Abstract
The structural flexibility of RNA interference (RNAi)-triggering nucleic acids suggests that the design of unconventional RNAi trigger structures with novel features is possible. Here, we report a cross-shaped RNA duplex structure, termed quadruple interfering RNA (qiRNA), with multiple target gene silencing activity. qiRNA triggers the simultaneous down-regulation of four cellular target genes via an RNAi mechanism. In addition, qiRNA shows enhanced intracellular delivery and target gene silencing over conventional siRNA when complexed with jetPEI, a linear polyethyleneimine (PEI). We also show that the long antisense strand of qiRNA is incorporated intact into an RNA-induced silencing complex (RISC). This novel RNA scaffold further expands the repertoire of RNAi-triggering molecular structures and could be used in the development of therapeutics for various diseases including viral infections and cancer.
Keywords: polyethyleneimine (PEI), quadruple interfering RNA (qiRNA), RNA interference, small interfering RNA (siRNA)
INTRODUCTION
RNA interference (RNAi) is a post-transcriptional gene silencing mechanism that is evolutionarily conserved across diverse species (Hannon, 2002). RNAi-mediated specific silencing of endogenous gene expression in mammalian cells using small interfering RNA (siRNA) has become a powerful tool for both functional genomics and therapeutic applications (Lares et al., 2010).
Early work emphasized the structural rigidity of RNAi-triggering nucleic acid structures (Elbashir et al., 2001), but several recent studies have shown that the molecular structure is much more variable than originally proposed (Chang et al., 2011a). Moreover, many of the non-classical RNAi-triggers have improved features over the classical siRNAs, including increased potency, reduced nonspecific responses, and enhanced cellular delivery.
The simultaneous silencing of multiple target genes is important for the therapeutic targeting of diseases such as cancer and viral infections. Normal gene changes to oncogenes lead to cancer formation, and this genetic complexity enables cancer cells to escape mono-target therapy. This is also true for viruses due to the rapid mutation rate that occurs during viral genome replication. Therefore, RNAi-triggering molecules with multi-targeting activity could be useful in reducing escape mutations in anti-viral therapy or executing the combinatorial inhibition of oncogenes in anti-cancer therapy (Boden et al., 2003).
To this end, we have previously developed tripodal RNAi-triggering structures that simultaneously silence the expression of three target genes (Chang et al., 2012a; 2012b). These triggers, termed tiRNAs, also exhibited enhanced intracellular delivery compared with conventional siRNAs when complexed with jetPEI, a linear polyethyleneimine (PEI), a widely used cationic polymer carrier. The branched RNA duplex structure was recognized by RNAi machinery, which triggered efficient RNAi-mediated gene silencing.
Here, we introduce four-armed, cross-shaped RNA duplexes, termed quadruple interfering RNAs (qiRNAs), which can trigger the efficient and simultaneous silencing of four cellular target genes. Moreover, similar to tiRNAs, qiRNA-mediated gene silencing was superior to conventional siRNAs when complexed with jetPEI as a result of enhanced intracellular delivery efficiency. This cross-shaped RNA scaffold could be potentially utilized as a platform for therapeutics targeting diseases such as viral infections and cancer.
MATERIALS AND METHODS
Oligonucleotides
Chemically synthesized DNAs and RNAs were obtained from Bioneer or BMT and annealed according to the manufacturer’s protocol. RNA and DNA sequences used in the experiments are shown below:
(1) siRNA; siSurvivin sense: 5′-AAGGAGAUCAACAUUUUCA(dTdT)-3′; siSurvivin antisense: 5′-UGAAAAUGUUGAUCUCCUU(dTdT)-3′; siβ-catenin sense: 5′-GUAGCUGAUAUUGAUGGAC(dTdT)-3’; siβ-catenin antisense: 5′-GUCCAUCA AUAUCAGCUAC(dTdT)-3′; siSTAT3 sense: 5′-CGUCAUUAGCAGAAUCUCA(dTdT)-3′; siSTAT3 antisense: 5′-UGAGAUUCUGCUAAUGACG(dTdT)-3′; siMET sense: 5′-CGAGAUGAAUGUGAAUAUGAA-3′; siMET antisense: 5′-CAUAUUCACAUUCAUCUCGGA-3′. (2) qiRNA; First strand: 5′-UGAAAAUGUUGAUCUCCUUGUAGCUGAUAUUGAUGGAC-3′; Second strand: 5′-GUCCAUCAAUAUCAGCUACCGUCAUUAGCAGAAUCUCA-3′; Third strand: 5′-UGAGAUUCUGCUAAUGACGCGAGAUGAAUGUGAAUAUG-3′; Fourth strand: 5′-CAUAUUCACAUUCAUCUCGAAGGAGAUCAACAUUUUCA-3′. (3) qiDNA; First strand: 5′-TGAAAATGTTGATCTCCTTGTAGCTGATATTGATGGAC-3′; Second strand: 5′-GTCCATCAATATCAGCTACCGTCATTAGCAGAATCTCA-3′; Third strand: 5′-TGAGATTCTGCTAATGACGCGAGATGAATGTGAATATG-3′; Fourth strand: 5′-CATATTCACATTCATCTCGAAGGAGATCAACATTTTCA-3′. (4) qiRNA-OMe(16); First strand: 5′-UGAAAAUGUUGAUCUCCUUGUAGCUGAUAUUGAUGGAC-3′; Second strand: 5′-GUCCAUCAAUAUCAGCUACCGUCAUUAGCAGAAUCUCA-3′; Third strand: 5′-UGAGAUUCUGCUAAUGACGCGAGAUGAAUGUGAAUAUG-3′; Fourth strand: 5′-CAUAUUCACAUUCAUCUCGAAGGAGAUCAACAUUUUCA-3′; Positions of the 2′-O-methyl modified ribonucleotides are underlined.
