Abstract
Rheumatoid arthritis (RA) is a chronic, systemic inflammatory disease of unknown origin, which exhibits a complex heterogeneity in its pathophysiological background, resulting in differential responses to a range of therapies and poor long-term prognosis. RA synovial fibroblasts (RASFs) are key player cells in RA pathogenesis. Identification of DNA methylation biomarkers is a field that provides potential for improving the process of diagnosis and prognosis of various human diseases. We utilized a genome-wide technique, methylated DNA isolation assay (MeDIA), in combination with a high resolution CpG microarray for discovery of novel hypermethylated genes in RASFs. Thirteen genes (APEX1, EBF3, EGR2, EN1, IRX1, IRX6, KIF12, LHX2, MIPOL1, SGTA, SIN3A, TOLLIP, and ZHX2) with three consecutive hypermethylated probes were isolated as candidate genes through two CpG microarrays. Pyrosequencing assay was performed to validate the methylation status of TGF-β signaling components, EBF3 and IRX1 genes in RASFs and osteoarthritis (OA) SFs. Hypermethylation at CpG sites in the EBF3 and IRX1 genes was observed with a high methylation index (MI) in RASFs (52.5% and 41.4%, respectively), while a lower MI was observed in OASFs and healthy SFs (13.2% for EBF3 and 4.3% for IRX1). In addition, RT-PCR analysis showed a remarkable decrease in their mRNA expression in the RA group, compared with the OA or healthy control, and their reduction levels correlated with MI. The current findings suggest that methylation-associated down-regulation of EBF3 and IRX1 genes may play an important role in a pathogenic effect of TGF-β on RASFs. However, further clinical validation with large numbers of patients is needed in order to confirm our findings.
Keywords: EBF3, hypermethylation, IRX1, rheumatoid arthritis, synovial fibroblast
INTRODUCTION
Rheumatoid arthritis (RA) is a common chronic inflammatory and multi-factorial disorder affecting 1% of the adult population worldwide; it cannot be cured and its personal, social, and economic costs are substantial (Firestein et al., 2008). It has a complex heterogeneity in its pathophysiological background, resulting in differential responses to a range of therapies and poor long-term prognosis (Firestein et al., 2008), suggesting that a sub-classification of RA using molecular biomarkers could be helpful in prediction of clinical outcomes of therapies. Of particular interest, there is evidence pointing to geographical difference in occurrence or outcome of RA (Carmona et al., 2010). Although the etiology of RA is not yet fully understood, environmental and genetic factors play a role in pathogenesis and they can be responsible for different clinical pictures and response to therapy (Molnar-Kimber and Kimber, 2012). RA synovial fibroblasts (RASFs) (also known as FLSs, fibroblast-like synoviocytes) exhibit abnormal behavior in RA and are thought to be the key effector cells responsible for synovial inflammation, contributing to destruction of bone and cartilage (Huber et al., 2006; Neumann et al., 2010). RASFs are also involved in production of inflammatory cytokines, molecular mediators of inflammation, and matrix-degrading enzymes.
Significant progress has been made in identification of more than 30 genes associated with RA (Bax et al., 2011), however, the precise molecular mechanisms underlying pathogenesis of RA remain unclear. Transcriptional deregulation in RASFs lacking specific genetic mutations suggests that epigenetic mechanisms may be involved. Global hypomethylation and promoter-specific hypermethylation are believed to be associated with conditions such as cancer, atherosclerosis, and autoimmune diseases (Rakyan et al., 2011). Considerable evidence of epigenetic changes, particularly altered patterns of DNA methylation, in RA has been reported (Ballestar 2011; Trenkmann et al., 2010). Of particular importance, changes in DNA methylation, leading to changes in gene expression in RASFs, shaping the aggressive phenotype of these cells has been reported (Karouzakis et al., 2009; Ospelt et al., 2011). Most recent studies have focused on hypomethylation of inflammatory cytokine genes affecting gene regulation in RA (Fu et al., 2011; Hashimoto et al., 2009; Lin et al., 2012; Nile et al., 2008). However, our understanding of promoter hypermethylation at multiple loci is limited to the few genes analyzed to date. Herein, in order to improve our understanding of the molecular complexity of RA and to identify epigenetic biomarkers for diagnosis, patient stratification and prognostication, we investigated a whole-genome methylation in SFs of Korean RA patients using methylated DNA isolation assay (MeDIA)-mediated CpG microarray analysis and the methylation status of two candidate methylation genes was evaluated in RASFs, SFs of osteoarthritis (OA) patients, and healthy controls (HC).
