tumor and non-tumor tissues. Using nano-UPLC-MSE proteomics, we identified 31 proteins containing 37 oxidation-sensitive cysteines. These proteins were observed with IAM-binding cysteines in non-tumoral region more than tumoral region of CRC patients. Then using the Ingenuity pathway program, we evaluated the cellular canonical networks connecting those proteins. Within the networks, proteins with multiple connections were related with organ morphology, cellular metabolism, and various disorders. We have thus identified networks of proteins whose redox status is altered by oxidative stress, perhaps leading to changes in cellular functionality that promotes tumorigenesis.
Keywords: colorectal cancer, cysteine oxidation, iodoacetamide, protein network, shotgun proteomics
INTRODUCTION
Colorectal cancer (CRC) is one of the most frequently occurring malignancies, worldwide (Weitz et al., 2005). The risk of developing CRC increases with advancing age, and although CRCs can develop sporadically, one may also inherit an increased risk of developing the disease. In addition, chronic inflammatory bowel diseases such as ulcerative colitis and Crohn’s disease may also increase ones risk of developing CRC (Parkin, 2006; Weitz et al., 2005). The activated inflammatory cells seen in these conditions induce expression of oxidant-generating enzymes such as NADPH oxidase, myeloperoxidase and inducible nitric oxide synthase, leading to the production of high levels of both ROS and reactive nitrogen species (RNS) (Roessner et al., 2008). However, the most important sources of ROS in cancer are the electron transport chains in the mitochondria and endoplasmic reticulum. Pre-neoplastic and cancer cells are metabolically active and require high levels of ATP to maintain their higher-than-normal proliferation rates; consequently, energy production in the cells’ mitochondrial respiratory chains is high, which leads to increases in ROS production (Pelicano et al., 2004). Like all cancers, CRC is a congregation of abnormal cells and represents an imbalance between cell renewal and cell death. Following this basic concept, much effort has gone into discovering methods to reduce the proliferation of the malignant cells, including antioxidant therapies. In particular, anti-oxidant enzymes may inhibit the promotion of carcinogenesis. Because their expression is reduced in many malignant tumors, their supplementation is thought by some to be potentially therapeutic (Mates and Sanchez-Jimenez, 2000; Roessner et al., 2008; Stone et al., 2004). However, the use of antioxidants for cancer prevention and therapy has proven controversial, highlighting the need for a better understanding of the molecular environments of malignant cells.
Reactive oxygen species (ROS) can be acutely toxic to cells, due in large part to their participation in oxidation reactions leading to the inactivation of biomolecules (Finkel, 2003; Veal et al., 2007). In addition, ROS generated in response to various growth factors and cytokines may contribute to human carcinogenesis (Lee and Lee, 2006; Ziech et al., 2011), as the accumulation of oxidative damage to cellular enzymes, structural proteins and membranes increases susceptibility to tumor initiation (Leonard and Carroll, 2011). And through reversible oxidation of sensor components, ROS function as second messengers in signaling cascades (Finkel, 2003; Veal et al., 2007), exerting effects on tumor growth, cellular survival and metastasis (Roessner et al., 2008). However, ROS mediated protein oxidation was not validated completely in tumor yet. In addition, physiological levels of hydrogen peroxide can stimulate cellular proliferation and survival among tumor cells (Burdon, 1995; Burdon et al., 1996), and ROS may play important roles in mediating loss of growth control (Behrend et al., 2003). It is therefore not surprising that cancer cells are frequently found to be in a condition of redox imbalance, reflecting an alteration in the homeostasis between pro-oxidants and antioxidants that results in an elevation in oxidant levels within the cell’s surroundings (Acharya et al., 2010).
Reactive cysteine residues within proteins are the main targets of oxidative modification due to their low pKa and high nucleophilicity (Bulaj et al., 1998), which are dependent on the charges of the adjacent amino acids (Choi et al., 2001). The vulnerability of cysteines to both reversible and irreversible oxidation is a concern when considering the regulation of ROS-responsive pathways (Janssen-Heininger et al., 2008; Winterbourn and Hampton, 2008) because it is now clear that a major component of ROS-linked modulation of cellular signaling is mediated via the active regulation of protein function through reversible thiol (R-SH) modification. In many proteins, the first product of cysteine oxidation by hydroperoxides is cysteine sulfenic acid (R-SOH), which undergoes rapid condensation with either another protein thiol (as an intra- or intermolecular interaction) or a small molecule thiol, like glutathione or cysteine, to form a disulfide bond. Disulfide bonds can stabilize proteins and associations within protein complexes; protect against irreversible inactivation; modify structures to create, destroy or modulate functional sites; and ultimately regulate the enzymatic and/or transcriptional activity of proteins (Garcia-Santamarina et al., 2011). In certain cases, the initial R-SOH may itself be stabilized within the protein microenvironment (Poole and Nelson, 2008). Alternatively, R-SOH is vulnerable to irreversible hyperoxidation to sulfinic (R-SO2H) or sulfonic acid (R-SO3H), which may also contribute to signaling the cellular redox state (Phalen et al., 2006).
In an attempt to better understand the relationship between oxidative stress and tumorigenesis, we performed comparative shotgun proteomics using the membrane fractions from tissues collected from 10 CRC patients. To identify proteins susceptible to cysteine oxidation, we used nano-UPLC MSE to confirm the patterns of IAM-labeled cysteines between CRC and non-tumor tissues. This enabled us to identify 31 susceptible proteins whose networks are related to tumorigenesis.
MATERIALS AND METHODS
Sample specimens
All chemicals were purchased from Sigma-Aldrich or Invitrogen. CRC samples were collected with informed consent from patients at the Chonnam National University Hwasun Hospital-National Biobank of Korea. The patients ranged from 32 to 76 years of age. The specimens included tissue samples from five stage II CRCs and five stage III CRCs resected in 2008 and 2009. The tissue samples were collected from the tumors within 30 min after surgical resection, snap frozen in liquid nitrogen, and stored at −80°C. Histological diagnoses were made for all CRC patients following microscopic examination of hematoxylin/ eosin-stained frozen sections by a board of certified pathologists (Table 1) (Yang et al., 2012a). This study was approved by the Institutional Review Board of Chonnam National University Hwasun Hospital (Korea).
Table 1.
Clinical information for the 10 CRC patients
| No. | Gendera | Age | Tumor stage | Pathological stage | Histology grade | Local.b | Necrosis (%) |
|---|---|---|---|---|---|---|---|
| 1 | M | 64 | II | T3N0M0 | G3 (Poorly) | Hepatic flexure | 20 |
| 2 | M | 32 | II | T3N0M0 | G2 (Moderately) | Transverse colon | X |
| 3 | F | 72 | II | T4N0M0 | G2 (Moderately) | Ascending colon | X |
| 4 | M | 66 | II | T3N0M0 | G2 (Moderately) | Rectal cancer | X |
| 5 | F | 86 | II | T3N0M0 | G3 (Poorly) | Hepatic flexure | X |
| 6 | M | 69 | III | T2N1M0 | G1 (Well) | Rectal cancer | X |
| 7 | M | 76 | III | T3N1M0 | G1 (Well) | Sigmoid colon | X |
| 8 | M | 64 | III | T3N1M0 | G2 (Moderately) | Sigmoid colon | 5 |
| 9 | F | 71 | III | T3N1M0 | G2 (Moderately) | Sigmoid colon | 10 |
| 10 | F | 75 | III | T3N2M0 | G1 (Well) | Ascending colon | 5 |
M, male; F, female
Local., tumor localization
Nano-UPLC MSE shotgun proteomics
For proteomics analysis, frozen tissue samples were homogenized in 800 μl of chilled buffer [220 μM iodoacetamide (IAM), 1 mM EDTA, 0.1 mM AEBSF, 2 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin A and 0.1 mM PMSF in 1× PBS (pH 7.4)] and then incubated for 4 h at 4°C. To remove the cytosolic fraction containing lipids and soluble proteins, the homogenized samples were centrifuged at 5, 000 rpm for 30 min at 4°C. The pellets were then washed 3 times with chilled PBS and centrifuged for 10 min at 13, 000 rpm at 4°C, after which the pellets were dissolved in 200 μl of urea buffer and subjected to five sonication cycles, each consisting of 5 s of sonication followed by a 5 s interval. The resultant suspensions were centrifuged at 13, 000 rpm for 30 min at 4°C, and the supernatants were collected. The protein concentration in each sample was determined using a Bradford assay (Bio-Rad). Aliquots containing 200 μg of protein were subjected to 10% SDS-PAGE, and the resultant bands were stained with Coomassie Blue R-250, after which the stained gels were washed 3 times with distilled water prior to tryptic digestion.
