Abstract
We performed activation tagging screen to isolate abscisic acid (ABA) response mutants. One of the mutants, designated ahs10 (ABA-hypersensitive10), exhibited ABA-hypersensitive phenotypes. TAIL-PCR analysis of the mutant revealed that T-DNA was inserted in the promoter region of the Arabidopsis gene, At2g01430, which encodes a homeodomain-leucine zipper protein ATHB17. Subsequent expression analysis indicated that ATHB17 was activated in ahs10. To recapitulate the mutant phenotypes, we prepared ATHB17 OX lines and investigated their phenotypes. The results showed that ATHB17 confers ABA-hypersensitivity and drought tolerance. On the contrary, ATHB17 knockout lines were ABA-insensitive and drought-sensitive, further demonstrating that ATHB17 is involved in ABA and water-stress responses. Interestingly, the ATHB17 effect on seedling growth in the presence of ABA was observed only during the postgermination seedling establishment stage, suggesting that it functions during a narrow developmental window of early seedling growth.
Keywords: abscisic acid (ABA), activation tagging, drought tolerance, HD-Zip protein
INTRODUCTION
Homeodomain-leucine zipper (HD-Zip) proteins belong to a subclass of homeodomain (HD) proteins that are characterized by the presence of a leucine zipper (Zip) motif downstream of the HD (Ariel et al., 2007; Harris et al., 2011; Mukherjee et al., 2009). The HD in HD-Zip proteins is involved in DNA binding, whereas the Zip domain is involved in dimerization of the proteins. Thus, HD-Zip proteins are transcription factors that function as homo- or hetero-dimers.
HD-Zip proteins are unique to the plant kingdom (Schena and Davis, 1994; Schena et al., 1993). Genome-wide analysis of the HD proteins (Mukherjee et al., 2009) revealed that there are 48 HD-Zip proteins in the Arabidopsis genome. In the rice genome, 47 HD-Zip proteins have been identified, and a similar number of HD-Zip proteins are also found in other plant genomes (Mukherjee et al., 2009). The Arabidopsis HD-Zip proteins are classified into four different classes based on their amino acid sequence similarity: HD-Zip I, HD-Zip II, HD-Zip III, and HD-Zip IV. Several of the 16 member HD-Zip IV proteins are known to be developmental regulators (Nakamura et al., 2006). ATHB10/GLABRA2, PDF2, and ATML1 are involved in epidermal cell (e.g, root hair and trichome cells) differentiation. Similarly, HDG11 is involved in trichome development, and ANL2 affects root development as well as anthocyanin accumulation in leaf subepidermal cells. HD-Zip III subfamily proteins, which consist of 5 family members (i.e., ATHB14/PHB, ATHB9/PHV, REV/IFL1, ATHB15/CAN, and ATHB8), are also involved in the regulation of developmental processes such as embryo patterning, meristem formation, leaf polarity, and vascular development (Prigge et al., 2005). On the other hand, the HD-Zip I subfamily is composed of 17 members, and many of them are known to modulate plant growth in response to developmental and environmental cues (Harris et al., 2011; Henriksson et al., 2005). For instance, ATHB7, ATHB12, and ATHB13 affect plant growth under abiotic stress conditions (Olsson et al., 2004; Son et al., 2010). ATHB6, ATHB5, and ATHB20 are regulators of a subset of ABA response (Barrero et al., 2010; Himmelbach et al., 2002; Johannesson et al., 2003). ATHB1 and ATHB16 regulate light-dependent processes such as leaf development and hypocotyl elongation (Aoyama et al., 1995; Wang et al., 2003). Relatively little is known about the roles of the 10 member HD-Zip II proteins, except for only a few (Ciarbelli et al., 2008; Harris et al., 2011). ATHB2/HAT4, HAT1 and HAT2 function in shade avoidance, photoperiodic control of growth, or auxin-mediated morphogenesis (Ciarbelli et al., 2008; Sawa et al., 2002). Another HD-Zip II protein, ATHB4, has also been reported to have similar function (Sorin et al., 2009).