The sequence of tiRNA used in this study has previously been reported (Chang et al., 2012a).
Native gel electrophoresis
Each RNA variant was separated on a 10% (w/v) nondenaturing polyacrylamide gel, stained with ethidium bromide (EtBr) and visualized using UV transillumination.
Cell culture and RNA transfection
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (FBS). Cells were seeded in 12-well plates 24 h before transfection at 60% confluency in complete medium without antibiotics. siRNAs and qiRNA were transfected using jetPEI reagent (Polyplus) with five nitrogen residues of jetPEI per DNA phosphate (N/P = 5), according to the manufacturer’s protocol.
HEK293T cells were grown in DMEM (Welgene) supplemented with 10% FBS (Welgene). The day before transfection, cells were seeded in 6-well plates to achieve approximately 50% confluency by the time of transfection. Cells were co-transfected with 10 nM of qiRNA and 1.25 μg of p3xFlag-hAGO2 using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. For the siRNA control, 10 nM of each constituent siRNA (siSurvivin, siβ-catenin, siSTAT3, and siMET) was combined to adjust the final concentration of the RNA duplex moiety to 40 nM.
Quantitative real-time PCR (qRT-PCR)
Total RNAs were extracted from cell lysates using the Isol-RNA Lysis Reagent kit (5 Prime). They were then used as templates for cDNA synthesis, which was performed using the ImProm-II™ Reverse Transcription System (Promega) according to the manufacturer’s protocol. Target gene expression levels were analyzed by qRT-PCR using a StepOne real-time PCR system (Applied Biosystems) according to the manufacturer’s protocol. The primer sequences for each gene were as follows: GAPDH-forward: 5′-GAGTCAACGGATTTGGTCGT-3′; GAPDH-reverse: 5′-GACAAGCTTCCCGTTCTCAG-3′; Survivin-forward: 5′-GCACCACTTCCAGGGTTTAT-3′; Survivin-reverse: 5′-CTCTGGTGCCACTTTCAAGA-3′; β-catenin-forward: 5′-ATGTCCAGCGTTTGGCTGAA-3′; β-catenin-reverse: 5′-TGGTCCTCGTCATTTAGCAG-3′; STAT3-forward: 5′-GATCCAGTCCGTGGAACCAT-3′; STAT3-reverse: 5′-ATAGCCCATGATGATTTCAGCAA-3′; MET-forward: 5′-TGGTGCAGAGGAGCAATGG-3′; MET-reverse: 5′-CATTCTGGATGCGTGTTTCC-3′.
The 5′- rapid amplification of cDNA ends (RACE)
Eighteen hour after siRNA transfection into HeLa cells, total RNAs were extracted using the Isol-RNA Lysis Reagent kit (5 Prime). Total RNAs (7 μl) were ligated to 0.25 μg of the GeneRacer RNA oligo and reverse transcribed using GeneRacer oligo dT and the Superscript III RT kit (Invitrogen). RNA oligoligated mRNAs were amplified using gene-specific primers (see below) and the resulting PCR products were cloned into the T&A cloning vector (RBC) before being sequenced using the M13 forward primer.