MATERIALS AND METHODS
Tissue specimens and genomic DNA isolation
Synovial tissue was obtained from seven patients with RA, four patients with OA, and two patients with joint trauma (healthy control specimens) undergoing joint replacement surgery at Daegu Catholic University Hospital. Informed consent was obtained from all patients, and RA patients fulfilled the American Rheumatism Association criteria for classification of the disease (Arnett et al., 1988). For cell culture, tissue was digested and SFs were grown as previously described (Stanczyk et al., 2008). Patients with other underlying disease that might affect the general condition of patients were excluded from the study. Mean age of patients in the RA group was 58.00 ± 6.66 years (yrs) and did not differ significantly from that of the OA group (62.00 ± 5.48 yrs) and the healthy control group (58.50 ± 2.12 yrs). A review of the baseline characteristics of recruited patients is shown in Table 1. Genomic DNA was extracted from cultured SFs using the QIAamp DNA kit and blood kit protocol (Qiagen, Germany). DNA concentration and purity was determined by measuring absorbance at 260 nm and 280 nm using spectrophotometer.
Table 1.
Baseline characteristics of study population
| Parameters, mean (SD) | RA (n = 7) | OA (n = 4) | Healthy control (n = 2) |
|---|---|---|---|
| M/F | 1/6 | 1/3 | 1/1 |
| Age, yrs | 58.00 (6.66) | 62.00 (5.48) | 58.50 (2.12) |
| RF, IU/ml | 138.47 (124.0) | NA | NA |
| Anti-CCP, U/ml | 188.39 (172.63) | NA | NA |
| CRP, mg/l | 28.33 (21.39) | NA | NA |
| ESR, mm | 38.43 (12.96) | NA | NA |
| DAS28 score | 4.03 (1.04) | NA | NA |
| Mean disease duration, years | 7.61 (4.36) | NA | NA |
RA, Rheumatoid arthritis; OA, Osteoarthritis
MeDIA-mediated CpG microarray analysis
MeDIA was performed as previously described (Moon et al., 2009) and all procedures are shown in Fig. 1. A minimized methyl-DNA binding domain (MBD) tagged by histidine (MBD2bt) of human MBD2b, which provides high binding sensitivity to methylated DNA was used. In brief, 2 μg of MBD2bt protein was incubated for 8 h at 4°C on a rocking platform with 500 ng of sonicated genomic DNA from RASFs and OASFs. The enriched methylated DNA was amplified using a whole genome amplification kit (Sigma, USA) as recommended by the manufacturer. The amplified products from RASFs and OASFs were labeled with Cy5-dUTP and Cy3-dUTP, respectively, using the BioPrime® Total for the Agilent® aCGH system (Invitrogen, USA). Next, the dye-labeled DNA samples were mixed and hybridized to the human 244K CpG island (CGI) microarrays (Agilent, USA) using conditions specified by the manufacturer. Two independent CpG microarray analyses were performed for RA and OA SFs. Preprocessing of raw data and normalization steps were performed using Gene-Spring 7.3.1 software. The signal intensity of the dye-labeled DNA samples was interpreted using a high resolution microarray scanner (Agilent). For selection of target genes, when comparing RA and OA SFs, it was considered methylation-positive when at least three adjacent probes allowing a one probe gap within the CpG islands (CGIs) in the promoter regions scored a fold-difference factor of ≥ 5.0.