Here, we report the function of ATHB17, which is a HD-Zip II protein. In the course of our activation tagging screen to isolate ABA response mutants, we isolated a mutant that was hypersensitive to ABA. Subsequent analysis showed that ATHB17 expression was activated in the mutant, and we present the data demonstrating that ATHB17 is involved in a subset of ABA response.
MATERIALS AND METHODS
Plant growth and mutant isolation
Arabidopsis plants (ecotype Columbia, Col-0, and Landsbergerecta, Ler) were used throughout this study. Plants were grown under long day condition (16-h-light/8-h-dark cycle) at 22°C. For aseptic growth, seeds were treated with 70% ethanol for 1 min and with 30% bleach for 1 min and were then washed with sterile water several times before planting. For soil growth, seeds were placed on the 1:1:1 (by weight) mixture of vermiculite, perlite and peat moss, which were irrigated with 0.1% Hyponex (Hyponex Co., USA). The seed pots were then placed at 4°C for 5 days in the dark to break residual seed dormancy and transferred to the normal growth condition. Unless stated otherwise, plants were watered once a week.
The generation of activation-tagged transgenic plants and screen for ABA response mutants have been described before (Park et al., 2011). For the analysis of the ahs10 mutant phenotypes shown in Fig. 1, we employed one of the heterozygous sublines, because homozygous lines set seeds very poorly. The T-DNA insertion site in the ahs10 mutant was determined by TAIL-PCR (Liu et al., 1995). The insertion site was then confirmed by sequencing the DNA fragment amplified using TDNA border sequences and gene-specific primers. For the RT-PCR analysis of ahs10 in Fig. 1E, the primer set, FP1 and RP1, was used. Other primer sequences used in the mutant analysis are available upon request.
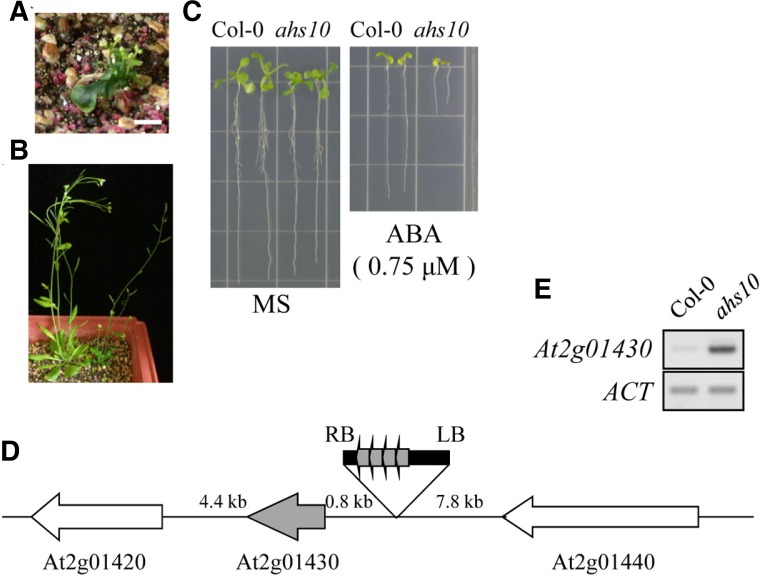
Fig. 1.
Isolation of the ABA-hypersensitive mutant ahs10. (A) Homozygous ahs10 grown in soil for 5 weeks. (B) Heterozygous ahs10 plants grown in soil for 5 weeks. (C) ABA sensitivity of ahs10. Wild type (Col-0) or ahs10 was grown in a MS medium with or without ABA (0.75 μM). (D) T-DNA insertion site in ahs10 is shown schematically. RB and LB indicate border sequences, and the 35S enhancers are indicated by boxed arrows. (E) RT-PCR analysis of ATHB17 expression in ahs10. ACT, actin-1.