For efficient trypsinization, the separated proteins were excised by extracting 10 slices of the gel for each patient sample. The gel slices were then chopped into 1 mm3 pieces and destained using five 20-min incubations in 50% acetonitrile and 50 mM ammonium bicarbonate. Once all the stain was removed, the gel pieces were dehydrated in 100% acetonitrile and then dried. The pieces from each gel were then reduced with 10 mM DTT and alkylated with 55 mM N-ethylmaleimide (NEM) in 100 mM ammonium bicarbonate. Following tryptic digestion (2 μg/sliced gel; Promega) for 16 h at 37°C, the peptides were recovered and extracted from the gel pieces using 5% formic acid and 50% acetonitrile. After extraction, the peptides were dried in a vacuum centrifuge and combined into one tube. The combined samples were desalted using a solid phase Oasis HLB C18 microelution plate (Waters, Inc., USA) and then stored at −80°C until subjected to nano-UPLC coupled Q-TOF tandem mass spectrometry.
Protein separations were accomplished using a nano-ACQUITY Ultra Performance LC Chromatography™ System (Waters Corporation, USA) equipped with a 75 μm × 250 mm nano-ACQUITY UPLC 1.7 μm BEH300 C18 RP column and a 180 μm × 20 mm Symmetry C18 RP 5 μm enrichment column. Trypsinized peptides (5 μl) were loaded onto the enrichment column in mobile phase A (3% acetonitrile in water with 0.1% formic acid). A step gradient was then used at a flow rate of 300 nL/min. The steps included a 3–40% gradient of mobile phase B (97% acetonitrile in water with 0.1% formic acid) run over 95 min, followed by a 40–70% gradient of mobile phase B run over 20 min, and finally a sharp increase to 80% B over 10 min. Sodium formate (1 μmol/min) was used to calibrate the TOF analyzer in the range of m/z 50–2, 000, and [Glu1]-fibrinopeptide (m/z 785.8426) was run at 600 nl/min for lock mass correction. During data acquisition, the collision energies of the low-energy mode (MS) and high-energy mode (MSE) were set to 4 eV and 15–40 eV energy ramping, respectively. One cycle of the MS and MSE modes of acquisition was performed every 3.2 s. During each cycle, MS spectra were acquired for 1.5 s with a 0.1 s interscan delay (m/z 300–1, 990), and ions exceeding 50 counts were selected for MSE fragmentation (m/z 50–2, 000) in the collision cell.
The continuum LC-MSE data were processed and searched using the IDENTITYE algorithm in PLGS (ProteinLynx Global Server) version 2.3.3 (Waters). MS/MS spectra were reconstructed by combining all masses with identical retention times (Supplementary Table S1). Each sample was analyzed in triplicate runs. The data acquired by alternating low and high energy modes in the LC-MSE were automatically smoothed, background subtracted, centered, deisotoped and charge state reduced, after which alignments of the precursor and fragmentation data were combined with the retention time tolerance (± 0.05 min) using PLGS software. Processed ions were mapped against the IPI human database (version 3.44) using the following parameters: parent ion tolerance, 100 ppm; fragment ion mass tolerance, 0.2 Da; and missed cleavage, 1; and variable modification containing Carbamidomethylation (IAM binding) and N-ethylmaleimide (NEM). The individual MS/MS spectra for peptides with molecular weight search scores lower than 40 (expect value < 0.015) in PLGS were inspected manually and included in the statistics only if a series of at least four continuous y or b ions were observed (Yang et al., 2012b). Peptide identification was performed using the trypsin digestion rule with one missed cleavage. As a result, protein identification was completed with arrangement of at least two peptides. All proteins identified on the basis of the IDENTITYE algorithm were in keeping with > 95% probability. The false positive rate for protein identification was set at 5% in the databank search query option, based on the automatically generated reversed database in PLGS 2.3.3. Protein identification was also based on the assignment of at least two peptides comprised of seven fragments or more.
Bioinformatics analysis
Ingenuity Pathway Analysis (IPA version 9.0; Ingenuity Systems Inc.) was used to perform knowledge-based network analysis of comparative proteomics data (www.ingenuity.com). To functionally categorize identified proteins, p-values were used to show the ratio between the number of identified proteins classified and the total number of proteins in the linked function referenced by the software.
Antibody production
The epitopes of the identified proteins were as follows: COL6A1, CSFECQPARGPPGLR and TPSB2, QVATAPHTFPAPS (Anygen Co., Korea). The peptides were prepared as previously described (Yang et al., 2012b). The peptides (2.5 mg/5 mice) were coupled to 5 mg of keyhole limpet hemocyanin (Pierce Biotechnology) by incubation overnight at 4°C in the presence of 1.25 mg sulfo-SMCC (Pierce Biotechnology) in 1× PBS (pH 7.4). Blood was collected 1 week after the third booster injection, and the antisera were extracted.
Western blotting and immunoprecipitation with BIAM labeling
To assess patterns of oxidation, samples (∼50 mg) of frozen tumor tissue and tissue from non-tumor regions were homogenized in 500 μl of chilled labeling buffer [20 mM Tris-HCl (pH 7.5), 100 mM NaCl 1 mM EDTA, 0.1 mM AEBSF, leupeptin (2.0 μg/ml), and aprotinin (1.9 μg/ml)] containing 40 μM N-(biotinoyl)-N’-(iodoacetyl) ethylenediamine (biotinylated iodoacetamide, BIAM) (Molecular Probes, USA) and incubated for 4 h at 4°C. The buffer was bubbled with nitrogen for 1 h to remove the oxygen. The labeling reaction was quenched by adding 4 mM dithiothreitol (DTT), after which the mixtures were centrifuged at 5, 000 rpm for 30 min at 4°C, and the supernatant were discarded. The pellets were washed three times with 1× PBS, and then dissolved in urea buffer (10 mM Tris-HCl, pH7.4, 7 M urea, 2 M thiourea, 0.49% CHAPS, and protease inhibitor mixture). The resolved proteins were transferred to PVDF membranes (Bio-Rad Laboratories), which were then incubated with horseradish peroxidase (HRP)-conjugated streptavidin (Pierce Biotechnology Inc.) and developed using an ECL system (iNtRON, Korea).
After the incubation with BIAM, 200 μg of membrane protein were incubated overnight with Streptavidin-conjugated Sepharose beads (GE Healthcare, Sweden) at 4°C on a shaker. The beads were washed three times with NET buffer [20 mM Tris-HCl (pH7.5), 100 mM NaCl, 1 mM EDTA, 0.5% Triton X-100] and boiled at 95°C for 10 min. The IP proteins were loaded on 12% SDS-PAGE gels and Western blotting was performed. Antibodies against COL6A1 (described in “Antibody Production”) and TPSB2 (described in “Antibody Production”) were incubated for overnight and developed using an ECL system (Yang et al., 2012a). And PDIA3 (SPA-585, Stressgen®) was purchased from Enzo Life Sciences and used according to the manufacture’s protocol. Densitometry values of the bands were evaluated for statistical significance with SigmaPlot software (version 10.0, Systat Software Inc.).
