Abstract
The mammalian target of rapamycin (mTOR) is an evolutionally conserved kinase which exists in two distinct structural and functional complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). Of the two complexes, mTORC1 couples nutrient abundance to cell growth and proliferation by sensing and integrating a variety of inputs arising from amino acids, cellular stresses, energy status, and growth factors. Defects in mTORC1 regulation are implicated in the development of many metabolic diseases, including cancer and diabetes. Over the past decade, significant advances have been made in deciphering the complexity of the signaling processes contributing to mTORC1 regulation and function, but the mechanistic details are still not fully understood. In particular, how amino acid availability is sensed by cells and signals to mTORC1 remains unclear. In this review, we discuss the current understanding of nutrient-dependent control of mTORC1 signaling and will focus on the key components involved in amino acid signaling to mTORC1.
Keywords: amino acids, glucose, glutamine, mTORC1, nutrient
INTRODUCTION
An essential ability of both single cells and mutlicellular organisms is to sense nutrient fluctuations in the environment and to adjust their consumption of nutrients accordingly. This ability enables cells to survive during periods of nutrient deficiency, and to grow and proliferate when nutrients are plentiful. Adaptation of cells in response to changes in nutrient availability is achieved by carefully coordinating energy-consuming anabolic processes with energy-producing catabolic processes. At the center of this balancing act is the mTOR signaling pathway, which was originally named the mammalian target of rapamycin, but has been officially renamed the mechanistic target of rapamycin (mTOR) signaling pathway. In nutrient-rich conditions, mTOR is activated and drives cell growth by stimulating a series of anabolic processes that include protein, lipid, and nucleotide synthesis, and by inhibiting degradative catabolic processes such as autophagy. Conversely, nutrient-deficient conditions trigger the rapid inhibition of mTOR to limit its stimulatory functions on anabolism. Instead, catabolic processes are activated to produce sufficient energy and nutrients to maintain minimal biological processes required for survival (Howell and Manning, 2011; Jewell and Guan, 2013; Wullschleger et al., 2006). Accordingly, any mutations in the components of the mTOR pathway that coordinate these responses can lead to metabolic or inflammatory disorders, and often promote tumorigenesis (Yecies and Manning, 2011; Zoncu et al., 2011b). Not surprisingly, disregulation of mTOR signaling has been observed in many human diseases, including cancer and diabetes, making mTOR an attractive therapeutic target for numerous clinical applications (Cornu et al., 2013; Howell and Manning, 2011; Inoki et al., 2012; Laplante and Sabatini, 2012). In this regard, it is important to thoroughly understand the molecular mechanisms underlying how nutrients regulate the mTOR pathway, and subsequently how the cell balances its growth and survival in accordance with the cell’s nutrient state. In this review, we first summarize the basics of mTOR signaling, including the regulation of mTOR by growth factors. We then discuss the current understanding of the molecular mechanisms by which mTOR is regulated by nutrients such as glucose and amino acids.
Organization and functions of the mTOR complexes
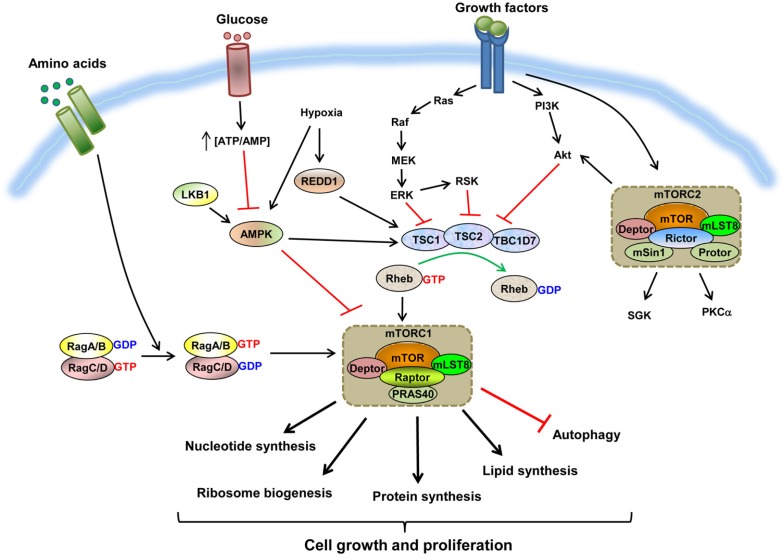
mTOR is an atypical serine/threonine protein kinase that belongs to the superfamily of phosphatidylinositide-3 kinase related-kinases (PI3KK). mTOR exists as two distinct multiprotein complexes termed mTOR complex 1 (mTORC1) and 2 (mTORC2), which differ in their components, regulation, functions, and sensitivity to the compound rapamycin (Sengupta et al., 2010). The two mTOR complexes are evolutionally well conserved from yeast to humans (Wullschleger et al., 2006). mTORC1, which is acutely and directly inhibited by the allosteric inhibitor rapamycin, contains mTOR, regulatory associated protein of mTOR (Raptor), mammalian lethal with SEC13 protein 8 (mLST8; also known as GβL), Akt/PKB substrate 40 kDa (PRAS40), and DEP domain containing mTOR-interacting protein (Deptor) (Fig. 1). Raptor, the defining component of mTORC1, recruits substrates for phosphorylation, and is essential for all of mTORC1’s functions (Hara et al., 2002; Kim et al., 2002). PRAS40 and Deptor appear to be both suppressors and substrates of mTORC1 (Oshiro et al., 2007; Peterson et al., 2009; Sancak et al., 2007; Vander Haar et al., 2007; Wang et al., 2008), and mLST8 appears to be dispensable for mTORC1 activity (Guertin et al., 2006; Kim et al., 2003). While active, mTORC1 plays a key role in cell growth by promoting protein synthesis, ribosomal biogenesis, lipid and nucleotide synthesis, and by inhibiting autophagy (Fig. 1) (Ben-Sahra et al., 2013; Laplante and Sabatini, 2012; Ma and Blenis, 2009; Robitaille et al., 2013). The two best characterized mTORC1 substrates, ribosomal S6 kinase (S6K) and eukaryotic translation initiation factor 4E binding protein (4E-BP), mediate many of these mTORC1-regulated processes (Laplante and Sabatini, 2012; Ma and Blenis, 2009).
Fig. 1.
The mTOR signaling pathway. The key signaling pathways that regulate mTORC1 and mTORC2 and the composition of each mTOR complex are depicted. Multiple inputs from growth factors, amino acids, cellular energy status, and stress are integrated into mTORC1 through the mechanisms shown. When mTORC1 is active, it plays a major role in promoting cell growth and proliferation by stimulating various anabolic processes such as protein, lipid, and nucleotide synthesis and ribosome biogenesis, and by inhibiting catabolic processes such as autophagy. mTORC2 is regulated by growth factors through a poorly identified mechanism, but unlike mTORC1, it does not respond to other upstream signals derived from nutrients or stress.
In contrast with mTORC1, mTORC2 consists of mTOR, Raptor-independent companion of mTOR (Rictor), mSin1 (MAPKAP1), Protor (PRR5), mLST8, and Deptor, and controls actin polymerization and the activation of kinases such as Akt, SGK, and PKCα by phosphorylating their hydrophobic motif (Fig. 1) (Oh and Jacinto, 2011). Similar to Raptor, Rictor appears to recruit substrates to mTOR for phosphorylation, thus explaining the differential selection of substrates by mTORC1 and mTORC2 (Jacinto et al., 2004; Sarbassov et al., 2004). Meanwhile, mSin1 and mLST8 are necessary for mTORC2’s complex integrity and its catalytic function, respectively (Frias et al., 2006; Guertin et al., 2006; Jacinto et al., 2006; Yang et al., 2006). Deptor also suppresses mTORC2 signaling (Peterson et al., 2009). The functional role of Protor is currently unclear (Pearce et al., 2007; Woo et al., 2007). Unlike mTORC1, mTORC2 is not sensitive to rapamycin, although prolonged treatment with rapamycin can indirectly inhibit mTORC2 by preventing mTORC2 assembly in some but not all cell types (Sarbassov et al., 2006; Zeng et al., 2007).