Generation of transgenic lines and phenotype analysis
To construct the ATHB17 OX lines, the entire coding region of HD-Zip17 was amplified using the primer set FP2 and RP2. The DNA fragment was then digested with Xba I and SacI and was cloned into pBI121 (Jefferson et al., 1987), which was prepared by removing the GUS coding after the XbaI-SacI digestion. We recovered ten T3 generation homozygous lines, and, for phenotype analysis, T4 generation amplified seeds from representative lines were used.
To prepare the promoter-GUS reporter construct, 2.5 kb of the 5′ flanking sequence of ATHB17 was amplified using the primers FP3 and RP3 and was cloned into the BamH1 and SalI sites of pBI101.2 after digestion of the vector with the same enzymes.
The constructs were introduced into A. tumefaciens strain GV3101, and Arabidosis plants, Col-0 for the promoter-GUS lines and Ler for the OX lines, were transformed by the method of Bechtold and Pelletier (1998).
The KO lines (SALK_075942 and SALK_128560) were obtained from the Arabidopsis Stock Center. The seeds were amplified, and homozygous knockout sublines were recovered from plants whose progeny segregated with a 3:1 ratio of kanamycin resistance and kanamycin susceptible seeds. T-DNA insertion at the annotated position was confirmed by genomic PCR and subsequent sequencing of the amplified fragments. For the expression analysis of ATHB17 shown in Fig. 3B, the primers FP1 and RP1 were used.
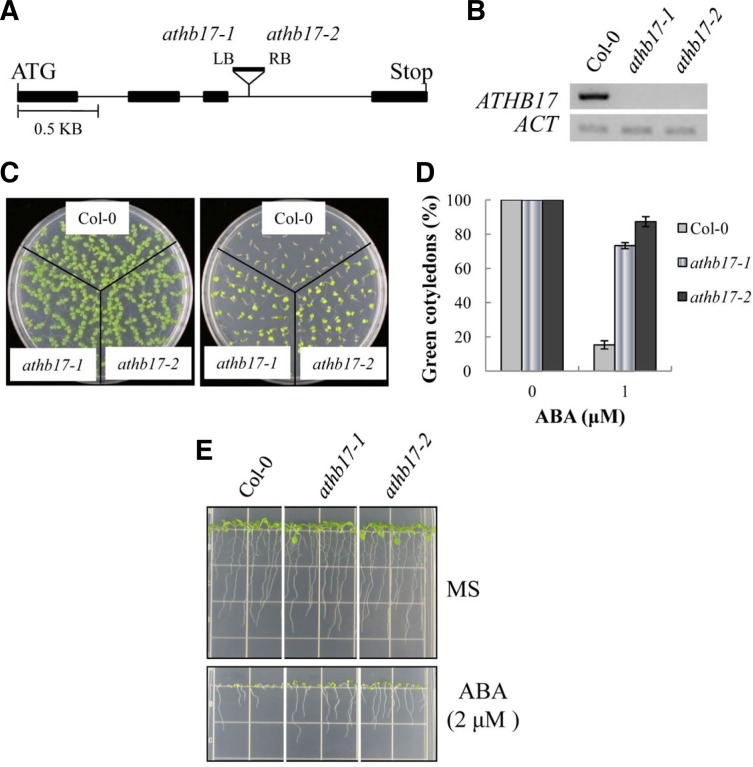
Fig. 3.
Phenotypes of the ATHB17 knockout (KO) line. (A) Schematic diagram depicting the T-DNA insertion site in the KO lines (athb17-1 and athb17-2). The filled bars represent exons, whereas the thin lines indicate introns. (B) ATHB17 transcript levels in the KO lines were determined by semi-quantitative RT-PCR. (C) Seedlings grown in ABA-free MS medium (left panel) or in MS medium containing 1 μM ABA for 7 days. (D) Cotyledon greening efficiency of the seedlings in (C). The values represent the mean of three independent experiments (n = 50 each), and the small bars represent standard errors. (E) Seedlings were grown in a vertical position for 8 days to examine root growth.