RESULTS
Oxidative modification in the membrane fraction of CRC tissue
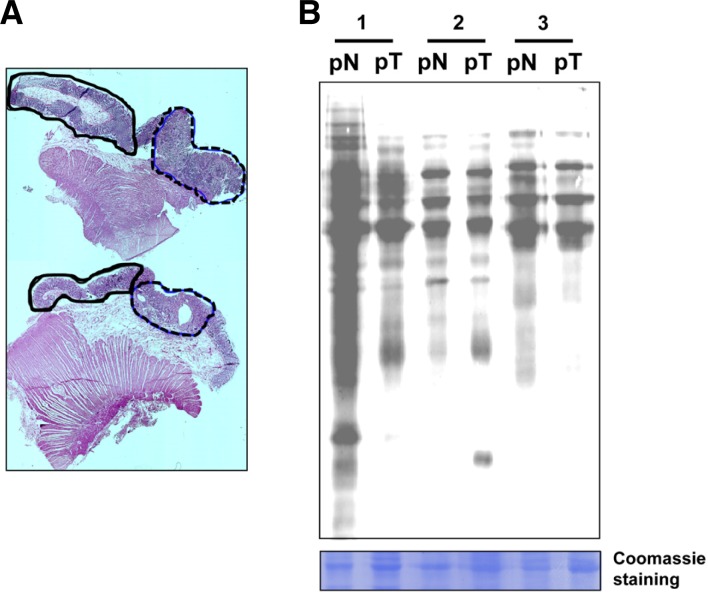
To compare the extent of oxidative modification between CRC and non-tumor tissues, we assessed cysteine oxidation using BIAM as an alkylating agent specific for reactive thiols. After separating the tumor from non-tumor tissue specimens based on histological staining (Fig. 1A), we observed that the membrane fractions of the tumor tissues (pT) showed less BIAM labeling than the non-tumor tissue (pN) (Fig. 1B). This indicates that some proteins were more susceptible to cysteine oxidation within colorectal tumors than in adjacent non-tumor tissue of patient.
Fig. 1.
Detection of proteins containing oxidation-sensitive cysteine residues. (A) Histological staining of tissues from CRC patients. The solid lines, non-tumoral regions; the dotted lines, tumoral regions. (B) Modification of cysteines in the membrane fraction of tumor and non-tumor tissues detected using BIAM-streptavidin immunoblotting. Samples were run in triplicate, and Coomassie-stained proteins served as a loading control. pN, non-tumor tissues of CRC patients; pT, tumor tissues of CRC patients.
We next applied a nano-UPLC-MSE proteomics system to identify the oxidation-sensitive cysteines (Supplementary Fig. S1). Nano-UPLC-MSE proteomics is suitable for large-scale protein analysis and for identifying protein modifications in complex samples containing proteins whose frequency among cells exhibits high dynamic range (Xu et al., 2008; Yang et al., 2012b). In this scheme, IAM was first used to label the endogenous free thiol groups exclusive of disulfide bonds and hyperoxides, after which we performed in-gel digestion using trypsin. The relative levels of the identified peptides were then determined after measuring the peak intensities of individual peptide masses obtained in separate chromatography runs. The validity of the relative quantitation was improved by analyzing the samples in triplicate runs, and by verifying the available information through replication and determination of confidence levels for the protein identification. Within the 10 CRC and non-tumor tissue samples (Table 1), we detected 62, 515 peptides in the membrane fraction (Supplementary Table S1), which were efficiently identified at a confidence level ≥ 95% (Supplementary Table S2). Using this nano-UPLC MS proteomics approach, we were able to detect many more proteins containing some form of modified cysteine than were detected using gel-based proteomics.
We found 37 peptides from 31 proteins that contained oxidatively modified cysteines in the membrane fractions of the 10 CRC specimens (Supplementary Table S3). To identify these peptides, we first preferentially selected those that contained more IAM-labeled (free reduced) cysteines within non-tumor tissues than the paired tumor tissues; based on our earlier findings, we anticipated that cysteines within tumor tissues were more likely to be oxidized. Using this criterion, we sorted proteins observed in at least two CRC samples. We found that among the tissue specimens, cysteines were more reduced in non-tumor tissues than CRC and more oxidized in the CRC tissues than the non-tumor tissues. A subset of the identified proteins is listed in Table 2.
Table 2.
Proteins containing oxidation-sensitive cysteines in tumor and non-tumor tissues from CRC patients
| IPI No. | Protein name | PLGS score | Cys. locala | Sequenceb | No. of IAM binding (total 10 case)c | |
|---|---|---|---|---|---|---|
|
| ||||||
| pN | pT | |||||
| IPI00479743 | A26C1A | 576.02 | 355 | EYAVSSHHHVICQLLSDYK | 8 | 5 |
| IPI00796881 | ACTG1 | 505.51 | 130 | RPCSSRPFPSHFLPGLISDI | 9 | 5 |
| IPI00025416 | ACTG2 | 1147.49 | 219 | LCYVALDFENEMATAASSSSLEK | 2 | N.D. |
| IPI00759776 | ACTN1 | 565.70 | 155 | FAIQDISVEETSAKEGLLLWCQR | 3 | N.D. |
| IPI00013508 | 1212.18 | 755 | ACLISLGYDIGNDPQK | 2 | 1 | |
| IPI00032137 | ACTN3 | 420.56 | 491 | CQAICDQWDNLGTLTQK | 8 | 3 |
| 420.56 | 594 | ICQTYGLRPCSTNPYITLSPQDINTK | 7 | 3 | ||
| IPI00746049 | ANKRD30B | 619.29 | 656 | AESPDKDGLLKPTCGR | 7 | 5 |
| IPI00797556 | ANXA2 | 147.13 | 208 | VSAELFALSFCFLVSLWIVYIVI | 2 | N.D. |
| IPI00014625 | CLCA1 | 411.41 | 251 | ASIMFAQHVDSIVEFCTEQNHNK | 2 | N.D. |
| IPI00291136 | COL6A1 | 562.93 | 595 | GPEGPQGPPGHQGPPGPDECEILDIIMK | 4 | 3 |
| 643.36 | 612 | CGPIDLLFVLDSSESIGLQNFEIAKDFVVK | 3 | 1 | ||
| IPI00556127 | DGKZ | 158.02 | 206 | EASVPLGTVVVPGDSDLELCR | 2 | N.D. |
| IPI00555900 | FKSG30 | 375.90 | 258 | CPEALFQPCFLGMESCGIHK | 10 | 9 |
| IPI00031522 | HADHA | 208.95 | 98 | SAVLISSKPGCFIAGADINMLAACK | 2 | 1 |
| 208.95 | 748 | QFTPCQLLADHANSPNK | 2 | N.D. | ||
| IPI00845339 | HSPA1A/B | 171.33 | 604 | KELEQVCNPIISGLYQGAGGPGPGGFGAQGPK | 3 | 1 |
| IPI00893099 | HSPA1L | 181.60 | 20 | GIAIGIDLGTTYSCVGVFQHGK | 2 | N.D. |
| IPI00339269 | HSPA6 | 199.38 | 625 | LYGGPGVPGGSSCGTQAR | 2 | 1 |
| IPI00166866 | IGHA1 | 178.60 | 20 | GVQCEVQLVESGGGVVRPGGSLR | 3 | N.D. |
| IPI00785084 | IGHV4-31 | 207.84 | 117 | MNSVTAADTAVYFCAR | 3 | N.D. |
| IPI00009865 | KRT10 | 377.09 | 26 | SGGGGGGGGCGGGGGVSSLR | 3 | N.D. |
| IPI00747707 | KRT17 | 210.40 | 12 | AAALGVLMGCWLEVR | 6 | 3 |
| IPI00888063 | KRT18P33 | 272.83 | 171 | SGDWDTLPSLQAHSSSVLAVPSGTAQTSGDLEQAPFCLLEAHSTR | 4 | 2 |
| 272.83 | 475 | GDCVCPPRPFSDVVK | 2 | N.D. | ||
| IPI00290078 | KRT4 | 185.87 | 15 | MTSVGVFSDMLNGCGKDGLVPR | 3 | N.D. |
| IPI00514817 | LMNA | 494.64 | 7 | QPLLCLGNLEDARER | 5 | 4 |
| IPI00000105 | MVP | 290.52 | 283 | TGEEWLVTVQDTEAHVPDVHEEVLGVVPITTLGPHNYCVILDPVGPDGK | 6 | N.D. |
| IPI00395772 | MYH9 | 578.31 | 376 | QELEEICHDLEAR | 2 | 1 |
| IPI00025252 | PDIA3 | 311.34 | 245 | FIQENIFGICPHMTEDNKDLIQGK | 2 | N.D. |
| IPI00299571 | PDIA6 | 242.86 | 14 | LVWVCRPLAPVEVPANISSDFQPCSPTSPAHSLSR | 8 | 1 |
| 242.86 | 64 | SPIMYPSTTMANAPGLVSCTFFLAVNGLYSSSDDVIELTPSNFNR | 7 | 5 | ||
| IPI00007188 | SLC25A5 | 118.45 | 258 | GTDIMYTGTLDCWRK | 2 | N.D. |
| IPI00010274 | TPSAB1 | 147.89 | 60 | YWMHFCGGSLIHPQWVLTAAHCLGPDVK | 4 | N.D. |
| IPI00419942 | TPSB2 | 154.49 | 219 | IVRDDMLCAGNTR | 2 | N.D. |
| IPI00007752 | TUBB2C | 403.91 | 13 | MREIVHLQAGQCGNQIGAK | 2 | N.D. |
| IPI00013847 | UQCRC1 | 314.55 | 155 | AVELLGDIVQNCSLEDSQIEK | 4 | N.D. |
Cys local indicates the cysteine position within the protein sequence.