Fig. 1.
Scheme of genome-wide screening in RASFs using MeDIA-mediated CpG microarray analysis. (A) Two pooled DNA samples from either two RASFs or two OASFs were amplified, and labeled with Cy5-dUTP and Cy3-dUTP, respectively. The dye-labeled DNA samples were mixed and hybridized to the human 244K CpG island (CGI) microarrays. Two independent chip microarrays (OA1 and OA2 versus RA1 and RA2, OA1 and OA2 versus RA3 and RA4) were performed. A Cy3-dUTP labeled OA DNA was represented as a green color, while Cy5-dUTP labeled RA DNA as a red color. (B) CpG microarray of genomic DNA in SFs. When comparing RASFs and OASFs, it was considered methylation-positive when at least three adjacent probes allowing a probe gap within the CGIs in the promoter regions scored a fold-difference factor of ≥5.0.
Bisulfite treatment and pyrosequencing (PS)
The PS method was used for quantitative analysis of the methylation status of individual CpG motifs in EBF3 and IRX1 genes (Tost and Gut, 2007). Following treatment of genomic DNA with bisulfite using an EZ-DNA methylation Gold Kit (Zymo Research, USA), according to the manufacturer’s instructions, it was amplified through performance of PCR using a forward primer and a reverse primer, enabling conversion of the PCR product to a single-stranded DNA template suitable for PS. All samples were heated to 95°C for 5 min, and then amplified for 45 cycles of 95°C (45 s), 58°C (45 s), and 72°C (45 s), followed by a final extension at 72°C (5 min). The quality of the PCR products was established on 2% agarose gel with ethidium bromide staining. After purification of PCR product using Se-pharose beads on a PyroMark Vacuum Prep Workstation (Qia-gen), PS was performed according to the manufacturer’s specifications with a sequencing primer using the PyroMark Q96MD System (Qiagen). A summary of the forward, reverse, and sequencing primers used in this analysis is shown in Table 2. A mean methylation index (MI) was calculated from the mean of the methylation percentage for all observed CpG sites. To set the controls for PS, we used CpGenome™ Universal methylated and unmethylated DNA (Chemicon, USA) that were consistently positive or negative with stable levels of methylation. We measured each gene in triplicate and used the average in the statistical analyses.
Table 2.
Primer set for Pyrosequencing and RT-PCR analysis
| Gene | Primer | Sequence (5′ → 3′) | Amplicon size (bp) |
|---|---|---|---|
| Pyrosequencing | |||
| EBF3 | Forward | GGTTGGATATTTTAGGAAGTGTGT | 259 |
| Reverse | TTCCACTCTCRACAAACACTAAT | ||
| Sequencing | TGATAATAATGTTTGTTTGG | ||
| IRX1 | Forward primer | AATAYGGGAAGAAATTGGAAGGTT | 142 |
| Reverse primer | AACCAAAACCACCTCCTCTTTCTC | ||
| Sequencing | AATTTTATAGATGGGTTAGT | ||
| RT-PCR | |||
| EBF3 | Forward | CAGATATCGTGCCTCTTAGTGC | 403 |
| Reverse | TCTCAGTGCGTCCATGTGAGC | ||
| IRX1 | Forward | GCTCTTCGGCAGCGACAC | 260 |
| Reverse | GCTCTGGGGCCTCCTTTG |
bp, base pairs; R = A + G; Y = C + T
Total RNA isolation and reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from six clinical samples using TRIzol (Invitrogen) according to the manufacturer’s instructions. Residual genomic DNA was digested with RNase-free DNase (Invitrogen). First strand cDNA was reverse-transcribed from 2 μg of total RNA in a total volume of 20 μl using oligo (dT) and a SuperScript pre-amplification kit (Invitrogen). The resulting cDNA was amplified by AmpliTaq Gold (PE Applied Biosystems, USA) using gene-specific primers whose sequences are described in Table 2. Amplified products were separated on 2% agarose gels, visualized using ethidium bromide, and photographed.