Phenotype analysis of transgenic plants was carried out as described previously (Kang et al., 2002; Lee et al., 2009; Park et al., 2011). For aseptic growth, seeds were treated as described above, placed at 4 C for 3–5 days, and were plated on MS medium solidified with 0.8% Phytoagar. The MS medium was supplemented with 1% sucrose and, for ABA tests, various concentrations of ABA was added to the MS medium. For drought test, ten day-old, soil-grown plants were withheld from water until plants lost turgor completely (usually 10–14 days). The treated plants were then re-watered, and survival rates were determined by counting plants that continue to grow. The same number of wild type and transgenic plants were grown on the same tray to minimize experimental variations.
RNA isolation and expression analysis
RNA was isolated employing the Qiagen RNeasy plant mini kit. Northern blot analysis was performed as described before (Kang et al., 2002). RNA was treated with DNase I to remove possible contaminating DNA before RT-PCR. The first strand cDNA was synthesized using Superscript III (Invitrogen) accoring to the supplier’s instructions. For semi-quantitative coupled reverse transcription - polymerase chain reactions (RT-PCR), cDNA amplification was carried out within a linear range. For Real-time RT-PCR, SsoFast EvaGreen supermix was used to amplify the first strand cDNA in a Bio-Rad CFX96 Real-time PCR Systems (Bio-Rad). Quantitation was carried out using the CFX96 Real-time PCR Systems software. Actin-1 was employed as a reference gene. Primer sequences used were as follows: FP1 and RP1 for ATHB17 and FP4 and RP4 for actin.
Histochemical GUS assay and subcellular localization
Histochemical GUS assay was carried out according to Jefferson et al. (Jefferson, 1987). Plants were immersed in a solution containing 1 mM X-gluc (5-bromo-4-chloro-3-indolyl-β-glucuonic acid) in staining buffer (100 mM sodium phosphate, pH 7.0, 0.1 mM EDTA, 0.5 mM ferricyanide, 0.5 mM ferrocyanide, and 0.1% Triton X-100). Staining was performed at 37°C for 24 h. At the end of staining, chlorophyll was removed from the plant tissues by immersing them in 50%, 75% and 95% ethanol sequentially.
For the subcellular localization experiment, the coding region of ATHB17 was amplified using the primers FP5 and RP5 and, after digestion with SacI, was cloned into p35S-FAST/EYFP in frame with the EYFP coding region. Tobacco (Nicotiana benthamiana) leaves were co-infiltrated with the Agrobacterium strains (C58C1) containing the fusion construct and p19, respectively, according to Witte et al. (2004). The tobacco epidermal cells were observed 40 hr after infiltration with a fluorescence microscope (Olympus BX51).
RESULTS
Isolation of an ABA-hypersensitive mutant
We conducted an activation tagging screen to isolate ABA response mutants. A library of activation-tagged lines consisted of approximately 25,000 independent transgenic lines was prepared, and mutants with altered ABA response were isolated as described before (Park et al., 2011).
One of the mutants, referred to as ahs10 (ABA-hypersensitive 10), is shown in Fig. 1. The original mutant exhibited growth retardation, and subsequent analysis of its progeny showed that homozygous mutant plants exhibited severe dwarf phenotypes (Fig. 1A) and set seeds poorly. Consequently, heterozygous plants were employed for the mutant phenotype analysis (Fig. 1B). As shown in Fig. 1C, growth of the mutant seedlings was more severely inhibited than wild type plants by ABA, indicating that they are hypersensitive to ABA.