C represents an IAM-labeled cysteine.
Number is a detected sample case out of total 10 cases (Table 1).
N.D., not detected.
The selected proteins were classified as cytoskeleton, extracellular proteins, stress-inducible proteins (e.g., heat shock proteins), and protein disulfide isomerase (chaperone), which are oxidation-sensitive proteins whose activity is regulated through oxidation of specific cysteines (Table 2). Based on IPA analysis, their top molecular functions were classified as cardiovascular system function, organ morphology, metabolism, and small molecule biochemistry based on cellular growth/proliferation and cellular assembly/organization (Table 3). For example, two cysteines (Cys491 and Cys595) in ACTN3 (IPI00032137, alpha actinin-3) were more often observed in a reduced (IAM-binding) form in non-tumor tissues (pN; 8/10 cases, Cys491; 7/10 cases, Cys595) than CRC patients (pT; each 3/10 cases, Cys491 and Cys595) (Table 2). Other specifically oxidized cysteines observed in most of CRC samples included Cys14 (pN:pT = 8:1 out of 10 cases) and Cys64 (pN:pT = 7:5 out of 10 cases) of PDIA6 (IPI00299571, protein disulfide isomerase family A, member 6). Many proteins were more reduced (IAM-binding) in non-tumor tissues from stage II CRC patients than from non-tumor tissues from stage III patients or stage II and III tumors (data not shown, Supplementary Table S2). These results suggest that a variety of proteins related to cellular maintenance and signaling are susceptible to cysteine oxidation that alters their functionality during tumorigenesis.
Table 3.
Network functions associated with candidate proteins containing oxidation-sensitive cysteines from tumor and non-tumor tissues from CRC patients
| Moleculesa | Score | No. of molecules | Relative networks |
|---|---|---|---|
| ACTG1, ACTG2, Actin, ACTN1, Akt, ANXA2, C1orf182, CD3, CENPW, COL6A1, Cytokeratin, DGKZ, Dnajb1-Hsp70, DNAJC22, DNAJC24, DNAJC25, DNAJC27, ERK, ERK1/2, estrogen receptor, F Actin, Hsp70, HSP, HSPA6, HSPA1A/HSPA1B, HSPA1L, Jnk, KRT4, KRT10, KRT17, LMNA, MVP, MYH9, NFkB (complex), SLC25A5 | 42 | 16 | Cardiovascular system development and Function, Organ morphology, Auditory disease |
| ACTN3, C2orf47, Ca2+, CAT, CLCA1, EGFLAM, EGFR, FLG2, H2AFJ, HADHA, IGHA1, KCNG1, KCTD9, KIAA0391, MCOLN3, PDIA3, PDIA6, PLCD3, PLCH1, PLCH2, POTEE/POTEF, S100PBP, SH3BGRL3, SH3BGRL, SLC24A6, TMCO3, TMEM55A, TMEM55B, TOP1MT, TPSAB1/TPSB2, TTC7A, TUBB8, TUBB4B, UBC, UQCRC1 | 26 | 11 | Carbohydrate metabolism, Lipid metabolism, Small molecule biochemistry |
Bold indicates proteins identified in Table 1.
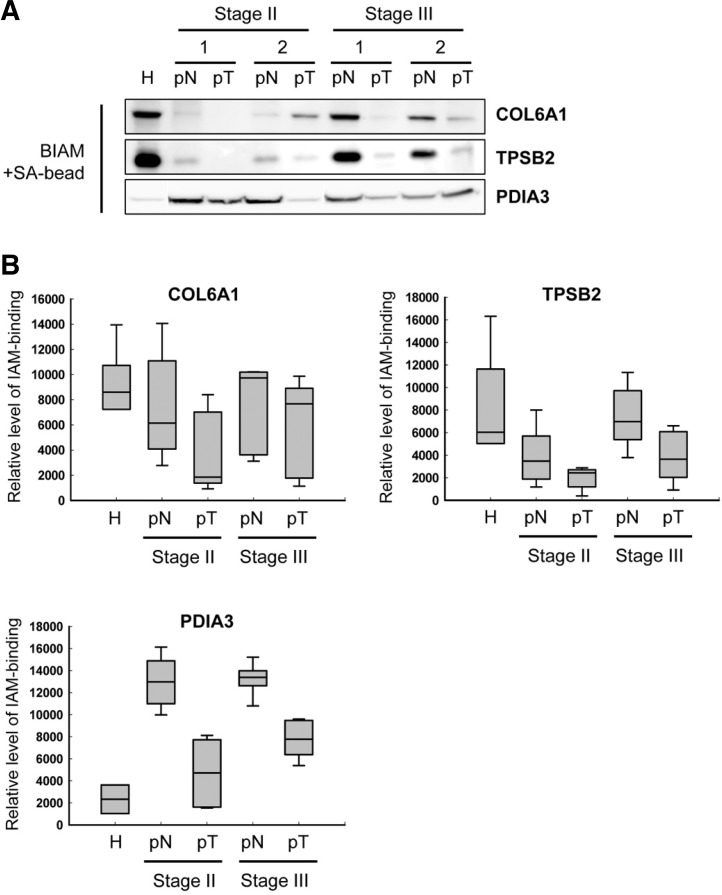
To verify the results of the proteomics analysis, we compared the results of the proteomics-based resolution with BIAM-streptavidin immunoprecipitation (IP). IP analysis showed that COL6A1 (IPI00291136, collagen, type VI, alpha 1) and TPSB2 (IPI00419942, tryptase beta 2) were more oxidized in membrane fraction of tumor; were reduced IAM binding in tumoral tissues than non-tumoral tissue of CRC patients and normal region of healthy subject (Fig. 2). And also, two proteins were highly reduced in normal condition than in tissue from CRC patients. Interestingly, COL6A1 and TPSB2 were significantly oxidized in stage II patients than stage III patients, and even in non-tumoral tissue of CRC patient. PDIA3 (IPI00025252, protein disulfide isomerase family A, member 3) was also oxidized in membrane fraction of tumor than non-tumoral tissue of CRC patients (Fig. 2). And, this protein has similar oxidative level between stage II and stage III patients. But, PDIA3 was reduced in non-tumoral tissue of CRC patients rather than healthy normal condition (H). Based on proteomic data (Table 2), these proteins may contain reduced cysteine: Cys595 (pN:pT = 4:3 out of 10 cases) and Cys612 (pN:pT = 3:1 out of 10 cases) in COL6A1, Cys219 (pN:pT = 2:N.D. out of 10 cases) in TPSB2, and Cys245 (pN:pT = 2:N.D. out of 10 cases) in PDIA3. Thus, a variety of proteins might be sensitive to cysteine oxidation, and be affected by tumorigenesis.