Statistical analysis
SPSS for windows 18.0 (SPSS, USA) was used in performance of statistical analyses. The Linear Mixed Model test was used for comparison of intergroup differences. The chi-squared (χ2) test was applied for identification of the correlation between the methylation index and clinical parameters. Statistical significance was defined as p < 0.05.
RESULTS AND DISCUSSION
Genome-wide screening and identification of candidate hypermethylation targets
Two independent CpG microarray analyses were performed for RASFs and OASFs (Fig. 1A). A total of 1,714 probes were detected as hypermethylated targets with more than five-fold differences in signal intensity (represented as red color in Fig. 1B). Abundance of hypomethylated probes was much greater than that of hypermethylated probes, supporting the working hypothesis that a hypomethylating milieu in growth factors and receptors, inflammatory cytokines, matrix-degrading enzymes, and adhesion molecules, contributes to the chronicity of RA (Karouzakis et al., 2009). Moreover, promoter hypomethylation and upregulation of these genes have extensively studied in RA (Fu et al., 2011; Hashimoto et al., 2009; Lin et al., 2012; Nile et al., 2008), but our understanding of CGI hypermethylation at multiple loci is limited to the few genes analyzed to date. We have therefore focused on the hypermethylated candidate targets. Thirteen genes (APEX1, EBF3, EGR2, EN1, IRX1, IRX6, KIF12, LHX2, MIPOL1, SGTA, SIN3A, TOLLIP, and ZHX2) with three consecutive hypermethylated probes were finally selected as candidate genes (Table 3). The signal intensity of the dye-labeled DNA samples at individual CpG motifs was interpreted using a high-resolution microarray scanner and visualized as a graph in Fig. 2. The methylation difference of the individual CpG motif was directly compared by ratio of signals from MeDIA-enriched DNA of RASF and OASFs. The stability of methylation patterns and its direct link to disease phenotype indicates that DNA methylation may prove to be a superior, promising biomarker for many applications (Tost, 2010). Therefore, our identification of aberrantly methylated genes can improve the clinical management of RA by facilitating earlier diagnosis of disease and providing more accurate prognostic information.
Table 3.
Genes list to show differential methylation in RASFs and their function
| Gene symbol | GenBank | UniGene | Product | Description |
|---|---|---|---|---|
| APEX1 | NM_001641 | Hs.73722 | APEX nuclease 1 | Apurinic/apyrimidinic endonuclease 1 |
| EBF3 | NM_001005463 | Hs.591374 | Early B-cell factor 3 | DNA-binding transcription factor COE3 |
| EGR2 | NM_000399 | Hs.1395 | Early growth response 2 | E3 SUMO-protein ligase, Zinc finger protein Krox-20 |
| EN1 | NM_001426 | Hs.271977 | Engrailed homeobox 1 | Homeobox protein engrailed-1 |
| IRX1 | NM_024337 | Hs.424156 | Iroquoishomeobox 1 | Iroquois homeobox protein 1 |
| IRX6 | NM_024335 | Hs.369907 | Iroquoishomeobox 6 | Iroquois homeobox protein 6 |
| KIF12 | NM_138424 | Hs.28149 | Kinesin family member 12 | Kinesin-like protein 12 |
| LHX2 | NM_004789 | Hs.445265 | LIM homeobox 2 | LIM homeobox protein 2 |
| MIPOL1 | NM_138731 | Hs.660396 | Mirror-image polydactyly 1 | Mirror-image polydactyly gene 1 protein |
| SGTA | NM_003021 | Hs.203910 | Small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha | Small glutamine-rich TPR-containing protein α |
| SIN3A | NM_015477 | Hs.513039 | SIN3 transcriptional regulator homolog A | Histone deacetylase complex subunit Sin3 homolog A |
| TOLLIP | NM_019009 | Hs.368527 | Toll interacting protein | Toll-interacting protein, adapter protein |
| ZHX2 | NM_014943 | Hs.377090 | Zinc fingers and homeobox 2 | Zinc fingers and homeobox protein 2, α-fetoprotein regulator 1 |
Fig. 2.