To determine the T-DNA insertion site in the mutant, we carried out TAIL-PCR (Liu et al., 1995). The result showed that T-DNA was inserted in the 5′ flanking region of the gene At2g01430 (Fig. 1D), which encodes a homeodomain-leucine zipper (HD-Zip) protein, ATHB17. The T-DNA insertion position in the mutant suggested that ATHB17 expression might be activated. Subsequent RT-PCR analysis to determine the expression level of ATHB17 showed that its expression was indeed enhanced in the ahs10 mutant compared with its wild type level (Fig. 1E). Thus, our analysis indicated that overexpression of ATHB17 caused the ABA-hypersensitive phenotype of the ahs10 mutant.
Overexpression of ATHB17 confers ABA hypersensitivity during the postgermination seedling establishment stage
To recapitulate the ATHB17 overexpression (OX) phenotypes observed in the ahs10 mutant, we generated ATHB17 OX lines, in which the coding region of the gene was expressed under the control of the CaMV 35S promoter. Ten homozygous transgenic lines were recovered, and after preliminary analysis, two representative lines (Fig. 2A) were employed for phenotype analysis.
Fig. 2.
Phenotypes of the ATHB17 overexpression (OX) lines. (A) Expression levels of ATHB17 in the OX lines. (B) OX line plants grown in soil for 5 weeks. The numbers indicate line numbers. (C) Seedlings grown in an ABA-free medium (left panel) or in the presence of 0.5 μM ABA (right panel) for 5 days. (D) Cotyledon greening efficiency of seedlings shown in (C). Experiments were done in triplicates (n = 50 each). The small bars indicate standard errors. (E) Seedlings grown in an ABA-free medium (top panel) or in the presence of 0.75 μM ABA (bottom panel) for 7 days in a vertical position.
Like ahs10, ATHB17 OX lines exhibited dwarfism, i.e., both seedlings and mature plants were smaller than the untransformed wild type plants, although the overall growth pattern was normal (Fig. 2B). To assess the effect of ATHB17 OX on ABA response, we determined the ABA sensitivity of the transgenic lines. Seeds were germinated and grown in a medium containing low concentration of ABA, and seedling growth was compared with that in the control medium without ABA (Figs. 2C and 2D). The result showed that shoot growth of the transgenic lines measured by cotyledon greening efficiency was severely inhibited in the presence of 0.5 μM ABA. Under the same growth condition, wild type seedling growth was also inhibited, but the degree of inhibition was much less compared with those of the transgenic lines. In a similar experiment, root elongation of the transgenic lines was also more severely inhibited by ABA than wild type plants (Fig. 2E). Thus, ATHB17 OX lines were ABA-hypersensitive during the early seedling growth stage.
To examine whether the altered ABA response is dependent on developmental stages, we assessed the ABA response of the transgenic plants at three different developmental stages: germination, seedling establishment (i.e., cotyledon expansion/greening and root formation), and root elongation stages. We did not observe clear phenotypes in germination or root elongation assays (data not shown), indicating that ATHB17 overexpression affected only postgermination seedling establishment process.
ATHB17 knockout lines are ABA-insensitive
To confirm that ATHB17 is involved in the ABA response, we analyzed ATHB17 knockout (KO) lines. Two KO lines, SALK_75492 and SALK_128560, were obtained from the Arabidopsis stock center. In both KO lines, T-DNA was inserted into the third intron (Fig. 3A), and ATHB17 expression is abolished completely (Fig. 3B).
The KO lines grew normally and did not exhibit discernible growth phenotypes. However, the KO plants exhibited an altered ABA response. For instance, cotyledon expansion and greening of the wild type seedlings was severely inhibited in the presence of 1 μM ABA (Figs. 3C and 3D), i.e., only 15% of them turned green. By contrast, most cotyledons (73% and 87%, respectively) of the ATHB17 KO seedlings turned green. Similarly, root growth of the KO seedlings was less sensitive to ABA inhibition than wild type plants (Fig. 3E). The results indicated that ATHB17 KO lines are less sensitive to ABA inhibition than wild type plants during the seedling establishment stage.