Survivin gene-specific primer: 5′-CAACAGGCACCTGCCAAGTCCACA- 3′; Survivin gene-specific nested primer: 5′-AGCACCCGCTGCACAGGCAGAAGCA-3′; β-catenin gene-specific primer: 5’-GTACGTACAATAGCAGACACCATCT-3′; β-catenin gene-specific nested primer: 5′-TAGTGGGATGAGCAGCATCAAACTGT-3′; STAT3 gene-specific primer: 5′-GCCGTTGTTGGATTCTTCCATGTTC-3′; STAT3 gene-specific nested primer: 5′-GTTTCTAAACAGCTCCACGATTCTCT-3’; MET gene-specific primer: 5′-GAACCTCCGACTGTATGTCAGCAG-3′; MET gene-specific nested primer: 5′-GTACTCAGCAACCTTCTGAAGGTCTT-3′.
Intracellular uptake analysis
To visualize the cellular uptake of RNAs, HeLa cells were transfected with Cy3-labeled siRNA variants using jetPEI (N/P = 5). At indicated times after transfection, cells were harvested and washed twice with phosphate-buffered saline (PBS) before being visualized using a fluorescent microscope (Olympus). The level of intracellular uptake was quantified using the NucleoCounter NC-3000 (Chemometec).
Stability test of the siRNA/PEI complex
Serum stability of siRNA and qiRNA variant RNA (0.1 nmole) was assessed by incubation in 30 μl of 10% FBS solution. Eight μl of each sample was taken at the indicated time points and immediately frozen at −70°C. Before the loading of the RNA/PEI complex, each sample was incubated with 1 μl (5 μg/μl) of heparin sodium salt (Sigma Aldrich) for 10 min. A 7 μl aliquot of each sample was then separated on a 10% (wt/vol) nondenaturing polyacrylamide gel, stained with EtBr, and visualized by UV transillumination.
Ago2 incorporation assay
Twenty-four hour post-transfection, cells were washed twice with ice-cold 1× PBS and harvested by scraping. The cells were resuspended in NET-2 buffer [100 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% NP-40, 1 mM DTT, 1× protease inhibitor cocktail (Roche), 1 U/ml RNaseOUT (Invitrogen)] and incubated on ice for 20 min. The lysate was further sonicated on ice for 1 min and subjected to centrifugation at 15,000 rpm at 4°C for 10 min. The supernatant was incubated with 20 μl of anti-Flag antibody-conjugated agarose beads (Sigma) with constant rotation at 4°C for 2 h. The beads were washed four times with NET-2 buffer and treated with TRI Reagent (Ambion) to extract coimmunoprecipitated RNAs. The RNAs recovered from input or Flag-immunoprecipitate samples were resolved on a 15% urea-polyacrylamide gel, transferred to a Hybond NX membrane (GE Healthcare), and crosslinked using the EDC-mediated method. DNA oligonucleotides complementary to the guide strand of siSurvivin (5′-AAGGAGATCAACATTTTCA-3′), let-7a-1 (5′-AACTATACAACCTACTACCTCA-3′), or U6 snRNA (5′-TTGCGTGTCATCCTTGCGCAGG-3′) were radiolabeled at the 5′ ends and used as probes for Northern blotting. The membrane was exposed to a phosphorimager and read using the BAS-2500 system (Fujifilm).
Inhibition of RNA induced silencing complex (RISC) loading by aurintricarboxylic acid (ATA)
Twenty-four hour after seeding, HeLa cells were treated with ATA (50 μM), incubated for 24 h, then transfected with siRNA or qiRNA (50 nM) for an additional 24 h. Gene silencing activities were analyzed by qRT-PCR.
RESULTS AND DISCUSSION
Structure of quadruple interfering RNA (qiRNA) with four siRNA units
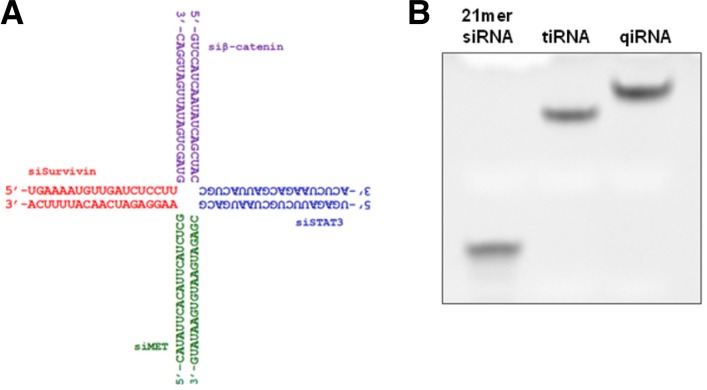
qiRNA was constructed by annealing four chemically synthesized 38 nt single-stranded (ss) RNAs. This qiRNA structure harbors the four 19 bp siRNA units siSurvivin, siβ-catenin, siSTAT3 and siMET, which target Survivin, β-catenin, STAT3 and c-MET mRNAs, respectively (Fig. 1A). The qiRNA structure was designed so that the 5′-end of the antisense strand of each siRNA unit was orientated towards the outside, as our previous studies showed that this arrangement maintains the gene silencing activity of the individual siRNA units in longer RNA duplexes (Chang et al., 2007; 2012b). Native gel electrophoresis confirmed the integrity of the qiRNA structure, as it showed a single band migrating slower than the siRNA or tiRNA (which is composed of three 38 nt ssRNAs; Chang et al., 2012a) (Fig. 1B).