Signal intensity of 13 hypermethylated genes in RASFs and OASFs. The signal intensity of the dye-labeled DNA samples at each of three adjacent probes was interpreted using a high resolution microarray scanner. Intensities of DNA of OASFs and RASFs were represented in green and red color, respectively. Numbers on X-indicate the relative probe positions from the transcription start site (TSS, +1).
Clinical validation of DNA hypermethylation and mRNA expression of EBF3 and IRX1 genes in RASFs
The methylation status of EBF3 and IRX1 genes was quantitatively determined in seven RASFs and four OASFs through performance of PS analysis. Accurate and reproducible estimates of methylated cytosine content were obtained in 100% tested samples and their pyrograms are shown in Fig. 3. We measured each gene in triplicate and used the average in the statistical analyses. In the case of the EBF3 gene, the MI (51.14%) of the RA group was significantly higher, compared with the OA (15.19%) or HC group (8.28%) (p < 0.0001), whereas no significant difference was observed between the OA and HC group (p = 0.11) (Fig. 4A). The MI of the IRX1 gene was also significantly higher in the RA group (40.77%) than in the OA (4.64%) or HC group (4.05%) (p < 0.0001, Fig. 4A). However, hypermethylation of the EBF3 and IRX3 genes did not show correlation with clinicopathologic features, including age, disease duration, and DAS28 score of RA patients (data not shown), which may be due to the small size of the sample. Next, we examined the expression levels of EBF3 and IRX1 mRNA in six clinical samples. RT-PCR analysis showed a remarkable decrease of their mRNA expression in the RA1 and RA2 samples with higher MI, compared to the OA or HC samples with unmethylated alleles, but no noticeable reduction was found in RA3 sample with lower MI (Fig. 4B), suggesting that hypermethylation of the EBF3 and IRX1 genes may be closely associated with their transcriptional silencing in RASF. However, conduct of further studies to confirm the methylation and transcriptional silencing of the IRX1 and EBF3 genes in large numbers of samples is warranted.
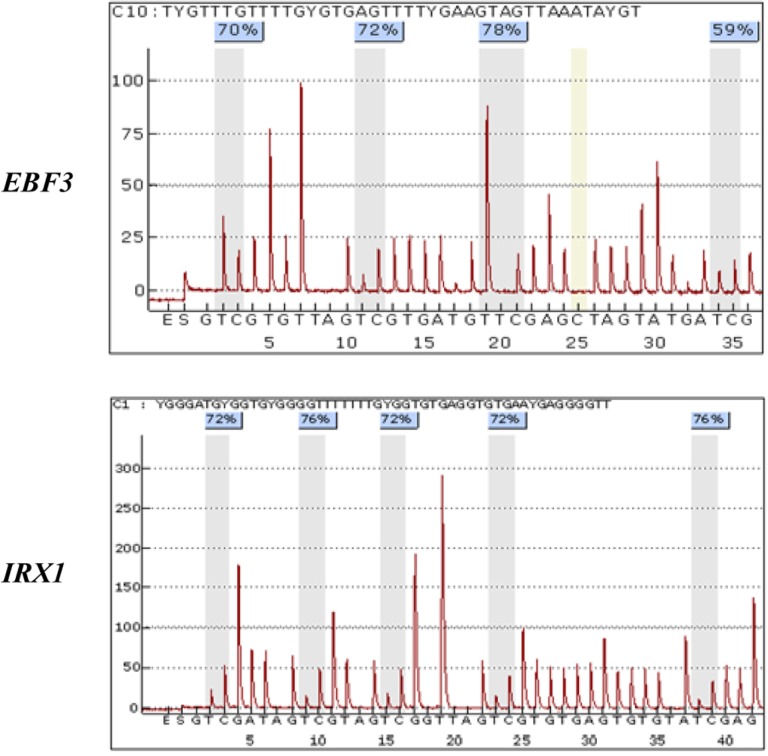
Fig. 3.