Growth factor signaling to mTORC1 via the TSC-Rheb axis
While our understanding of the mechanism of mTORC2 regulation is very limited, studies on the mechanism of mTORC1 regulation have provided a much clearer albeit a more complicated picture. mTORC1 is regulated by a variety of upstream signals including growth factors and nutrients such as amino acids and glucose. Among the multiple upstream signals, growth factor-regulation of mTORC1 has been extensively characterized (Fig. 1). Growth factors bind to specific receptor tyrosine kinases, which lead to activation of the PI3K-Akt and/or Ras-MAPK pathways (Mendoza et al., 2011). These pathways activate mTORC1 primarily by phosphorylating and inhibiting tuberous sclerosis complex 2 (TSC2; also known as tuberin) (Ballif et al., 2005; Inoki et al., 2002; Manning et al., 2002; Roux et al., 2004; Tee et al., 2003a), which in complex with TSC1 (also known as hamartin) and TBC (Tre2-Bub2-Cdc16)1 domain family member 7 (TBC1D7), functions as a GTPase activating protein (GAP) for the small GTPase Rheb (Dibble et al., 2012; Inoki et al., 2003a; Tee et al., 2003b). Rheb is active in its GTP-bound form like other small GTPases and indispensable for mTORC1 activation in response to all stimuli. The TSC complex stimulates the intrinsic GTPase activity of Rheb, thereby increasing the rate at which Rheb transitions from its active GTP-bound state to its inactive GDP-bound state. As a result, growth factors drive mTORC1 activation by increasing the Rheb-GTP to Rheb-GDP ratio through inhibition of TSC2 GAP activity (Fig. 1). Despite the necessity of Rheb in mTORC1 activation, the mechanistic details of how Rheb-GTP activates mTORC1 remain controversial, although the strongest evidence supports activation through direct interaction (Long et al., 2005a; Sancak et al., 2007). Likewise, the molecular consequences of TSC2 phosphorylation still remain somewhat unclear due to lack of evidence that the in vitro GAP activity of TSC2 is indeed inhibited by phosphorylation. It is more likely that phosphorylation of TSC2 may induce its dissociation from TSC1 and/or lead to its mislocalization from the endomembranes where Rheb exists, thereby increasing Rheb-GTP and activating mTORC1.
Glucose and energy regulation of mTORC1 via AMPK-dependent pathways
Most anabolic processes require energy in the form of ATP to carry out enzymatic reactions to generate sufficient lipids, nucleotides, and proteins for cell growth and division. mTORC1 senses the energetic status of the cell to modulate such energy-consuming anabolic processes, and is thus inhibited under energetic stress conditions to ensure cell survival.
In most actively dividing cells, glucose is the primary energy source (DeBerardinis et al., 2008). Glucose is initially broken down through glycolysis, which is a series of enzymatic reactions that produces two 3-carbon molecules of pyruvate per glucose molecule. Pyruvate can either be secreted as lactic acid or enter into the tricarboxylic acid (TCA) cycle for mitochondrial respiration. The overall glycolytic reaction produces 2 net molecules of ATP while aerobic glycolysis can generate 36 molecules of ATP per molecule of glucose. Accordingly, deprivation of glucose or inhibition of glycolysis or mitochondrial respiration causes a rapid reduction in intracellular ATP levels, leading to an increase in both the ADP/ATP and AMP/ATP ratios. A serine/threonine kinase called AMP-dependent protein kinase (AMPK) directly senses increases in both ratios, particularly the AMP/ATP ratio, and is a crucial cellular energy sensor that is found in nearly all eukaryotes (Hardie et al., 2012; Shackelford and Shaw, 2009). AMPK is a heterotrimeric complex consisting of one catalytic (α) and two regulatory (β and γ) subunits, and in addition to being activated by binding of ADP or AMP to the γ subunit, phosphorylation at threonine 172 in the activation loop by the serine/threonine kinase LKB1 is required for activation (Lizcano et al., 2004; Shaw et al., 2004; Woods et al., 2003).
Dennis et al. (2001) originally proposed that mTOR directly senses intracellular ATP levels due to its high Km for ATP, however, a more recent report suggests that unlike non-complexed mTOR, the Km of mTORC1 for ATP is in the low micromolar range, too low to act directly as an ATP sensor (Tao et al., 2010). Indeed, recent studies from several groups have suggested that mTORC1 indirectly senses low energy through multiple mechanisms that are mainly convergent on AMPK (Fig. 1). Under nutrient-deprived conditions resulting in energy depletion, AMPK is activated and transmits energetic stress signals to mTORC1 through direct phosphorylation of TSC2 and Raptor. AMPK inhibits mTORC1 by phosphorylating TSC2 at serine 1345, likely leading to the activation of its GAP activity and thus inactivation of Rheb (Inoki et al., 2003b). In addition to the inhibition through TSC2, AMPK prevents mTORC1 activation in a TSC-independent manner by directly phosphorylating Raptor, which leads to the association of Raptor with 14-3-3 (Gwinn et al., 2008). Other stress conditions such as hypoxia also cause energetic stress and are thus capable of inhibiting mTORC1 activation in part through the AMPK–TSC2 pathway. Concomitantly, hypoxia can also induce the expression of the regulated in development and DNA damage responses 1 (REDD1) gene, which suppresses mTORC1 activation through a TSC-dependent mechanism involving release of TSC2 from its growth factor-induced association with inhibitory 14-3-3 proteins (Fig. 1) (Brugarolas et al., 2004; DeYoung et al., 2008; Reiling and Hafen, 2004; Sofer et al., 2005). Based on its role in sensing and signaling energetic stress, AMPK appears to function as a key integrator of inputs from cellular energetic stresses to inhibit mTORC1 in both TSC-dependent and -independent manners. Interestingly, however, recent observations have demonstrated that inhibition of mitochondrial respiration is able to suppress mTORC1 activity even in AMPKα1/α2−/− cells (Kalender et al., 2010; Kim et al., 2013), revealing an alternative mechanism of mTORC1 pathway inhibition independent of AMPK under energy-deprived conditions, which will be discussed later.
Regulation of mTORC1 by amino acids
As mentioned previously, in order to balance anabolic and catabolic processes, single cells and multicellular organisms need to tightly coordinate their usage of energy as well as amino acids. Since the initial observation that linked amino acid levels to TOR activity was demonstrated in the budding yeast by Hall and colleagues (Barbet et al., 1996), the linkage between TOR and amino acids has been firmly established by observations in Drosophila, where deletion of dTOR led to growth arrest and features of nutrient starvation, and in mammals, where depletion of amino acids in CHO-IR cells led to rapid inhibition of mTORC1 (Hara et al., 1998; Oldham et al., 2000; Zhang et al., 2000). However, unlike other regulatory signaling to mTORC1, the mechanism of amino acids sensing and signaling to mTORC1 has remained a mystery. What is known is that leucine and arginine are the two most potent amino acids implicated in mTORC1 activation, as deprivation of either amino acid most closely phenocopies total amino acid deprivation in inhibiting mTORC1 (Hara et al., 1998). In addition, amino acids seem to regulate mTORC1 through a TSC-Rheb-independent pathway, as growth factors do not activate mTORC1 efficiently in amino acid-deprived conditions and total amino acid deprivation can suppress mTORC1 signaling even in Tsc2−/− cells where mTORC1 is hyperactivated (Smith et al., 2005). Very recently, our understanding of amino acid signaling mechanisms that control mTORC1 signaling has improved due to a number of discoveries of essential components of the amino acid pathways to mTORC1.