Fig. 1.
Structure of qiRNA. (A) Structure of a quadruple-interfering RNA (qiRNA) that targets Survivin, β-catenin, STAT3 and MET mRNAs. The qiRNA was constructed by annealing four 38 nt ssRNAs. (B) Gel electrophoresis analysis of qiRNA.
qiRNA triggers specific and potent target gene silencing
We first tested whether this qiRNA nucleic acid structure could trigger specific target gene silencing via RNAi. The assembled qiRNA was transfected into HeLa cells using jetPEI, a widely used transfection reagent for nucleic acid delivery (Aigner, 2006; Urban-Klein et al., 2005), and the mRNA levels of four target genes and GAPDH (internal control) were measured by qRTPCR.
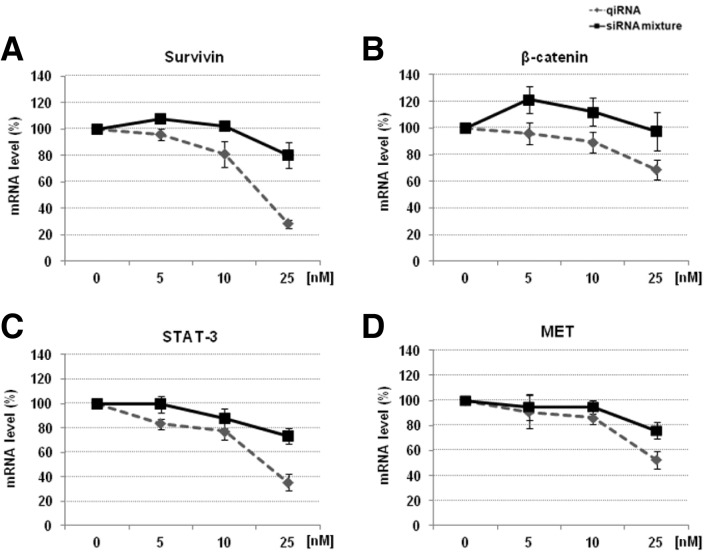
The target gene silencing activity of a mixture of four siRNAs (siSurvivin/siβ-catenin/siSTAT3/siMET) was found to be rather poor when transfected with jetPEI (Fig. 2), as observed in our previous experiments with tiRNAs (Chang et al., 2012a; 2012b). This was to be expected as linear PEI is not an optimal transfection reagent for siRNA (Lee et al., 2010). In contrast, qiRNA induced more potent target gene silencing than the siRNA mixture, with the most significant difference observed at a concentration of 25 nM (Fig. 2). These results demonstrate that the qiRNA structure not only induces simultaneous silencing of four endogenous target genes, but also shows enhanced silencing efficiency compared with conventional siRNAs when complexed with a cationic polymer delivery vehicle.
Fig. 2.
qiRNA simultaneously silences four target genes with increased activity over an siRNA mixture. Gene silencing activity of the qiRNA that targets Survivin, β-catenin, STAT3 and MET mRNAs. The small interfering RNA (siRNA) mixture and qiRNA were transfected into HeLa cells using jetPEI (N/P = 5) at the indicated concentration; the medium was exchanged after 3 h. (A) Survivin, (B) β-catenin, (C) STAT3 and (D) MET mRNA levels were analyzed by qRT-PCR 24 h after transfection and plotted relative to GAPDH (control) mRNA levels. Data represent the mean ± SD of three independent experiments.