Fig. 2.
Detection of proteins containing IAM binding cysteine by immunoprecipitation using BIAM-streptavidin beads. (A) A representative gel image of IAM-binding level of COL6A1, TPSB2 and PDIA3. The cysteine oxidation of COL6A1, TPSB2 and PDIA3 was confirmed in the membrane fraction of normal subject, non-tumoral, and tumoral tissues (see the information in Table 2). Samples were run in triplicate. (B) Histograms of IAM-binidng level of COL6A1, TPSB2, and PDIA3. The graph is shown along with the numeric data obtained by densitometry analysis (n = 5). H, normal tissue of healthy subject; pN, non-tumor tissues of CRC patients; pT, tumor tissues of CRC patients.
Reciprocal networks among cysteine oxidation-sensitive proteins
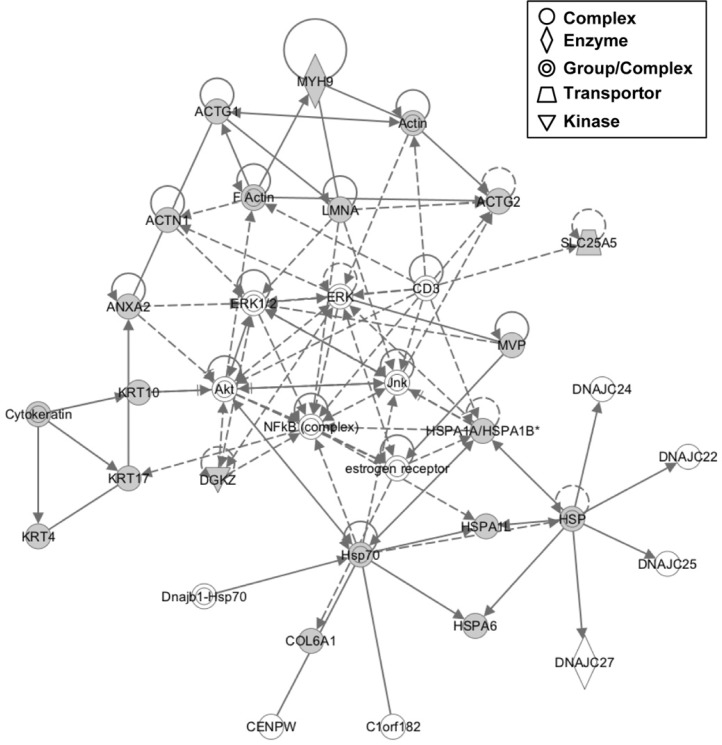
Comparative proteomic data were applied to Ingenuity network analysis to assess the functional connections among proteins containing oxidation-sensitive cysteines. A multidirectional interaction network among the identified target proteins was established for the most significantly related functions. The components of the main network were associated with various disorders (Table 3). A representative network is shown in Fig. 3. Networks among oxidation-sensitive proteins also affected various cellular maintenance pathways involving cell junctions, cytoskeletal networks and several metabolisms (Table 3). In addition, dense networks among oxidation-sensitive proteins were developed to represent significantly connected biological functions involved in the pathogenesis of various diseases/disorders (Fig. 3). These interactions exhibited associations with cancer, neurological disease, hereditary disorders and hematological disease. Next, we analyzed upstream regulator of cysteine oxidation-sensitive proteins under knowledge-based network. The analysis showed direction or indirection change of our targets activated or inhibited by upstream regulator (Table 4). Consequently, we focused endogenous chemical factors and growth factor among various chemical regulators that contain chemical reagent, drug, and toxicant (Table 4). The cellular functionality of oxidative sensitive proteins may be affected by proliferation controlling-stimulants, such as beta-estradiol and TGFB1, in colorectal cancer. Taken together, our network analysis appears to highlight the potential for proteins containing oxidation-sensitive cysteines to be significantly affected during the pathogenesis of CRC.
Fig. 3.
Reciprocal network annotation among proteins containing oxidation-sensitive cysteines determined using IPA. Candidate proteins containing oxidation-sensitive cysteines (Tables 2 and 3) are shown as gray-filled figures; solid lines, protein-protein interactions; solid arrow lines, expression/membership; dotted arrow lines, expression/activation.
Table 4.
Upstream regulator analysis of oxidation-sensitive proteins by Ingenuity system
| Molecule type | Upstream regulator | p-value of overlapa | Target molecules in dataset |
|---|---|---|---|
| Chemical-endogenous mammalian | Hydrogen peroxide | 1.72E-03 | ACTG2, ANXA2, HSPA1A/HSPA1B, PDIA3 |
| beta-Estradiol | 3.82E-03 | ACTG2, HSPA1A/HSPA1B, KRT10, KRT17, KRT4, LMNA, PDIA3 | |
| UDP-D-glucose | 8.75E-03 | PDIA3 | |
| P1, P4-Di(adenosine-5′) tetraphosphate | 1.17E-02 | TPSAB1/TPSB2 | |
| Mannose | 1.60E-02 | PDIA3 | |
| Butyric acid | 1.69E-02 | ANXA2, MVP, PDIA3 | |
| Sucrose | 2.75E-02 | PDIA3 | |
| S-adenosylmethionine | 3.31E-02 | ACTG2 | |
| Prostaglandin D2 | 3.60E-02 | HSPA1A/HSPA1B | |
| Growth factorb | TGFB1 (includes EG:21803) | 1.26E-05 | ACTG2, ACTN1, ANXA2, COL6A1, HSPA1A/HSPA1B, KRT10, KRT17, MYH9, TPSAB1/TPSB2, TUBB4B |
| INHA | 4.11E-04 | ACTG2, COL6A1, LMNA | |
| ANGPT2 | 4.36E-04 | HSPA1A/HSPA1B, PDIA3, PDIA6 | |
| INHBA | 1.41E-03 | ACTG2, ACTN1, TPSAB1/TPSB2 | |
| PDGFC | 1.31E-02 | ACTG2 | |
| MDK | 1.45E-02 | ACTG2 | |
| VEGFA | 3.60E-02 | LMNA, SLC25A5 | |
| JAG1 | 3.74E-02 | ACTG2 | |
| MSTN | 3.88E-02 | ACTG2 |
The overlap p-value is a statistically significant overlap between the dataset genes and the genes that are regulated by regulator. It is calculated using Fisher’s Exact Test, and significance is generally attributed to p-values < 0.01.
TGFB1, transforming growth factor, beta 1; INHA, alpha-inhibin; ANGPT2, angiopoietin 2; INHBA, inhibin, beta A; PDGFC, platelet-derived growth factor C; MDK, midkine (neurite growth-promoting factor 2); VEGFA, vascular endothelial growth factor A; JAG1, jagged 1; MSTN, myostatin.