The role of the TSC complex and Rheb in amino acid signaling to mTORC1
Given that various signals such as growth factors and energy levels converge on the TSC complex to regulate its GAP activity toward Rheb and thus mTORC1 signaling, and that amino acid regulation of mTORC1 may be largely independent of the TSC complex, several groups have examined the role of amino acids in controlling mTORC1 signaling through a TSC-Rheb independent pathway by utilizing Tsc1−/− or Tsc2−/− cells. The conclusion from these studies is that leucine deprivation, which inhibits mTORC1 in Tsc+/+ cells, fails to inhibit mTORC1 in Tsc−/− cells. However, deprivation of total amino acids does repress mTORC1 activity in Tsc−/− cells, although the repression is not as significant as in Tsc+/+ cells (Gao et al., 2002; Smith et al., 2005). This has led to various proposals including possible dual amino acid inputs into mTORC1 through both TSC-dependent and -independent mechanisms. Direct measurements of the Rheb-GTP levels in response to changes in amino acid levels have led to rather confounding results. Amino acids were not seen to have an effect on the GTP levels of overexpressed Rheb, but amino acid deprivation and subsequent stimulation did affect the GTP levels of endogenous Rheb (Long et al., 2005b; Nobukuni et al., 2005; Roccio et al., 2006). Whether the lack of effect on the overexpressed Rheb is an artifact resulting from too much expression remains to be determined, as overexpressed proteins beyond the cell’s normal control capacity often escape endogenous regulation. Conversely, the change in the GTP levels of endogenous Rheb was reported only once and hence remains to be validated. Therefore, it has yet to be clarified if amino acid signaling to mTORC1 involves regulation of Rheb or not, and, if it does, if this regulation of Rheb is TSC-dependent or -independent. However, with the observation that amino acid deprivation in Tsc2−/− cells does not decrease endogenous Rheb-GTP significantly but can inhibit mTORC1 (Nobukuni et al., 2005; Roccio et al., 2006), it is more likely that amino acids are able to transduce signals to mTORC1 at least through a pathway in parallel with the TSC-Rheb pathway.
Rag GTPases are essential mediators of amino acid signaling to mTORC1
The molecular mechanisms of how amino acids are sensed and signal to mTORC1 is a very active area of investigation. A major breakthrough in the elucidation of this signaling cascade came from the discovery by the laboratories of Kun-Liang Guan, using an RNAi strategy in drosophila S2 cells, and David Sabatini, using a biochemical approach, that the Rag GTPases were crucial mediators of amino acid signaling to mTORC1 (Kim et al., 2008; Sancak et al., 2008). These discoveries set the stage for several important reports detailing how amino acids contribute to mTORC1 activation in animal cells (Efeyan et al., 2012).
The Rag family of GTPases contains four members, consisting of RagA, RagB, RagC, and RagD, which are evolutionally well conserved throughout all eukaryotes (Hirose et al., 1998; Schurmann et al., 1995; Sekiguchi et al., 2001). RagA and RagB are very similar in amino acid sequence, are functionally redundant, and are homologous to yeast Gtr1p, whereas RagC and RagD are homologous to yeast Gtr2p and are also similar and functionally redundant. In yeast, Gtr1p forms a heterodimer with Gtr2p (Nakashima et al., 1999). Similarly, RagA or RagB forms a stable heterodimer with RagC or RagD, leading to four possible dimer combinations (Sekiguchi et al., 2001). The recently solved crystal structure of the yeast Gtr1p-Gtr2p heterodimer suggests that the unusually long C-terminal domain of each Rag protein is crucial for the stable dimeric formation, while the N-terminal domain on each Rag protein is responsible for the GTPase function (Gong et al., 2011; Jeong et al., 2012).
The two reports from the Sabatini and Guan groups (Kim et al., 2008; Sancak et al., 2008) showed that expression of the dominant-negative mutant form of RagA/B (fixed in a guanine nucleotide-free form) almost completely inhibited mTORC1 even in amino acid-sufficient conditions, while expression of the dominant-active mutant of RagA/B (fixed in the GTP-bound form) was sufficient to maintain mTORC1 activity even in amino acid-deprived conditions. In contrast, expression of any mutants of RagC/D did not affect mTORC1 activity significantly. These findings indicate that the nucleotide-bound state of RagA/B, rather than RagC/D, is the primary determinant for mTORC1 activation, and also imply that amino acids regulate the nucleotide-bound state of RagA/B (Fig. 1). Consistent with this, Sabatini and colleagues showed that amino acid stimulation indeed increased the GTP charging of endogenous RagB (Sancak et al., 2008). Although RagC and RagD were also essential for mTORC1 activation, they may contribute to mTORC1 activation by increasing the stability of RagA/B through the formation of dimers, as the effects of expression of the RagA/B mutants on mTORC1 were enhanced when RagC/D was coexpresssed. Interestingly, when a dominant-active RagA/B and a dominant-negative RagC/D (theoretically, RagA/B-GTP·RagC/D-GDP) were coexpresssed, in comparison to other combinations, mTORC1 was maximally activated in amino acid-deprived conditions, while coexpression of a dominant-negative RagA/B and a dominant-active RagC/D-GTP (RagA/B-GDP RagC/D-GTP) strongly suppressed mTORC1 in amino acid-sufficient conditions. These results suggest that amino acids may regulate the nucleotide state of RagC/D as well as RagA/B, and indeed molecular mechanisms for each regulation has recently been proposed (Bar-Peled et al., 2012; Han et al., 2012).
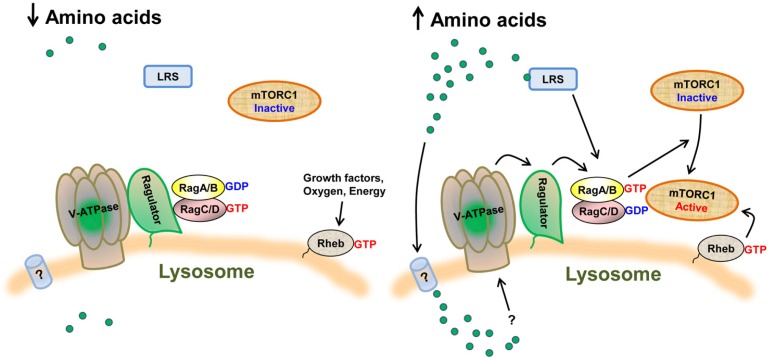
Unlike Rheb, Rag GTPases are unable to directly activate mTORC1 kinase activity (Sancak et al., 2008), which led Sabatini and colleagues to test the role of the Rags in the spatial regulation of mTORC1. Indeed, amino acids and Rag GTPases were found to control mTORC1 signaling by regulating its intracellular localization; under amino acid deficiency or depletion of the Rags, mTORC1 was localized diffusely throughout the cytoplasm, while under amino acid sufficiency or expression of the dominant-active RagA or RagB, the activated Rag complex bound directly to Raptor and recruited mTORC1 to the lysosome where Rheb appears to be localized. In line with these data, they established a model in which amino acids stimulate the GTP charging of the RagA/B, which transmit amino acid signals to mTORC1 by recruiting mTORC1 to the lysosome where it is activated by the lysosome-associated Rheb (Fig. 2) (Sancak et al., 2008; 2010). According to this model, the signaling inputs from the amino acid-Rag pathway and the growth factor-Rheb pathway are in parallel and converge on mTORC1 at the lysosome to effectively promote its activation. This model also explains why growth factors cannot activate mTORC1 efficiently in the absence of amino acids and vice versa. However, due to lack of a validated Rheb antibody in the field, it is currently unclear where endogenous Rheb is localized. Several previous reports using exogenously expressed Rheb have suggested that Rheb can be localized on the various endomembrane structures including endoplasmic reticulum, Golgi complex, endosome, and/or lysosome, but commonly not on the plasma membrane (Buerger et al., 2006; Hanker et al., 2010; Saito et al., 2005; Takahashi et al., 2005). Therefore, it remains to be further confirmed whether endogenous Rheb is localized on the lysosomal surface and also whether the lysosome is the sole membrane structure for mTORC1 activation by Rheb.