qiRNA triggers gene silencing via an RNAi mechanism
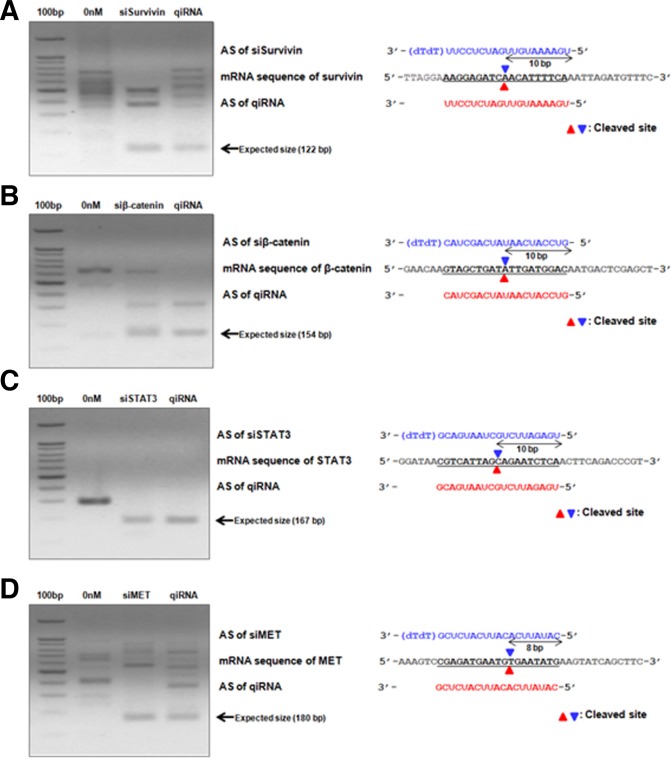
To confirm whether qiRNA triggers gene silencing via an RNAi mechanism similar to conventional siRNAs, we performed 5′-RACE to analyze the cleavage sites of each target mRNA caused by qiRNA. 5′-RACE PCR products from qiRNA-transfected cells were similar in size to those from siRNA mixture-transfected cells (Fig. 3). For a more detailed analysis of cleavage sites, 5′-RACE PCR products from each reaction were cloned and three independent clones from each reaction were sequenced. Both the siRNA mixture and the qiRNA cleaved target mRNAs at the same site: 8 or 10 nt downstream from the 5′-end of the antisense strand of each siRNA unit (Fig. 2). These results suggest that the qiRNA triggers target gene silencing by the same RNAi pathway utilized by conventional siRNAs.
Fig. 3.
qiRNA works via an RNAi mechanism. The 5′-RACE assays were performed using a 21-bp siRNA mixture or qiRNA-transfected cells. (A) PCR products of a 5′-RACE assay amplified using Survivin gene-specific primers. The Survivin antisense sequence of qiRNA is shown in red and cleaved sites are marked with red arrowheads. (B) PCR products of a 5′-RACE assay amplified using β-catenin gene-specific primers, STAT3 gene-specific primers (C), and MET gene-specific primers (D).
To further confirm the RNAi-mediated gene silencing by qiRNA, we examined the effect of ATA, a small molecule that inhibits the loading of siRNA to endogenous Ago2 (Tan et al., 2012), on the gene silencing activity of qiRNA. HeLa cells were treated with 50 μM ATA for 24 h before RNA transfection. As shown in Supplementary Fig. S1, the qiRNA gene silencing activity for four target genes was completely abolished following ATA pretreatment, suggesting that ATA inhibits the loading of qiRNAs to Ago2. These results again support the idea that the gene silencing activity of qiRNA is mediated by an RNAi mechanism.
jetPEI-complexed qiRNA shows increased cellular delivery compared with siRNA
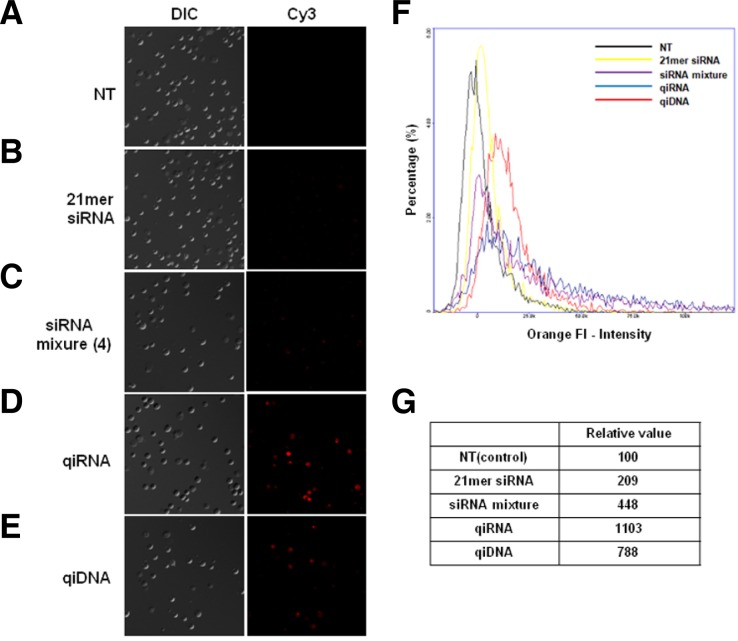
The enhanced gene silencing potency of the qiRNA/jetPEI complex compared with the siRNA/jetPEI complex might be caused by increased cellular delivery. To test this hypothesis, we compared the jetPEI-mediated delivery efficiency of Cy3-labeled siRNA and qiRNA using fluorescence microscopy. As shown in Figs. 4B and 4C, HeLa cells incubated with Cy3-labeled single siRNA or a siRNA mixture (siSurvivin/siβ-catenin/ siSTAT3/siMET) showed weak fluorescence. In contrast, Cy3-labeled qiRNA showed strong fluorescence intensity with scattered spots in intracellular regions (Fig. 4D). These results demonstrate that the qiRNA structure is more efficiently delivered to cells than siRNA.