Pyrograms of EBF3 and IRX1 genes in RASFs. The methylation status of the two candidate genes was determined quantitatively on the PyroMark Q96MD System using the PS assay. The letters on the axis represent the order of dispensation; E, enzyme mix; S, substrate; A, G, C, and T, nucleotides. Shaded bars encompassing T/C (T, conversion of unmethylated C to U; C, non-conversion of methylated C) pairs indicate individual interrogated CpG motifs. The methylation of each CpG site was calculated as a percentage of C incorporation.
Fig. 4.
Methylation index and mRNA expression of EBF3 and IRX1 genes in healthy control SFs, OASFs and RASFs. (A) The methylation status of EBF3 and IRX1 genes was determined on genomic DNA obtained from six clinical samples by PS. We measured each gene in triplicate and used the average in the statistical analyses. MI, methylation index; HC, healthy control; OA, osteoarthritis; RA, rheumatoid arthritis. (B) Expression of EBF3 and IRX1 mRNAs was performed by RT-PCR on total RNA obtained from some representative samples. Amplification of GAPDH was used as an internal loading control.
Homeobox gene IRX1 is a member of the Iroquois family of genes, involved in patterning and regionalization of embryonic tissues in both vertebrates and invertebrates (Cavodeassi et al., 2001). IRX1 has also been identified as a tumor suppressor gene to be hypermethylated in several types of cancer (Bennett et al., 2008; Guo et al., 2010). EBF3 belongs to the EBF/Olf/Collier family of transcription factors, with specific roles in differentiation and maturation of several cell lineages, including B-progenitor lymphoblasts, neuronal precursors, and osteoblast progenitors (Liberg et al., 2002). In addition, aberrant DNA methylation and tumor suppressive activity of the EBF3 gene have been observed in various cancers (Liao, 2009). Of particular interest, the most prominent feature of RA is the progressive destruction of articular cartilage and bone, which is orchestrated by activated RASFs. These aggressive cells share many characteristics with tumor cells, with up-regulated expression of proto-oncogenes and pro-migratory adhesion molecules, increased production of pro-inflammatory cytokines and matrix-degrading enzymes (Huber et al., 2006), and increased resistance to apoptosis (Baier et al., 2003). In addition, RASFs can undergo a process resembling epithelial-mesenchymal transition (EMT), a phenomenon that occurs during carcinogenesis (Steenvoorden et al., 2006). Notably, IRX1 and EBF3 are downstream targets of SMAD2 and SMAD3, components of the transforming growth factor-β (TGF-β) pathway, respectively (Amir et al., 2006; Ferguson et al., 2001). Indeed, TGF-β and its receptor are up-regulated in RASFs, compared to OASFs, resulting in significantly enhanced expression of matrix-degrading enzymes and inflammatory mediators (Pohlers et al., 2007; Rosengren et al., 2010). TGF-β is also important for induction of fibrosis associated with RA (Pohlers et al., 2009). Taken together, it is reasonable to hypothesize that hypermethylation of tumor suppressor genes IRX1 and EBF3 by TGF-β signaling could lead to uncontrolled proliferation of SFs, apoptosis resistance, and gain of mesenchymal characteristics during RA progression. Therefore, further study will be required in order to clarify this issue.
In summary, although the current investigation was limited by a small number of samples and a virtual lack of direct interactions at molecular levels, results of this study have demonstrated that the CGIs in the EBF3 and IRX1 genes are specifically methylated to down-modulate their mRNA expression in rheumatoid synovial fibroblasts, which may shape the activated phenotype of these cells in RA pathogenesis.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011-0007659), and a grant from the Research Institute of Medical Science, Catholic University of Daegu (2009).