Fig. 2.
Amino acid signaling to mTORC1. The key mechanisms by which amino acids activate mTORC1 on the lysosomal surface are depicted. Under amino acid deficiency, the inactive state of the v-ATPase-Ragulator complex is unable to activate Rag GTPases on the lysosomal surface, thus mTORC1 cannot be recruited to the lysosome. In addition, LRS is inactive due to low level of leucine. Under amino acid sufficiency, the v-ATPase-Ragulator complex is activated through a lysosomal ‘inside-out’ mechanism, which in turn promotes the GTP-charging of RagA/B via a GEF function of the Ragulator. Activated Rags then recruit mTORC1 to the lysosomal surface, where mTORC1 can be activated by Rheb by integrating amino acid signals with other upstream signals converging on the Rheb GTPase. Under conditions of leucine availability, activated LRS signals to the Rags through the cytoplasmic face of the lysosome.
Amino acid regulation of Rag GTPases requires the Ragulator complex at the lysosome
Most small G proteins are localized to their target membrane structures via a prenylated CAAX motif (C is cysteine, A is typically an aliphatic residue, and X is any amino acid), which is located at their C-terminus (Ahearn et al., 2012). The Rag GTPases, however, lack such a motif, and it was therefore originally not known how the Rags were able to recruit mTORC1 to the lysosomal membrane. Sabatini and colleagues provided a possible explanation when they identified a complex of proteins that interacts with and is responsible for targeting the Rags to the lysosomal surface. This complex was termed “Ragulator” and was originally characterized as a trimeric complex consisting of p18 (LAMTOR1), p14 (LAMTOR2), and MAPK scaffold protein 1 (MP1; also termed LAMTOR3) (Sancak et al., 2010), but was soon expanded to a pentameric complex that also includes C7orf59 (LAMTOR4) and hepatitis B virus X interacting protein (HBXIP; also termed LAMTOR5) (Bar-Peled et al., 2012). Lipid modification of one Ragulator component, p18, through N-terminal myristoylation and palmitoylation, appears to keep the pentameric complex at the lysosomal surface. Their study showed that genetic deletion or RNAi-mediated depletion of any of the Ragulator components was sufficient to disrupt Rag lysosomal localization and consequently the translocation of mTORC1 to the lysosome in response to amino acids. Therefore, Ragulator appears to be essential for tethering the Rags to the lysosomal surface, thereby providing the docking site for mTORC1 at the lysosome under amino acid-sufficient conditions.
In addition to its role in targeting the Rags to the lysosomal surface, Ragulator also functions as a guanine nucleotide exchange factor (GEF) toward RagA/B in the presence of amino acids, promoting the exchange of GDP with GTP and thereby triggering activation of RagA/B GTPases (Bar-Peled et al., 2012). Although none of the Ragulator components possess a canonical domain homologous to any known GEF catalytic domains, they observed that Ragulator preferentially interacts with the nucleotide free forms of RagA/B over the nucleotide bound ones, which is a characteristic of a GEF-GTPase interaction, and showed that the pentameric complex by itself displayed in vitro GEF activity toward RagA/B, but not toward RagC/D. Interestingly, somewhat consistent with this role, the strength of the Rag-Ragulator interaction was dependent on amino acids; Rag binding affinity to Ragulator was greatly reduced by amino acid stimulation when compared to their binding affinity under amino acid deficiency. These findings suggest that when amino acids are limiting, Rag GTPases are tethered at the lysosomal surface in the inactive state via their tight interaction with Ragulator. Upon amino acid stimulation, the Rags get activated through the GEF function of Ragulator, leading to decreased binding affinity, which may allow the Rags to expose an effector domain required for mTORC1 recognition and anchoring to the lysosome (Fig. 2). However, since intracellular localization of Rag GTPases was reported to be very distinctive depending on their nucleotide-bound state (Sekiguchi et al., 2001), and the interaction between the Rags and the Ragulator was greatly reduced when RagA/B is constitutively active (Bar-Peled et al., 2012), it remains to be determined whether the Rag GTPases can be dissociated from the Ragulator. Likewise, it remains unclear what proteins function as a GAP for RagA/B that would antagonize the Ragulator GEF function by stimulating the intrinsic GTPase activity of RagA/B.
In yeast, the Gtr1p-Gtr2p heterodimer was shown to be localized to the vacuole (the yeast equivalent of the lysosome) as part of the EGO complex that includes Ego1p and Ego3p (Gao and Kaiser, 2006). The EGO complex plays a role in sorting of a general amino acid transporter to the vacuole (Gao and Kaiser, 2006), and later, as in mammalian cells, the Gtr proteins were found to be essential for TORC1 activation by amino acids (Binda et al., 2009). However, their role in recruiting TORC1 doesn’t seem to be conserved because the vacuolar localization of TORC1 was not affected by amino acids (Binda et al., 2009). Although there is no significant sequence similarity between the EGO and the Ragulator components, the recent study that solved the crystal structure of Ego3p revealed that it is structurally similar to the two mammalian Ragulator components, MP1 and p14 (Kogan et al., 2010; Kurzbauer et al., 2004; Zhang et al., 2012). Like p18, Ego1p was also found to be myristoylated and palimitoylated (Ashrafi etal., 1998; Kogan et al., 2010). These findings suggest that the role of the Ragulator in tethering the Rags to the lysosome may be structurally conserved in yeast as the EGO proteins also tether the Gtrs to the vacuole. However, the GEF function of the Ragulator doesn’t seem to be conserved in the EGO complex, as Vam6p (a yeast homologue of the mammalian vacuolar protein sorting 39) was proposed to function as a GEF for Gtr1p in yeast (Binda et al., 2009).