DISCUSSION
With the rapid advancement of proteomic technologies, there has been an increasing number of reports assessing global changes in the redox state of thiol proteins in oxidatively stressed cells (Garcia-Santamarina et al., 2011; Leonard and Carroll, 2011; Nagahara et al., 2009). Moreover, mass spectrometry (MS) using chemical indicators sensitive to thiol oxidation has recently been employed in functional investigations of posttranslational modification, such as redox proteomics, (Charles et al., 2007; Choi et al., 2006; Fu et al., 2009; Qin et al., 2009; Yang et al., 2012b). With the advent proteomics, it has become possible to identify specific proteins that are susceptible to oxidative modification.
Although a variety of methodologies have been used to assess the redox state of proteins, they all generally entail quantifying the incorporation of labeled IAM or maleimide derivatives that alkylate cysteine residues, or monitoring the structural modifications of oxidized proteins. In the present study, we used IAM to detect endogenous free cysteinyl thiols in the membrane fraction of specimens of primary CRC tissue and paired specimens of non-tumor tissue. We then identified 31 proteins containing 37 peptides with cysteines susceptible to oxidation (Table 2). Notably, these proteins were more likely to be reduced (IAM-binding) in non-tumor than tumor tissue. The affected proteins included cytoskeletal proteins (actin, tubulin, collagen and keratin), stress-inducible proteins (HSP and PDI) and metabolism-related proteins (CLCA1, HADHA, and SLC25A5), which are all regulated largely through structural changes. For example, Cys406 in the C-terminal domain of PDIA3 affects the protein’s DNA-binding activity through a redox-dependent conformational change (Grillo et al., 2007). But, it is not yet discussed about Cys245 in PDIA3. Redox proteomics analysis recently showed actin to be a key target of oxidative stress, as the cysteines in actin are highly susceptible to oxidation (Dalle-Donne et al., 2001), which likely induces cytoskeletal rearrangements in mammalian cells (Farah et al., 2011; Milzani et al., 2000). The best-studied examples of actin oxidation are in erythrocytes from patients with sickle cell anemia, which can be considered a disease of redox imbalance (Abraham et al., 2002; Coyle and Puttfarcken, 1993). In addition, Wang et al. (2010) reported that Trx 1 (thioredoxin-1) affects the redox state of actin and its polymerization in SH-SY5Y cells. In functionally active actin (i.e., polymerized fraction), most of the functional groups in cysteine, methionine and tryptophan residues are oxidized (Fedorova et al., 2010). However, little is known about the mechanisms by which actin’s redox state is regulated. Modification of the cysteines in tubulin, including oxidation to disulfides and S-glutathionylation/nitro-sation, has been identified in several proteomics studies using cell lines and tissue samples (Brennan et al., 2004; Cumming et al., 2004; Pamplona et al., 2005). In the porcine brain, oxidation of the cysteines of microtubule proteins by hypochlorous acid and chloramines is associated with a marked reduction in microtubule polymerization (Landino et al., 2011), which may promote further oxidative damage and cellular dysfunction.
Using IPA, canonical networks of identified proteins have been studied in the context of cancer, and 18 proteins containing oxidation-sensitive cysteines have been shown to associate with tumorigenesis (p-value = 2.94−4): ACTG1, ACTG2, ACTN1, ACTN3, ANXA2, COL6A1, HSPA1A/B, KRT4, KRT10, KRT17, LMNA, MYH9, PDIA3, PDIA6, SLC25A5, TPSAB1, TPSB2 and TUBB4B. Notably, epothilone B, a modulator of human TUBA1C/TUBB2C, is in a phase I clinical trial for the treatment of colon cancer (Chen et al., 2003). And although changes in the expression of COL6A1/COL6A2 have been associated with colon cancer (Saaf et al., 2007), no studies of the posttranslational modification of these proteins are yet available. Generally, ROS deregulate the redox homeostasis and promote tumor formation by initiating an aberrant induction of signaling networks that cause tumorigenesis. In this point, we analyzed the upstream stimulants of the oxidation sensitive proteins in Table 2. Of these, the upstream regulators thought to be related to oxidative stress in CRC are hydrogen peroxide (Chang et al., 2003), beta-estradiol (Miro et al., 2011; Olivieri et al., 2002), butyric acid (Domokos et al., 2010; Giardina et al., 1999), prostaglandin D2 (Dionne et al., 2010; Kitz et al., 2011), TGFB1 (Sampson et al., 2011), PDGFC (Werth et al., 2008), VEGFA (Monaghan-Benson and Burridge, 2009), and myostatin (Mastrocola et al., 2008). However, little is currently known, and most of the relation between factors and targets under oxidative stress in CRC is unclear.
It was recently reported that anticancer agents could act, at least in part, by inducing ROS and RNS (Adachi et al., 2007; Kozoni et al., 2007; Matsubara et al., 2003; Stone et al., 2004). Among the anticancer compounds that generate ROS/RNS are nitric oxide-donating nonsteroidal anti-inflammatory drugs (NONSAIDs), which are promising chemopreventive agents (Adachi et al., 2007; Kozoni et al., 2007; Rigas, 2007). NO-donating aspirin (NO-ASA) generates a state of oxidative stress that affects redox-sensitive signaling pathways, ultimately leading to elimination of neoplastic cells via apoptosis or necrosis (Gao et al., 2005). These compounds can also exert anti-cancer effects by inducing phase II enzymes such as NAD(P)H:quinone oxireductase (NQO), glutathione S-transferases (GSTs) and UDP-glucuronyltransferases (UGTs), and by modulating the Keap1-Nrf2 pathway (Gao et al., 2006), which leads to inhibition of proliferation, induction of apoptosis and inhibition of cell cycle phase transitions. NO-NSAIDs also have significant effects a variety of important signaling molecules, including NF-κB, Wnt, MAPK (mitogen activated protein kinase) and NOS (the various nitric oxide synthases) (Kozoni et al., 2007). Other anticancer compounds, including arsenic trioxide (As2O3), phosphoaspirin (a structurally similar anticancer derivative of aspirin), phosphosulindac (a derivative of sulindac) and even No-ASA, exert significant effects on the regulatory Trx system, which leads to redox-induced cell death (Sun and Rigas, 2008). But despite much effort, the precise molecular mechanisms underlying the effects of these anticancer compounds remain unknown. We anticipate that shedding light on the cellular molecular mechanisms governing redox-mediated effects will enable better understanding of the pharmacological activities of these compounds in CRC patients. Our screening for proteins containing oxidation-sensitive cysteines and network analysis furthers to this effort.
In summary, using functional proteomics, we have identified proteins containing cysteines whose oxidation is paralleled by CRC-related changes in biological function or loss of enzyme activity. The deleterious changes caused by cysteinyl oxidation might foster further production of ROS, increase oxidative damage to cells, and ultimately exacerbate tumorigenesis and cell cycle dysregulation, though the precise biological mechanisms remain unclear. The present study highlights an initial global approach to identifying a subproteome of potential targets; a more in depth analysis of each protein may yield specific information regarding the redox state and reactivity of individual cysteines and proteins.