Fig. 4.
qiRNA is more efficiently delivered into cells than siRNAs. The 3′ end of the Survivin sense strand in a 19-bp siRNA mixture (25 nM each) and a qiRNA (25 nM) were labeled with Cy3 and transfected into HeLa cells using polyethyleneimine (PEI). (A) Non-transfected HeLa cells showed no fluorescence intensity. HeLa cells transfected with Cy3-labeled 21-bp siRNA (B) and the four siRNA mixture (C) showed similarly weak fluorescence, but cells transfected with Cy3-labeled qiRNA (D), and qiDNA (E) showed strong fluorescence with scattered fluorescent spots. (F) The flow cytometry data using NC-3000. (G) Relative quantification analysis of (F).
We next tested the DNA counterpart of qiRNA (termed qiDNA) to determine whether the observed enhanced intracellular delivery is a feature of the cross-shaped nucleic acid structure, irrespective of the type of nucleic acid (DNA or RNA). qiDNA also showed increased intracellular delivery efficiency (Fig. 4E), so we conclude that the increased gene silencing activities of jetPEI-delivered qiRNA over conventional siRNA comes from the increased delivery efficacy which is a unique feature of the cross-shaped RNA structure.
jetPEI protects the qiRNA structure from degradation by nucleases
As qiRNA has a branched structure, its nuclease susceptibility might differ from that of linear siRNAs. To test the serum nuclease stability of the qiRNA structure, qiRNA or siRNA was incubated in 10% serum solution and visualized by polyacrylamide gel electrophoresis and EtBr staining. siRNA was gradually degraded upon incubation and was almost undetectable after 6 h (Supplementary Fig. S2A, upper panel). Interestingly, incubation of qiRNA in serum rapidly yielded smaller dsRNA products of a similar size to siRNA (Supplementary Fig. S2A, middle panel). This observation suggests that the naked qiRNA is vulnerable to nuclease attack. We were able to stabilize qiRNA structure by incorporating 16 consecutive 2′-OMe modified nucleotides in the middle of each qiRNA strand (qiRNA-OMe) (Supplementary Fig. S2A, lower panel), suggesting that the junction part is particularly susceptible to the nuclease attack.
However, when qiRNA was complexed with jetPEI, it maintained an intact form upon incubation with serum for up to 6 h (Supplementary Fig. S2B, lower panel), demonstrating that the PEI complex affords protection from nuclease attack within serum. This suggests that following transfection, qiRNA enters cells as an intact structure.
The 38 nt-long antisense strand of qiRNA is incorporated into Ago2
As we propose that the intact form of qiRNA is introduced into cells, it is conceivable that the 38 nt-long antisense strand of qiRNA is incorporated into RISCs to trigger RNAi. Thus we investigated whether the long antisense strand of qiRNA is stably incorporated into the Argonaute (Ago) protein in vivo to form a mature RISC. Because the naked qiRNA was vulnerable to nuclease attack, we used qiRNA-OMe for this experiment. qiRNA-OMe showed similar gene silencing activity compared with naked qiRNA (Supplementary Fig. S3), suggesting that the internal OMe modification does not significantly affect the RNAi activity of qiRNA. Following transfection of qiRNA-OMe into HEK293T cells together with the Flag-hAgo2 expression plasmid, RNAs associated with Flag-hAgo2 were extracted from the Flag-immunoprecipitate and subjected to Northern blot analysis to detect one guide strand of the qiRNA (Fig. 5). The four siRNA mixture constituent of the qiRNA was used as a control transfection. The guide strand of siSurvivin incorporated into Flag-hAgo2 was detected at ∼20 nt as expected, demonstrating the reliability of the assay. Notably, the major signal for the hAgo2-associated guide strand of the qiRNA was detected at ∼38 nt, which is to be expected if the strand is bound by hAgo2 in its intact form. This result demonstrates that 38 nt-long antisense strands of qiRNA can be incorporated into Ago2 and trigger RNAi in mammalian cells.