The roles of the v-ATPase and leucyl-tRNA synthetase in sensing amino acids
Although significant advancements have been made in understanding the molecular mechanisms of how amino acid signals are transmitted to mTORC1 through Rag GTPases, it is still unclear exactly how amino acid sensing is initiated, what molecules are responsible for this initiation, and whether such molecules physically interact with amino acids to detect their presence. As mentioned above, the Ragulator displays GEF activity in the presence of amino acids on the lysosomal surface, which in turn activates the Rags, leading to mTORC1 recruitment and activation on this organelle. Accordingly, Sabatini and colleagues have recently reported that the lysosome is the cellular compartment where amino acid sensing occurs. They proposed an “inside-out” model in which accumulation of amino acids inside the lysosomal lumen evokes the sensing signals, which are in turn delivered outside of the lysosome through the vacuolar H+-adenosine triphosphatase (v-ATPase), leading to activation of mTORC1 via the Ragulator-Rag pathway (Fig. 2) (Zoncu et al., 2011a). The v-ATPase was identified as another essential factor for mTORC1 activation in response to amino acids through screening with RNA interference to a number of known genes with roles in lysosomal function/biogenesis in Drosophila S2 cells. The v-ATPase is a large multisubunit proton pump crucial for maintaining the lysosomal function by lowering the pH; the peripheral cytosolic V1 domain is responsible for ATP hydrolysis, which in turn allows rotation of the integral membrane V0 domain to pump protons into the lysosomal lumen and thereby acidify it (Forgac, 2007). In their study, they demonstrated that depletion of v-ATPase subunits or chemical inhibition of its activity was sufficient to inhibit mTORC1 activation and its amino acid-stimulated lysosomal localization, illuminating its essential role in amino acid signaling to mTORC1. Further characterization revealed that the v-ATPase is implicated in mTORC1 activation by directly interacting with the Ragulator-Rag complex, more specifically with the Ragulator, on the lysosomal surface. Amino acid abundance weakens the binding affinity between the V1 domain of v-ATPase and the Ragulator, but not the binding between the V0 domain and Ragulator, whereas amino acid deficiency strengthens such binding. Moreover, chemical inhibition of the v-ATPase prevents amino acids from regulating these interactions, implying a role for the v-ATPase upstream of the Ragulator. These findings also imply that amino acids may promote the Ragulator GEF activity toward the Rags by altering its interaction with the v-ATPase. However, it is currently unknown what molecule resides upstream of v-ATPase in amino acid sensing and how precisely this upstream sensor would alter v-ATPase action toward the Ragulator.
In an effort to further clarify the role of v-ATPase in amino acid signaling to mTORC1 at the lysosome, Zoncu and colleagues established a cell free system by immunopurifying Rag GTPase-bound lysosomes. Using this system, they showed that the purified lysosomal fraction could recruit myc-raptor from the cytosolic fraction when amino acids were added to this cell free system, and that this recruitment was prevented in the presence of the v-ATPase inhibitor. These findings suggest that the lysosome appears to contain almost all of the required components to sense and signal amino acid availability to mTORC1. By measuring and modulating amino acid levels within the lysosome, they also demonstrated that accumulation of amino acids inside the lysosomal lumen was critical for the role of v-ATPase in promoting mTORC1 recruitment, implying that amino acid signaling to mTORC1 is unlikely to be through the cytoplasmic face. However, the lysosomal proton gradient driven by v-ATPase activity does not seem to be implicated in the lysosomal accumulation of amino acids, as treatment with an ionophore that disrupts the lysosomal proton gradient without affecting v-ATPase functions showed no inhibitory effect on the Rag-mTORC1 interaction induced by amino acids. These results indicate that amino acids appear to be sensed inside the lysosomal lumen, where they generate an initiating signal that communicates with v-ATPase, ultimately being transmitted to mTORC1 through an ‘inside-out’ mechanism. These findings also raise several questions such as what is the nature of the driving force that transports amino acids into the lysosome and what specific type of amino acid transporters are responsible for accumulation of amino acids inside lysosomes. In addition, as v-ATPase is well conserved in yeast, it remains an open question as to whether the yeast v-ATPase has a similar function as in mammalian cells.
Importantly, in addition to amino acid sensing at the lysosome, two recent studies suggest the existence of another amino acid sensing mechanism in both yeast and mammals, in which leucine availability is sensed by the leucyl-tRNA synthetase (LRS) (Bonfils et al., 2012; Han et al., 2012). LRS is the enzyme that charges leucine to its cognate tRNA and is thus essential for protein synthesis. Leucine is one of the most crucial amino acids implicated in amino acid-induced mTORC1 activation. The two studies showed that LRS is essential for amino acid signaling to mTORC1 by promoting activation of the Gtr/Rag GTPases, although the proposed mechanistic details are largely different between mammals and yeast. In mammals, LRS was proposed to be a GAP for RagD, but not RagC, and was implicated in mTORC1 activation in a leucine-dependent manner by stimulating the formation of the GDP-bound form of RagD, thereby promoting configuration of the active Rag heterodimer complex (RagA/B-GTP·RagD-GDP) (Han et al., 2012). In yeast, LRS was shown to preferentially bind to GTP-bound Gtr1 (the RagA/B homolog) instead of Gtr2 (the RagC/D homolog), preventing GTP hydrolysis, leading to TORC1 activation when leucine is present (Bonfils et al., 2012). Although further studies are needed to elucidate the precise role of LRS in relaying amino acid signals to TORC1, and to understand its contribution to mTORC1 signaling relative to the lysosome-based inside-out sensing model, these findings provide a possibility that amino acid sensing to control mTORC1 may occur at various cellular compartments and by different mechanisms.
Glutamine regulates mTORC1 signaling through multiple mechanisms
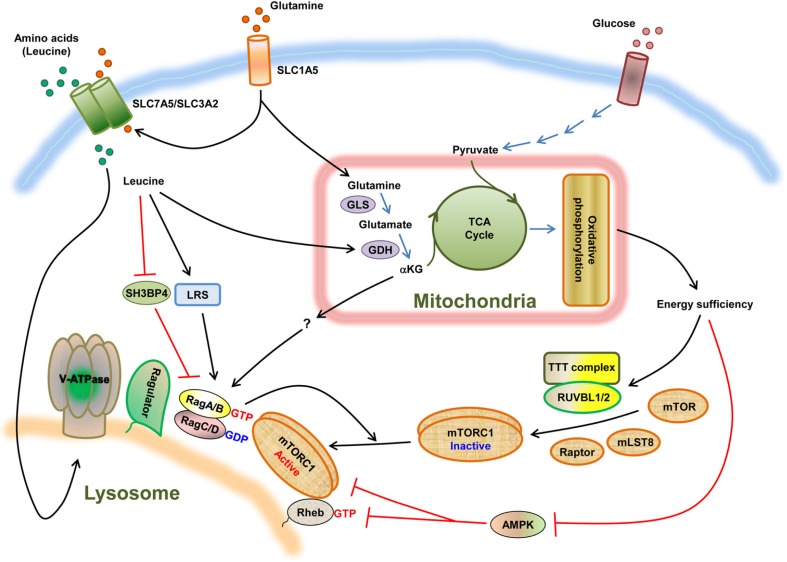
Glutamine is the most abundant amino acid in the blood and is metabolized in the mitochondria through two deamination reactions termed glutaminolysis; glutamine is deaminated by glutaminase (GLS) to produce glutamate, which is further deaminated by glutamate dehydrogenase (GDH) to produce α-ketoglutarate (αKG). αKG replenishes the TCA cycle. Glutamate also serves as a source for glutathione production. Thus, in addition to glucose, glutamine and its metabolites, participate in many biological functions important for cell growth including incorporation into proteins during protein synthesis, contribution to several biosynthetic reactions including amino acid, fatty acid and nucleotide synthesis, and engagement in redox reactions to sustain cellular energy (DeBerardinis et al., 2008). Given mTORC1’s central roles and the importance of glutamine in cell growth, glutamine has also been thought to be an important amino acid for mTORC1 signaling, along with leucine and arginine. Nicklin et al. (2009) proposed that glutamine transport into and out of the cell is required for leucine-induced mTORC1 activation. This study showed that in several cell types, intracellular glutamine, which can enter the cell via the Na+-dependent SLC1A5 transporter, is required for preloading the SLC7A5/ SLC3A2 bidirectional transporter, and the subsequent efflux of the loaded glutamine drives the uptake of essential amino acids including leucine, leading to leucine-induced mTORC1 activation (Fig. 3). This role of glutamine appears to be independent of glutaminolysis, as glutamate or αKG failed to recover the loss of glutamine. These findings suggest that glutamine promotes mTORC1 activity as a free amino acid at least in part by increasing the uptake of amino acids such as leucine.