REFERENCES
- Amir S, Wang R, Matzkin H, Simons JW, Mabjeesh NJ. MSF-A interacts with hypoxia-inducible factor-1alpha and augments hypoxia-inducible factor transcriptional activation to affect tumorigenicity and angiogenesis. Cancer Res. 2006;66:856–866. doi: 10.1158/0008-5472.CAN-05-2738. [DOI] [PubMed] [Google Scholar]
- Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- Baier A, Meineckel I, Gay S, Pap T. Apoptosis in rheumatoid arthritis. Curr Opin Rheumatol. 2003;15:274–279. doi: 10.1097/00002281-200305000-00015. [DOI] [PubMed] [Google Scholar]
- Ballestar E. Epigenetic alterations in autoimmune rheumatic diseases. Nat Rev Rheumatol. 2011;7:263–271. doi: 10.1038/nrrheum.2011.16. [DOI] [PubMed] [Google Scholar]
- Bax M, van Heemst J, Huizinga TW, Toes RE. Genetics of rheumatoid arthritis: what have we learned? Immunogenetics. 2011;63:459–466. doi: 10.1007/s00251-011-0528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett KL, Karpenko M, Lin MT, Claus R, Arab K, Dyckhoff G, Plinkert P, Herpel E, Smiraglia D, Plass C. Frequently methylated tumor suppressor genes in head and neck squamous cell carcinoma. Cancer Res. 2008;68:4494–4499. doi: 10.1158/0008-5472.CAN-07-6509. [DOI] [PubMed] [Google Scholar]
- Carmona L, Cross M, Williams B, Lassere M, March L. Rheumatoid arthritis. Best Pract Res Clin Rheumatol. 2010;24:733–745. doi: 10.1016/j.berh.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Cavodeassi F, Modolell J, Gomez-Skarmeta JL. The Iroquois family of genes: from body building to neural patterning. Development. 2001;128:2847–2855. doi: 10.1242/dev.128.15.2847. [DOI] [PubMed] [Google Scholar]
- Ferguson CA, Tucker AS, Heikinheimo K, Nomura M, Oh P, Li E, Sharpe PT. The role of effectors of the activin signalling pathway, activin receptors IIA and IIB, and Smad2, in patterning of tooth development. Development. 2001;128:4605–4613. doi: 10.1242/dev.128.22.4605. [DOI] [PubMed] [Google Scholar]
- Firestein GS, Budd RC, Edward D, Harris J, McInnes IB, Ruddy S, Sergent JS. Kelly’s Textbook of Rheumatology. 8th ed. Elsevier; 2008. [Google Scholar]
- Fu LH, Ma CL, Cong B, Li SJ, Chen HY, Zhang JG. Hypomethylation of proximal CpG motif of interleukin-10 promoter regulates its expression in human rheumatoid arthritis. Acta Pharmacol Sin. 2011;32:1373–1380. doi: 10.1038/aps.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Liu W, Pan Y, Ni P, Ji J, Guo L, Zhang J, Wu J, Jiang J, Chen X, et al. Homeobox gene IRX1 is a tumor suppressor gene in gastric carcinoma. Oncogene. 2010;29:3908–3920. doi: 10.1038/onc.2010.143. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Oreffo RO, Gibson MB, Goldring MB, Roach HI. DNA demethylation at specific CpG sites in the IL1B promoter in response to inflammatory cytokines in human articular chondrocytes. Arthritis Rheum. 2009;60:3303–3313. doi: 10.1002/art.24882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber LC, Distler O, Tarner I, Gay RE, Gay S, Pap T. Synovial fibroblasts: key players in rheumatoid arthritis. Rheumatology (Oxoford) 2006;45:669–675. doi: 10.1093/rheumatology/kel065. [DOI] [PubMed] [Google Scholar]
- Karouzakis E, Gay RE, Gay S, Neidhart M. Epigenetic control in rheumatoid arthritis synovial fibroblasts. Nat Rev Rheumatology. 2009;5:266–272. doi: 10.1038/nrrheum.2009.55. [DOI] [PubMed] [Google Scholar]