Acknowledgments
The authors declare that they have no competing interests. This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government [The Ministry of Education, Science and Technology (MEST)] (No. 2012-0009425), and by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare (A111455), Republic of Korea.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Abraham A, Bencsath FA, Shartava A, Kakhniashvili DG, Goodman SR. (2002). Preparation of irreversibly sickled cell beta-actin from normal red blood cell beta-actin. Biochemistry 41, 292–296 [DOI] [PubMed] [Google Scholar]
- Acharya A, Das I, Chandhok D, Saha T. (2010). Redox regulation in cancer: a double-edged sword with therapeutic potential. Oxid Med Cell Longev. 3, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi M, Sakamoto H, Kawamura R, Wang W, Imai K, Shinomura Y. (2007). Nonsteroidal anti-inflammatory drugs and oxidative stress in cancer cells. Histol Histopathol. 22, 437–442 [DOI] [PubMed] [Google Scholar]
- Behrend L, Henderson G, Zwacka RM. (2003). Reactive oxygen species in oncogenic transformation. Biochem Soc Trans. 31, 1441–1444 [DOI] [PubMed] [Google Scholar]
- Brennan JP, Wait R, Begum S, Bell JR, Dunn MJ, Eaton P. (2004). Detection and mapping of widespread intermolecular protein disulfide formation during cardiac oxidative stress using proteomics with diagonal electrophoresis. J Biol Chem. 279, 41352–41360 [DOI] [PubMed] [Google Scholar]
- Bulaj G, Kortemme T, Goldenberg DP. (1998). Ionization-reactivity relationships for cysteine thiols in polypeptides. Biochemistry. 37, 8965–8972 [DOI] [PubMed] [Google Scholar]
- Burdon RH. (1995). Superoxide and hydrogen peroxide in relation to mammalian cell proliferation. Free Radic Biol Med. 18, 775–794 [DOI] [PubMed] [Google Scholar]
- Burdon RH, Gill V, Alliangana D. (1996). Hydrogen peroxide in relation to proliferation and apoptosis in BHK-21 hamster fibroblasts. Free Radic Res. 24, 81–93 [DOI] [PubMed] [Google Scholar]
- Chang DK, Goel A, Ricciardiello L, Lee DH, Chang CL, Carethers JM, Boland CR. (2003). Effect of H(2)O(2) on cell cycle and survival in DNA mismatch repair-deficient and - proficient cell lines. Cancer Lett. 195, 243–251 [DOI] [PubMed] [Google Scholar]
- Charles RL, Schroder E, May G, Free P, Gaffney PR, Wait R, Begum S, Heads RJ, Eaton P. (2007). Protein sulfenation as a redox sensor: proteomics studies using a novel biotinylated dimedone analogue. Mol Cell Proteomics 6, 1473–1484 [DOI] [PubMed] [Google Scholar]
- Chen JG, Yang CP, Cammer M, Horwitz SB. (2003). Gene expression and mitotic exit induced by microtubule-stabilizing drugs. Cancer Res. 63, 7891–7899 [PubMed] [Google Scholar]
- Choi H, Kim S, Mukhopadhyay P, Cho S, Woo J, Storz G, Ryu SE. (2001). Structural basis of the redox switch in the OxyR transcription factor. Cell. 105, 103–113 [DOI] [PubMed] [Google Scholar]
- Choi KS, Park SY, Baek SH, Dey-Rao R, Park YM, Zhang H, Ip C, Park EM, Kim YH, Park JH. (2006). Analysis of protein redox modification by hypoxia. Prep Biochem Biotechnol. 36, 65–79 [DOI] [PubMed] [Google Scholar]
- Coyle JT, Puttfarcken P. (1993). Oxidative stress, glutamate, and neurodegenerative disorders. Science 262, 689–695 [DOI] [PubMed] [Google Scholar]
- Cumming RC, Andon NL, Haynes PA, Park M, Fischer WH, Schubert D. (2004). Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem. 279, 21749–21758 [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, Rossi R, Milzani A, Di Simplicio P, Colombo R. (2001). The actin cytoskeleton response to oxidants: from small heat shock protein phosphorylation to changes in the redox state of actin itself. Free Radic Biol Med. 31, 1624–1632 [DOI] [PubMed] [Google Scholar]
- Dionne S, Levy E, Levesque D, Seidman EG. (2010). PPARgamma ligand 15-deoxy-delta 12, 14-prostaglandin J2 sensitizes human colon carcinoma cells to TWEAK-induced apoptosis. Anticancer Res. 30, 157–166 [PubMed] [Google Scholar]
- Domokos M, Jakus J, Szeker K, Csizinszky R, Csiko G, Neogrady Z, Csordas A, Galfi P. (2010). Butyrate-induced cell death and differentiation are associated with distinct patterns of ROS in HT29-derived human colon cancer cells. Dig Dis Sci. 55, 920–930 [DOI] [PubMed] [Google Scholar]
- Farah ME, Sirotkin V, Haarer B, Kakhniashvili D, Amberg DC. (2011). Diverse protective roles of the actin cytoskeleton during oxidative stress. Cytoskeleton (Hoboken). 68, 340–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova M, Kuleva N, Hoffmann R. (2010). Identification of cysteine, methionine and tryptophan residues of actin oxidized in vivo during oxidative stress. J Proteome Res. 9, 1598–1609 [DOI] [PubMed] [Google Scholar]
- Finkel T. (2003). Oxidant signals and oxidative stress. Curr Opin Cell Biol. 15, 247–254 [DOI] [PubMed] [Google Scholar]
- Fu C, Wu C, Liu T, Ago T, Zhai P, Sadoshima J, Li H. (2009). Elucidation of thioredoxin target protein networks in mouse. Mol Cell Proteomics 8, 1674–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Liu X, Rigas B. (2005). Nitric oxide-donating aspirin induces apoptosis in human colon cancer cells through induction of oxidative stress. Proc Natl Acad Sci USA 102, 17207–17212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Kashfi K, Liu X, Rigas B. (2006). NO-donating aspirin induces phase II enzymes in vitro and in vivo. Carcinogenesis 27, 803–810 [DOI] [PubMed] [Google Scholar]
- Garcia-Santamarina S, Boronat S, Espadas G, Ayte J, Molina H, Hidalgo E. (2011). The oxidized thiol proteome in fission yeast--optimization of an ICAT-based method to identify H2O2-oxidized proteins. J Proteomics 74, 2476–2486 [DOI] [PubMed] [Google Scholar]
- Giardina C, Boulares H, Inan MS. (1999). NSAIDs and butyrate sensitize a human colorectal cancer cell line to TNF-alpha and Fas ligation: the role of reactive oxygen species. Biochim Biophys Acta 1448, 425–438 [DOI] [PubMed] [Google Scholar]
- Grillo C, D’Ambrosio C, Consalvi V, Chiaraluce R, Scaloni A, Maceroni M, Eufemi M, Altieri F. (2007). DNA-binding activity of the ERp57 C-terminal domain is related to a redox-dependent conformational change. J Biol Chem. 282, 10299–10310 [DOI] [PubMed] [Google Scholar]
- Janssen-Heininger YM, Mossman BT, Heintz NH, Forman HJ, Kalyanaraman B, Finkel T, Stamler JS, Rhee SG, van der Vliet A. (2008). Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic Biol Med. 45, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitz K, Windischhofer W, Leis HJ, Huber E, Kollroser M, Malle E. (2011). 15-Deoxy-Delta12, 14-prostaglandin J2 induces Cox-2 expression in human osteosarcoma cells through MAPK and EGFR activation involving reactive oxygen species. Free Radic Biol Med. 50, 854–865 [DOI] [PubMed] [Google Scholar]
- Kozoni V, Rosenberg T, Rigas B. (2007). Development of novel agents based on nitric oxide for the control of colon cancer. Acta Pharmacol Sin. 28, 1429–1433 [DOI] [PubMed] [Google Scholar]