Fig. 3.
Nutrient signaling to mTORC1. The involvement of glutamine in nutrient regulation of mTORC1 signaling is depicted. Glutamine, as a free amino acid, promotes amino acid-induced mTORC1 activation by enhancing the uptake of essential amino acids including leucine. In addition, through metabolism to αKG, glutamine is able to activate Rag GTPases through a leucine/GDH-dependent mechanism. Leucine can also signal to the Rags by activating LRS and/or inhibiting SH3BP4, which is proposed to inhibit leucine signaling to mTORC1 by preventing the formation of the active Rags (Kim et al., 2012). The metabolism of the critical nutrients glucose and glutamine contribute to generating cellular energy, which is required for the assembly of a functional, dimeric mTORC1 via the TTT-RUVBL1/2 complex and its subsequent amino acid/Rag-dependent lysosomal localization.
In contrast to the model proposed by Nicklin et al. where glutamine stimulates mTORC1 indirectly via leucine, recent findings from Duran et al. (2012) suggested that leucine stimulates mTORC1 indirectly through its effects on glutaminolysis. They demonstrated that glutamine, in combination with leucine, increased the GTP charging of exogenously expressed RagB, promoting mTORC1 activation by enhancing glutaminolysis and αKG production. Chemical inhibitors or depletion of GLS and GDH to block glutaminolysis decreased the GTP charging of the expressed RagB, mTORC1 lysosomal localization, and thus its activation in response to glutamine and leucine. Moreover, single treatment with a cell permeable αKG analog in amino acid-deprived conditions significantly restored mTORC1 lysosomal localization and activation. These results suggest that glutaminolysis and αKG production may be the key events for leucine to activate Rag-mTORC1 signaling (Fig. 3). Combined with the idea that leucine can function as an allosteric regulator of GDH through direct binding, thus enhancing conversion of glutamate into αKG, they propose a model where mTORC1 senses the fluctuations of glutamine and leucine together by sensing leucine-dependent production of αKG (Fig. 3). However, given that the allosteric activators of GDH including leucine do not seem to be essential for GDH catalytic activity (Fang et al., 2002), and that single treatment with leucine is sufficient to activate mTORC1 at least compared to that with glutamine in total amino acid-depleted conditions (Duran et al., 2012), it is more likely that leucine may promote mTORC1 activation through multiple mechanisms that involve not just enhancement of glutaminolysis but perhaps all mechanisms described above (Fig. 3).
Most experiments studying the role of glutamine in mTORC1 signaling have used the strategy of first depriving cells of all amino acids, followed by stimulation with both glutamine and leucine. This approach is likely used because while deprivation of leucine, in the presence of all other nutrients, significantly inhibits mTORC1 signaling in many cell types, even long-term deprivation of glutamine alone results in only a small inhibitory effect on mTORC1 (Duran et al., 2012; Hara et al., 1998). One possible explanation for the negligible effect of glutamine withdrawal on mTORC1 signaling in these studies was that low levels of glutamine could be compensated for by the presence of another nutrient. Blenis and colleagues showed that the redundant nutrient was not leucine but instead was another energetic source, glucose. Under conditions of glucose deficiency, glutamine was the redundant nutrient absolutely required to meet the energetic needs of cells with high mTORC1 activity (Choo et al., 2010). Not only was glutamine a critical nutrient source for this cell survival balancing act, glutamine and glucose were also shown to be essential nutrients for maintaining mTORC1 signaling (Kim et al., 2013). Kim et al. showed that the inhibitory effect of glutamine and glucose deprivation on mTORC1 could be compensated for by the presence of pyruvate and other TCA cycle intermediates, but not leucine. When both glucose and glutamine were depleted, as can happen during the early stages of tumor growth, before angiogenesis can provide an adequate nutrient supply, mTORC1 was strongly inhibited. Interestingly, however, this new mechanism worked independently of but in collaboration with AMPK-or TSC-dependent mechanisms. Moreover, ATP depletion by nutrient deprivation or inhibition of mitochondrial respiration prevented mTORC1 lysosomal localization even in the presence of amino acids, and this prevention was not recovered by expression of the dominant-active RagB. These findings indicated that replenishment of the TCA cycle by the metabolism of glutamine to αKG, glucose to pyruvate, and ATP production are necessary for mTORC1 lysosomal localization and thus its activation, but in this case through an AMPK-and Rag-independent mechanism (Fig. 3). By screening the mouse genome with a siRNA library in Tsc2−/− cells, Kim et al. discovered that the Tel2-Tti1-Tti2 (TTT)-RuvB-like 1 and 2 (RUVBL1/2) complex is a key regulator of mTORC1 signaling in a TSC-independent manner. This multiprotein complex is an AAA+ ATPase RUVBL-containing complex and has been shown to regulate the assembly and stability of the PI3KK containing proteins including mTORC1 (Horejsi et al., 2010; Hurov et al., 2010; Izumi et al., 2010; Venteicher et al., 2008). Consistent with this, energetic stress was found to disassemble the TTT-RUVBL1/2 complex and thus inhibit its ability to stabilize formation of an active dimeric mTORC1 (Yip et al., 2010). Furthermore, loss of the TTTRUVBL1/ 2 components or inhibition of the RUVBL ATPase activity was sufficient to suppress mTORC1 activation by preventing mTORC1 assembly and its lysosomal localization. Therefore, this study has discovered a role of the TTT-RUVBL1/2 complex in coupling the cell’s metabolic state to mTORC1 signaling by directly controlling mTORC1’s assembly and thus its lysosomal localization (Fig. 3). Therefore, as a free amino acid or as a metabolite through glutaminolysis, glutamine can promote amino acid-induced mTORC1 activation by facilitating the uptake of essential amino acids and by activating Rag GTPases through an unknown mechanism, respectively. In addition, by replenishing the TCA cycle, glutamine also contributes to generating energy sufficient conditions that are necessary for mTORC1’s functional assembly and its proper localization through a mechanism involving the TTT-RUVBL1/2 complex.