Fig. 5.
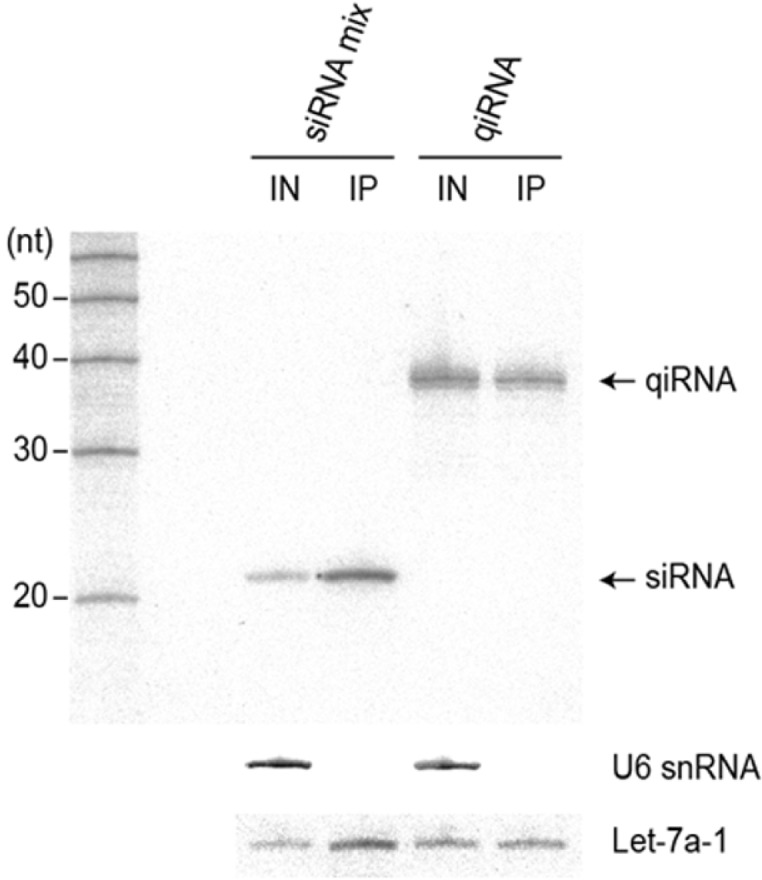
The long antisense strand of qiRNA is incorporated into RISC. Immunoprecipitation of Flag-hAGO2 in HEK293T cells transfected with qiRNA-OMe followed by northern blot analysis to detect the RNA strand targeting the Survivin gene. IN and IP represent input cell lysate (10%) and Flag-immunoprecipitate, respectively. Endogenous let-7a-1 and U6 snRNA serve as positive and negative controls for Ago immunoprecipitation, respectively.
The efficient delivery of RNAi moieties into cells is necessary for effective target gene silencing. However, conventional siRNAs bind inefficiently to cationic polymer vehicles such as jetPEI because of the smaller number of negative charges within the siRNAs. This can result in the exchange of siRNAs with the negatively charged proteoglycans at the cell surface upon transfection, resulting in compromised gene silencing activity. This problem can be overcome by increasing the total size of the RNAi-triggering molecular structure, for example, polymeric siRNA (Mok et al., 2010) or long interfering RNA (Chang et al., 2011b). We also previously showed that tiRNA, which harbors three siRNA units, triggers the simultaneous silencing of three target genes with increased delivery efficiency using jetPEI (Chang et al., 2012a; 2012b). This finding suggests that a branched RNA structure forms a more stable complex with jetPEI than conventional siRNA.
In this study, we designed another branched RNA structure, qiRNA, which behaves very similarly to tiRNA in terms of enhanced cellular delivery by jetPEI. Together, these observations strongly suggest the presence of a unique mode of interaction between branched small RNA molecules and jetPEI to form a stronger complex. Computational and further biophysical studies will elucidate the details of this.
The hAgo2 incorporation assay findings show that the qiRNA guide strand is stably loaded into hAgo2 in vivo in its full-length form (∼38 nt; Fig. 5). Although the typical guide RNA size incorporated into Ago-family proteins falls within ∼20–30 nt (Czech and Hannon, 2011), our data suggest that Ago proteins are structurally more flexible in accommodating guide RNAs than previously thought. Indeed, it has been reported that mammalian Ago2 forms a mature RISC using precursor microRNAs (pre-miRNAs) or even long unstructured single-stranded RNAs as guides (Diederichs and Haber, 2007; Tan et al., 2009). A conserved vertebrate miRNA, miR-451, is encoded by a 42-nt hairpin precursor, which is directly loaded into and processed by Ago2 to generate mature miR-451 (Cheloufi et al., 2010; Cifuentes et al., 2010; Yang et al., 2010).