- Landino LM, Hagedorn TD, Kim SB, Hogan KM. (2011). Inhibition of tubulin polymerization by hypochlorous acid and chloramines. Free Radic Biol Med. 50, 1000–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Lee HJ. (2006). Biphasic effects of dietary anti-oxidants on oxidative stress-mediated carcinogenesis. Mech Ageing Dev. 127, 424–431 [DOI] [PubMed] [Google Scholar]
- Leonard SE, Carroll KS. (2011). Chemical ‘omics’ approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol. 15, 88–102 [DOI] [PubMed] [Google Scholar]
- Mastrocola R, Reffo P, Penna F, Tomasinelli CE, Boccuzzi G, Baccino FM, Aragno M, Costelli P. (2008). Muscle wasting in diabetic and in tumor-bearing rats: role of oxidative stress. Free Radic Biol Med. 44, 584–593 [DOI] [PubMed] [Google Scholar]
- Mates JM, Sanchez-Jimenez FM. (2000). Role of reactive oxygen species in apoptosis: implications for cancer therapy. Int J Biochem Cell Biol. 32, 157–170 [DOI] [PubMed] [Google Scholar]
- Matsubara K, Komatsu S, Oka T, Kato N. (2003). Vitamin B6-mediated suppression of colon tumorigenesis, cell proliferation, and angiogenesis (review). J Nutr Biochem. 14, 246–250 [DOI] [PubMed] [Google Scholar]
- Milzani A, Rossi R, Di Simplicio P, Giustarini D, Colombo R, Dalle-Donne I. (2000). The oxidation produced by hydrogen peroxide on Ca-ATP-G-actin. Protein Sci. 9, 1774–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miro AM, Sastre-Serra J, Pons DG, Valle A, Roca P, Oliver J. (2011). 17beta-Estradiol regulates oxidative stress in prostate cancer cell lines according to ERalpha/ERbeta ratio. J Steroid Biochem Mol Biol. 123, 133–139 [DOI] [PubMed] [Google Scholar]
- Monaghan-Benson E, Burridge K. (2009). The regulation of vascular endothelial growth factor-induced microvascular permeability requires Rac and reactive oxygen species. J Biol Chem. 284, 25602–25611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahara N, Matsumura T, Okamoto R, Kajihara Y. (2009). Protein cysteine modifications: (1) medical chemistry for proteomics. Curr Med Chem. 16, 4419–4444 [DOI] [PubMed] [Google Scholar]
- Olivieri G, Novakovic M, Savaskan E, Meier F, Baysang G, Brockhaus M, Muller-Spahn F. (2002). The effects of beta-estradiol on SHSY5Y neuroblastoma cells during heavy metal induced oxidative stress, neurotoxicity and beta-amyloid secretion. Neuroscience 113, 849–855 [DOI] [PubMed] [Google Scholar]
- Pamplona R, Dalfo E, Ayala V, Bellmunt MJ, Prat J, Ferrer I, Portero-Otin M. (2005). Proteins in human brain cortex are modified by oxidation, glycoxidation, and lipoxidation. Effects of Alzheimer disease and identification of lipoxidation targets. J Biol Chem. 280, 21522–21530 [DOI] [PubMed] [Google Scholar]
- Parkin DM. (2006). The global health burden of infection-associated cancers in the year 2002. Int J Cancer 118, 3030–3044 [DOI] [PubMed] [Google Scholar]
- Pelicano H, Carney D, Huang P. (2004). ROS stress in cancer cells and therapeutic implications. Drug Resist Updat. 7, 97–110 [DOI] [PubMed] [Google Scholar]
- Phalen TJ, Weirather K, Deming PB, Anathy V, Howe AK, van der Vliet A, Jonsson TJ, Poole LB, Heintz NH. (2006). Oxidation state governs structural transitions in peroxiredoxin II that correlate with cell cycle arrest and recovery. J Cell Biol. 175, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole LB, Nelson KJ. (2008). Discovering mechanisms of signaling-mediated cysteine oxidation. Curr Opin Chem Biol. 12, 18–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Meng X, Wang Q, Tian S. (2009). Oxidative damage of mitochondrial proteins contributes to fruit senescence: a redox proteomics analysis. J Proteome Res. 8, 2449–2462 [DOI] [PubMed] [Google Scholar]
- Rigas B. (2007). The use of nitric oxide-donating nonsteroidal anti-inflammatory drugs in the chemoprevention of colorectal neoplasia. Curr Opin Gastroenterol. 23, 55–59 [DOI] [PubMed] [Google Scholar]
- Roessner A, Kuester D, Malfertheiner P, Schneider-Stock R. (2008). Oxidative stress in ulcerative colitis-associated carcinogenesis. Pathol Res Pract. 204, 511–524 [DOI] [PubMed] [Google Scholar]
- Saaf AM, Halbleib JM, Chen X, Yuen ST, Leung SY, Nelson WJ, Brown PO. (2007). Parallels between global transcriptional programs of polarizing Caco-2 intestinal epithelial cells in vitro and gene expression programs in normal colon and colon cancer. Mol Biol Cell 18, 4245–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson N, Koziel R, Zenzmaier C, Bubendorf L, Plas E, Jansen-Durr P, Berger P. (2011). ROS signaling by NOX4 drives fibroblast-to-myofibroblast differentiation in the diseased prostatic stroma. Mol Endocrinol. 25, 503–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone WL, Krishnan K, Campbell SE, Qui M, Whaley SG, Yang H. (2004). Tocopherols and the treatment of colon cancer. Ann N Y Acad Sci. 1031, 223–233 [DOI] [PubMed] [Google Scholar]
- Sun Y, Rigas B. (2008). The thioredoxin system mediates redox-induced cell death in human colon cancer cells: implications for the mechanism of action of anticancer agents. Cancer Res. 68, 8269–8277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal EA, Day AM, Morgan BA. (2007). Hydrogen peroxide sensing and signaling. Mol. Cell 26, 1–14 [DOI] [PubMed] [Google Scholar]
- Wang X, Ling S, Zhao D, Sun Q, Li Q, Wu F, Nie J, Qu L, Wang B, Shen X, et al. (2010). Redox regulation of actin by thioredoxin-1 is mediated by the interaction of the proteins via cysteine 62. Antioxid Redox Signal. 13, 565–573 [DOI] [PubMed] [Google Scholar]
- Weitz J, Koch M, Debus J, Hohler T, Galle PR, Buchler MW. (2005). Colorectal cancer. Lancet 365, 153–165. [DOI] [PubMed] [Google Scholar]
- Werth C, Stuhlmann D, Cat B, Steinbrenner H, Alili L, Sies H, Brenneisen P. (2008). Stromal resistance of fibroblasts against oxidative damage: involvement of tumor cell-secreted platelet-derived growth factor (PDGF) and phosphoinositide 3-kinase (PI3K) activation. Carcinogenesis 29, 404–410 [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Hampton MB. (2008). Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 45, 549–561 [DOI] [PubMed] [Google Scholar]
- Xu D, Suenaga N, Edelmann MJ, Fridman R, Muschel RJ, Kessler BM. (2008). Novel MMP-9 substrates in cancer cells revealed by a label-free quantitative proteomics approach. Mol Cell Proteomics 7, 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HY, Kwon J, Park HR, Kwon SO, Park YK, Kim HS, Chung YJ, Chang YJ, Choi HI, Chung KJ, et al. (2012a). Comparative proteomic analysis for the insoluble fractions of colorectal cancer patients. J Proteomics 75, 3639–3653 [DOI] [PubMed] [Google Scholar]
- Yang HY, Kwon J, Choi HI, Park SH, Yang U, Park HR, Ren L, Chung KJ, Kim YU, Park BJ, et al. (2012b). In-depth analysis of cysteine oxidation by the RBC proteome: advantage of peroxiredoxin II knockout mice. Proteomics 12, 101–112 [DOI] [PubMed] [Google Scholar]
- Ziech D, Franco R, Pappa A, Panayiotidis MI. (2011). Reactive oxygen species (ROS)--induced genetic and epigenetic alterations in human carcinogenesis. Mutat Res. 711, 167–173 [DOI] [PubMed] [Google Scholar]