CONCLUSION
A growing body of evidence has provided much evidence supporting a role for mTORC1 in sensing diverse environmental conditions and for its ability to link the availability of growth factors and nutrients such as amino acids and glucose to cell growth and proliferation. Many significant discoveries from a number of groups have greatly advanced our understanding of how a variety of signaling cascades derived from environmental cues converge on the mTORC1 pathway, leading to the current model of mTORC1 regulation. As described in this review, the molecular mechanisms by which growth factors transmit their signals to mTORC1 via the PI3K-Akt and/or Ras-MAPK pathways are well characterized. The energetic stress signaling cascades that inhibit mTORC1 via AMPK-dependent pathways are also well described. However, how nutrients, particularly amino acids, are sensed by and relay their signals to mTORC1 has been elusive. Identification of the Rag GTPases and the subsequent discoveries of the Ragulator and v-ATPase have greatly improved our understanding of how amino acids regulate the mTORC1 pathway. These findings also elucidate the importance of the lysosomal membrane as an mTORC1 signaling platform for integrating Rag-dependent amino acid inputs with other upstream inputs converging on Rheb. In addition, numerous proteins such as LRS, SH3BP4, p62, MAP4K3, Vps34, PLD, and RalA have been identified and proposed to be involved in amino acid signaling to mTORC1 (Duran et al., 2011; Gulati et al., 2008; Han et al., 2012; Kim et al., 2012; Maehama et al., 2008; Yan et al., 2010; Yoon et al., 2011), most of which are not discussed in this review due to space limitations but which are reviewed elsewhere (Jewell and Guan, 2013; Kim and Guan, 2011). These findings not only expand our knowledge of this signaling cascade but also reveal its complexity. Moreover, studies with glutamine demonstrate its dual roles in relaying amino acid and energy signals to mTORC1, leading to identification of AMPK-independent energy regulation of mTORC1. Despite these advances, much more work is needed to identify additional key regulators and completely decipher the mechanistic details of nutrient regulation of mTORC1 signaling. In addition to the remaining issues discussed above, if and how amino acids are sensed at multiple sites other than the lysosome remains unknown, as is the precise nature of amino acid sensors, whether these sensors have a preference for a certain type of amino acid such as leucine or arginine, and how these amino acids initiate signaling. Answering these questions and determining the precise roles of all of the identified components in amino acid signaling to mTORC1 will ultimately lead us to understand how the cell balances its growth and survival in accordance with the cell’s nutrient state, and provide additional therapeutic options for treating metabolic diseases associated with improper regulation of mTORC1 such as diabetes, muscle wasting diseases, neurodegenerative diseases, aging and cancer.
Acknowledgments
We thank the Blenis lab members for helpful discussions. We acknowledge support from the National Institute of Health (Grant#RO1CA46595 and RO1GM51405 to JB), the LAM Foundation (LAM082E0110 to JB) and the fellowship from National Cancer Institute to GB. We also apologize to authors whose important work we could not acknowledge due to limitations of space.
REFERENCES
- Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. (2012). Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol. 13, 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Farazi TA, Gordon JI. (1998). A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J Biol Chem. 273, 25864–25874 [DOI] [PubMed] [Google Scholar]
- Ballif BA, Roux PP, Gerber SA, MacKeigan JP, Blenis J, Gygi SP. (2005). Quantitative phosphorylation profiling of the ERK/p90 ribosomal S6 kinase-signaling cassette and its targets, the tuberous sclerosis tumor suppressors. Proc. Natl. Acad. Sci. USA. 102, 667–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. (2012). Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 150, 1196–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. (1996). TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell. 7, 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Howell JJ, Asara JM, Manning BD. (2013). Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 339, 1323–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C. (2009). The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell. 35, 563–573 [DOI] [PubMed] [Google Scholar]
- Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, De Virgilio C. (2012). Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol. Cell. 46, 105–110 [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr (2004). Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 18, 2893–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerger C, DeVries B, Stambolic V. (2006). Localization of Rheb to the endomembrane is critical for its signaling function. Biochem Biophys Res Commun. 344, 869–880 [DOI] [PubMed] [Google Scholar]
- Choo AY, Kim SG, Vander Heiden MG, Mahoney SJ, Vu H, Yoon SO, Cantley LC, Blenis J. (2010). Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol. Cell. 38, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu M, Albert V, Hall MN. (2013). mTOR in aging, metabolism, and cancer. Curr Opin Genet Dev. 23, 53–62 [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. (2008). The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11–20 [DOI] [PubMed] [Google Scholar]
- Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G. (2001). Mammalian TOR: a homeostatic ATP sensor. Science. 294, 1102–1105 [DOI] [PubMed] [Google Scholar]
- DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. (2008). Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 22, 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. (2012). TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol. Cell. 47, 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. (2011). p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol. Cell. 44, 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, Hall MN. (2012). Glutaminolysis activates Rag-mTORC1 signaling. Mol. Cell. 47, 349–358 [DOI] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Sabatini DM. (2012). Amino acids and mTORC1: from lysosomes to disease. Trends Mol Med. 18, 524–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Hsu BY, MacMullen CM, Poncz M, Smith TJ, Stanley CA. (2002). Expression, purification and characterization of human glutamate dehydrogenase (GDH) allosteric regulatory mutations. Biochem J. 363, 81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgac M. (2007). Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, Sabatini DM. (2006). mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 16, 1865–1870 [DOI] [PubMed] [Google Scholar]
- Gao M, Kaiser CA. (2006). A conserved GTPase-containing complex is required for intracellular sorting of the general aminoacid permease in yeast. Nat Cell Biol. 8, 657–667 [DOI] [PubMed] [Google Scholar]
- Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, Pan D. (2002). Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat Cell Biol. 4, 699–704 [DOI] [PubMed] [Google Scholar]
- Gong R, Li L, Liu Y, Wang P, Yang H, Wang L, Cheng J, Guan KL, Xu Y. (2011). Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev. 25, 1668–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. (2006). Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell. 11, 859–871 [DOI] [PubMed] [Google Scholar]
- Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G. (2008). Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 7, 456–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. (2008). AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 30, 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. (2012). Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 149, 410–424 [DOI] [PubMed] [Google Scholar]
- Hanker AB, Mitin N, Wilder RS, Henske EP, Tamanoi F, Cox AD, Der CJ. (2010). Differential requirement of CAAXmediated posttranslational processing for Rheb localization and signaling. Oncogene. 29, 380–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. (1998). Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 273, 14484–14494 [DOI] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. (2002). Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 110, 177–189 [DOI] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. (2012). AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 13, 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose E, Nakashima N, Sekiguchi T, Nishimoto T. (1998). RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. J. Cell Sci.. 111Pt 111–21 [DOI] [PubMed] [Google Scholar]
- Horejsi Z, Takai H, Adelman CA, Collis SJ, Flynn H, Maslen S, Skehel JM, de Lange T, Boulton SJ. (2010). CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol. Cell. 39, 839–850 [DOI] [PubMed] [Google Scholar]
- Howell JJ, Manning BD. (2011). mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends in Endocrinol. Metab. TEM. 22, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurov KE, Cotta-Ramusino C, Elledge SJ. (2010). A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 24, 1939–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. (2002). TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 4, 648–657 [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. (2003a). Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17, 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. (2003b). TSC2 mediates cellular energy response to control cell growth and survival. Cell. 115, 577–590 [DOI] [PubMed] [Google Scholar]
- Inoki K, Kim J, Guan KL. (2012). AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol. 52, 381–400 [DOI] [PubMed] [Google Scholar]
- Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S. (2010). AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci Signal. 3, ra27. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. (2004). Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. (2006). SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 127, 125–137 [DOI] [PubMed] [Google Scholar]