- Liao D. Emerging roles of the EBF family of transcription factors in tumor suppression. Mol Cancer Res. 2009;7:1893–1901. doi: 10.1158/1541-7786.MCR-09-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberg D, Sigvardsson M, Akerblad P. The EBF/Olf/Collier family of transcription factors: regulators of differentiation in cells originating from all three embryonal germ layers. Mol Cell Biol. 2002;22:8389–8397. doi: 10.1128/MCB.22.24.8389-8397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Hsieh SC, Lin YC, Lee CN, Tsai MH, Lai LC, Chuang EY, Chen PC, Hung CC, Chen LY, et al. A whole genome methylation analysis of systemic lupus erythematosus: hypomethylation of the IL10 and IL1R2 promoters is associated with disease activity. Genes Immun. 2012;13:214–220. doi: 10.1038/gene.2011.74. [DOI] [PubMed] [Google Scholar]
- Molnar-Kimber KL, Kimber CT. Each type of cause that initiates rheumatoid arthritis or RA flares differentially affects the response to therapy. Med Hypotheses. 2012;78:123–129. doi: 10.1016/j.mehy.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Moon YH, Oh TJ, Kim NY, Kim MS, An S. Methylated DNA isolation assay-mediated DNA methylation detection and whole-genome methylation profiling. Am Biotechnol Lab. 2009;27:23–25. [Google Scholar]
- Neumann E, Lefevre S, Zimmermann B, Gay S, Muller-Ladner U. Rheumatoid arthritis progression mediated by activated synovial fibroblasts. Trends Mol Med. 2010;16:458–468. doi: 10.1016/j.molmed.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Nile CJ, Read RC, Akil M, Duff GW, Wilson AG. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 2008;58:2686–2693. doi: 10.1002/art.23758. [DOI] [PubMed] [Google Scholar]
- Ospelt C, Reedquist KA, Gay S, Tak PP. Inflammatory memories: is epigenetics the missing link to persistent stromal cell activation in rheumatoid arthritis? Autoimmun Rev. 2011;10:519–524. doi: 10.1016/j.autrev.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Pohlers D, Beyer A, Koczan D, Wilhelm T, Thiessen HJ, Kinne RW. Constitutive upregulation of the transforming growth factor-b pathway in rheumatoid arthritis synovial fibroblasts. Arthritis Res Ther. 2007;9:R59–R69. doi: 10.1186/ar2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlers D, Brenmoehl J, Loffler I, Muller CK, Leipner C, Schultze-Mosgau S, Stallmach A, Kinne RW, Wolf G. TGF-beta and fibrosis in different organs - molecular pathway imprints. Biochim Biophys Acta. 2009;1792:746–756. doi: 10.1016/j.bbadis.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengren S, Corr M, Boyle DL. Platelet-derived growth factor and transforming growth factor beta synergically potentiate inflammatory mediator synthesis by fibroblast-like synoviocytes. Arthritis Res Ther. 2010;12:R65–R75. doi: 10.1186/ar2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, Detmar M, Gay S, Kyburz D. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheumat. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- Steenvoorden MM, Tolboom TC, van der Pluijm G, Lowik C, Visser CP, DeGroot J, Gittenberger-DeGroot AC, DeRuiter MC, Wisse BJ, Huizinga TW, et al. Transition of healthy to diseased synovial tissue in rheumatoid arthritis is associated with gain of mesenchymal/fibrotic characteristics. Arthritis Res Ther. 2006;8:R165–R164. doi: 10.1186/ar2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Mol Biotechnol. 2010;44:71–81. doi: 10.1007/s12033-009-9216-2. [DOI] [PubMed] [Google Scholar]
- Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- Trenkmann M, Brock M, Ospelt C, Gay S. Epigenetics in rheumatoid arthritis. Clin Rev Allergy Immunol. 2010;39:10–19. doi: 10.1007/s12016-009-8166-6. [DOI] [PubMed] [Google Scholar]