The position of the Ago-catalyzed cleavage site in the target mRNA is dictated by its distance from the 5′ end phosphate of the guide RNA, which is tightly anchored in a 5′-end binding pocket composed of residues from the MID domain and the PIWI domain (Elkayam et al., 2012; Jinek and Doudna, 2009; Schirle and MacRae, 2012). Given that the qiRNA directs cleavage of the target mRNA at the same position as the siRNA (Fig. 3), it is likely that the mode of Ago2 recognition of the 5′ end of the guide strand would be similar between qiRNA and siRNA.
In summary, our qiRNA study findings further expand the structural diversity repertoire of RNAi-triggering molecular structures and present more evidence of the flexibility of human RISCs in guiding RNA interactions. The multi-targeting feature of qiRNA together with the enhanced cellular delivery makes it an attractive candidate for the future development of novel RNAi therapeutics targeting multiple diseases such as viral infections or cancer.
Acknowledgments
This work was supported by a Global Research Laboratory grant from the Ministry of Education, Science and Technology (MEST) of Korea (no. 2008-00582) to D-K. L., by Korea Ministry of Environment as “EI project” (E211-41003-0007-0) to S. K., by a Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the MEST (2010-0022096) to C. S., and by the NRF to D. H.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Aigner A. Gene silencing through RNA interference (RNAi) in vivo: strategies based on the direct application of siRNAs. J Biotechnol. 2006;124:12–25. doi: 10.1016/j.jbiotec.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CI, Hong SW, Kim S, Lee DK. A structure-activity relationship study of siRNAs with structural variations. Biochem Biophys Res Commun. 2007;359:997–1003. doi: 10.1016/j.bbrc.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Chang CI, Andrade H, Dua P, Kim S, Li CJ, Lee DK. Structural diversity repertoire of gene silencing siRNAs. Nucleic Acid Ther. 2011a;21:125–131. doi: 10.1089/nat.2011.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CI, Lee TY, Dua P, Kim S, Li CJ, Lee DK. Long dsRNA-mediated RNA interference and immunostimulation: long interfering RNA (liRNA) as a potent anticancer therapeutics. Nucleic Acid Ther. 2011b;21:149–155. doi: 10.1089/nat.2011.0296. [DOI] [PubMed] [Google Scholar]
- Chang CI, Lee TY, Yoo JW, Shin D, Kim M, Kim S, Lee DK. Branched, tripartite-interfering RNAs silence multiple target genes with long guide strands. Nucleic Acid Ther. 2012a;22:30–39. doi: 10.1089/nat.2011.0315. [DOI] [PubMed] [Google Scholar]
- Chang CI, Lee TY, Kim S, Sun X, Hong SW, Yoo JW, Dua P, Kang HS, Kim S, Lee D, et al. Enhanced intracellular delivery and multi-target gene silencing triggered by tripodal RNA structures. J Gene Med. 2012b;14:138–146. doi: 10.1002/jgm.1653. [DOI] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A Dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. Small RNA sorting matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA. Dual role for Argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The structure of human Argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- Lares MR, Rossi JJ, Ouellet DL. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010;28:570–579. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Huh MS, Lee SK, Lee SJ, Chung HJ, Park JH, Oh YK, Choi KW, Kim KM, Kwon IC. Stability and cellular uptake of polymerized siRNA (poly-siRNA)/polyethylenimine (PEI) complexes for efficient gene silencing. J. Control Release. 2010;141:339–346. doi: 10.1016/j.jconrel.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Mok H, Lee SH, Park JW, Park TG. Multimeric small interfering ribonucleic acid for highly efficient sequence-specific gene silencing. Nat Mater. 2010;9:272–278. doi: 10.1038/nmat2626. [DOI] [PubMed] [Google Scholar]
- Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GS, Garchow BG, Liu X, Yeung J, Morris JP, 4th, Cuellar TL, McManus MT, Kiriakidou M. Expanded RNA-binding activities of mammalian Argonaute2. Nucleic Acids Res. 2009;37:7533–7545. doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GS, Chiu C, Garchow BG, Metzler D, Diamond SL, Kiriakidou M. Small molecule inhibition of RISC loading. ACS Chem Biol. 2012;7:403–410. doi: 10.1021/cb200253h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]