- Jeong JH, Lee KH, Kim YM, Kim DH, Oh BH, Kim YG. (2012). Crystal structure of the Gtr1p(GTP)-Gtr2p(GDP) protein complex reveals large structural rearrangements triggered by GTP-to-GDP conversion. J Biol Chem. 287, 29648–29653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell JL, Guan KL. (2013). Nutrient signaling to mTOR and cell growth. Trends Biochem Sci. 38, 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalender A, Selvaraj A, Kim SY, Gulati P, Brule S, Viollet B, Kemp BE, Bardeesy N, Dennis P, Schlager JJ, et al. (2010). Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab. 11, 390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guan KL. (2011). Amino acid signaling in TOR activation. Annu Rev Biochem. 80, 1001–1032 [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. (2002). mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 110, 163–175 [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM. (2003). GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell. 11, 895–904 [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. (2008). Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 10, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Stone M, Hwang TH, Kim YG, Dunlevy JR, Griffin TJ, Kim DH. (2012). SH3BP4 is a negative regulator of amino acid-Rag GTPase-mTORC1 signaling. Mol. Cell. 46, 833–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SG, Hoffman GR, Poulogiannis G, Buel GR, Jang YJ, Lee KW, Kim BY, Erikson RL, Cantley LC, Choo AY, et al. (2013). Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol. Cell. 49, 172–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan K, Spear ED, Kaiser CA, Fass D. (2010). Structural conservation of components in the amino acid sensing branch of the TOR pathway in yeast and mammals. J Mol Biol. 402, 388–398 [DOI] [PubMed] [Google Scholar]
- Kurzbauer R, Teis D, de Araujo ME, Maurer-Stroh S, Eisenhaber F, Bourenkov GP, Bartunik HD, Hekman M, Rapp UR, Huber LA, et al. (2004). Crystal structure of the p14/MP1 scaffolding complex: how a twin couple attaches mitogenactivated protein kinase signaling to late endosomes. Proc. Natl. Acad. Sci. USA. 101, 10984–10989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. (2012). mTOR signaling in growth control and disease. Cell. 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, et al. (2004). LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. (2005a). Rheb binds and regulates the mTOR kinase. Curr Biol. 15, 702–713 [DOI] [PubMed] [Google Scholar]
- Long X, Ortiz-Vega S, Lin Y, Avruch J. (2005b). Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem. 280, 23433–23436 [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. (2009). Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 10, 307–318 [DOI] [PubMed] [Google Scholar]
- Maehama T, Tanaka M, Nishina H, Murakami M, Kanaho Y, Hanada K. (2008). RalA functions as an indispensable signal mediator for the nutrient-sensing system. J Biol Chem. 283, 35053–35059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. (2002). Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell. 10, 151–162 [DOI] [PubMed] [Google Scholar]
- Mendoza MC, Er EE, Blenis J. (2011). The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 36, 320–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima N, Noguchi E, Nishimoto T. (1999). Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 152, 853–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. (2009). Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 136, 521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, et al. (2005). Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. USA. 102, 14238–14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WJ, Jacinto E. (2011). mTOR complex 2 signaling and functions. Cell Cycle. 10, 2305–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E. (2000). Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 14, 2689–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro N, Takahashi R, Yoshino K, Tanimura K, Nakashima A, Eguchi S, Miyamoto T, Hara K, Takehana K, Avruch J, et al. (2007). The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem. 282, 20329–20339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce LR, Huang X, Boudeau J, Pawlowski R, Wullschleger S, Deak M, Ibrahim AF, Gourlay R, Magnuson MA, Alessi DR. (2007). Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 405, 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. (2009). DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 137, 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling JH, Hafen E. (2004). The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 18, 2879–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. (2013). Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 339, 1320–1323 [DOI] [PubMed] [Google Scholar]
- Roccio M, Bos JL, Zwartkruis FJ. (2006). Regulation of the small GTPase Rheb by amino acids. Oncogene. 25, 657–664 [DOI] [PubMed] [Google Scholar]
- Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. (2004). Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc. Natl. Acad. Sci. USA. 101, 13489–13494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Araki Y, Kontani K, Nishina H, Katada T. (2005). Novel role of the small GTPase Rheb: its implication in endocytic pathway independent of the activation of mammalian target of rapamycin. J Biochem. 137, 423–430 [DOI] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. (2007). PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell. 25, 903–915 [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 320, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. (2010). Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. (2004). Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. (2006). Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 22, 159–168 [DOI] [PubMed] [Google Scholar]
- Schurmann A, Brauers A, Massmann S, Becker W, Joost HG. (1995). Cloning of a novel family of mammalian GTP-binding proteins (RagA, RagBs, RagB1) with remote similarity to the Ras-related GTPases. J Biol Chem. 270, 28982–28988 [DOI] [PubMed] [Google Scholar]
- Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. (2001). Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 276, 7246–7257 [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. (2010). Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell. 40, 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ. (2009). The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev. 9, 563–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. (2004). The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. USA. 101, 3329–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. (2005). The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 280, 18717–18727 [DOI] [PubMed] [Google Scholar]
- Sofer A, Lei K, Johannessen CM, Ellisen LW. (2005). Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 25, 5834–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Nakagawa M, Young SG, Yamanaka S. (2005). Differential membrane localization of ERas and Rheb, two Ras-related proteins involved in the phosphatidylinositol 3-kinase/mTOR pathway. J Biol Chem. 280, 32768–32774 [DOI] [PubMed] [Google Scholar]
- Tao Z, Barker J, Shi SD, Gehring M, Sun S. (2010). Steady-state kinetic and inhibition studies of the mammalian target of rapamycin (mTOR) kinase domain and mTOR complexes. Biochemistry. 49, 8488–8498 [DOI] [PubMed] [Google Scholar]
- Tee AR, Anjum R, Blenis J. (2003a). Inactivation of the tuberous sclerosis complex-1 and -2 gene products occurs by phosphoinositide 3-kinase/Akt-dependent and -independent phosphorylation of tuberin. J Biol Chem. 278, 37288–37296 [DOI] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. (2003b). Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 13, 1259–1268 [DOI] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. (2007). Insulin signalling to mTOR mediated by the Akt/ PKB substrate PRAS40. Nat Cell Biol. 9, 316–323 [DOI] [PubMed] [Google Scholar]
- Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. (2008). Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 132, 945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Harris TE, Lawrence JC., Jr (2008). Regulation of proline-rich Akt substrate of 40 kDa (PRAS40) function by mammalian target of rapamycin complex 1 (mTORC1)-mediated phosphorylation. J Biol Chem. 283, 15619–15627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SY, Kim DH, Jun CB, Kim YM, Haar EV, Lee SI, Hegg JW, Bandhakavi S, Griffin TJ. (2007). PRR5, a novel component of mTOR complex 2, regulates platelet-derived growth factor receptor beta expression and signaling. J Biol Chem. 282, 25604–25612 [DOI] [PubMed] [Google Scholar]
- Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. (2003). LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol. 13, 2004–2008 [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. (2006). TOR signaling in growth and metabolism. Cell. 124, 471–484 [DOI] [PubMed] [Google Scholar]
- Yan L, Mieulet V, Burgess D, Findlay GM, Sully K, Procter J, Goris J, Janssens V, Morrice NA, Lamb RF. (2010). PP2A T61 epsilon is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol. Cell. 37, 633–642 [DOI] [PubMed] [Google Scholar]
- Yang Q, Inoki K, Ikenoue T, Guan KL. (2006). Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 20, 2820–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yecies JL, Manning BD. (2011). mTOR links oncogenic signaling to tumor cell metabolism. J. Mol. Med. (Berl.). 89, 221–228 [DOI] [PubMed] [Google Scholar]
- Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. (2010). Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol. Cell. 38, 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon MS, Du G, Backer JM, Frohman MA, Chen J. (2011). Class III PI-3-kinase activates phospholipase D in an amino acid-sensing mTORC1 pathway. J Cell Biol. 195, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Sarbassov dos D, Samudio IJ, Yee KW, Munsell MF, Ellen Jackson C, Giles FJ, Sabatini DM, Andreeff M, Konopleva M. (2007). Rapamycin derivatives reduce mTORC2 signaling and inhibit AKT activation in AML. Blood. 109, 3509–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. (2000). Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14, 2712–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Peli-Gulli MP, Yang H, De Virgilio C, Ding J. (2012). Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1. Structure. 20, 2151–2160 [DOI] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. (2011a). mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 334, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. (2011b). mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]