Drought tolerance of ATHB17 OX and KO lines
ABA plays a key role in adaptive response to various abiotic stresses, especially to water stress response (Finkelstein, 2002; Xiong, 2002). We, therefore, examined the drought tolerance of ahs10 and the ATHB17 OX and KO lines. Plants were grown under normal condition for 10 days and then subjected to water deficit condition by withholding water until plants wilted. The survival rates of the plants were then scored after the treatment.
The result showed that the original mutant was droughttolerant. For instance, Figs. 4A and 4B show that the survival rate of the wild type plants was 17%, whereas the ahs10 survival rate was 66%. Similar results were obtained with the ATHB17 OX line (#18); whereas wild type survival rate was 10%, the survival rate of the OX line was 63% (Figs. 4C and 4D). Another OX line (#47) also exhibited a higher survival rate than the wild type (data not shown). By contrast, ATHB17 KO lines displayed lower survival rates under the water-deficit condition: Whereas the survival rate of the wild type plants was 55%, the mutant survival rate was approximately 8% (Figs. 4E and 4F). Thus, our results showed that ATHB17 OX lines are drought-tolerant, whereas its KO lines are drought-sensitive.
Fig. 4.
Drought tolerance of ahs10, ATHB17 OX and ATHB17 KO lines. 10-day-old seedlings were withheld from water for 10-13 days and were then watered. Their survival rates were determined 2 days after the re-watering. (A, C, and E) Representative ahs10, ATHB 17 OX line and ATHB17 KO line plants, respectively, after the re-watering. (B, D, and F) Survival rates of the plants in (A), (C) and (E), respectively. The values are the mean of three independent experiments (n = 20 each). The small bars represent standard errors.
We also investigated the salt tolerance of the ATHB17 OX and KO lines. As shown in Fig. 5A, growth of ahs10 seedlings was more severely inhibited than wild type plants in the presence of 125 mM NaCl. The result suggested that ATHB17 overexpression enhanced salt sensitivity. To verify the result, we determined the salt sensitivity of ATHB17 OX lines. Figures 5B and 5C show that the cotyledon greening efficiency of the transgenic lines (11% and 28% for #18 and #47, respectively) in the presence of an inhibiting amount of salt (i.e., 125 mM NaCl) was lower than that of the wild type seedlings (66%). By contrast, the cotyledon greening efficiency of ATHB17 KO lines (83% and 87%, respectively) was higher than that of the wild type seedlings (Figs. 5D and 5E). Thus, ATHB17 OX lines were hypersensitive to salt, whereas ATHB17 KO lines were insensitive to salt. The results indicate that ATHB17 is a positive regulator of salt response.
Fig. 5.
Salt tolerance of ahs10, ATHB17 OX and ATHB17 KO lines. The seeds were germinated and grown in the salt-free MS medium or in MS media with salt. (A) Salt tolerance of ahs10. Wild type (Col-0) and ahs10 seedlings were grown in MS medium or MS medium containing 125 mM NaCl for 10 days. (B, C) Salt tolerance of ATHB17 OX lines. Seedlings were grown for 4 days in MS medium or MS medium containing 125 mM NaCl, and seedlings with green cotyledons were counted. (D, E) Salt tolerance of ATHB17 KO lines. Seedlings were grown for 4 days in MS medium or MS medium containing 150 mM NaCl, and seedlings with green cotyledons were counted. Experiments were carried out in triplicates (n = 50), and the small bars in (C) and (E) indicate standard errors.
Expression patterns of ATHB17
The tissue-specific expression pattern of ATHB17 was determined by RT-PCR. As shown in Fig. 6A, ATHB17 was expressed at the highest level in roots. Induction patterns were also determined, and the result showed that ATHB17 expression was induced several fold by ABA, high salt, and mannitol (Fig. 6B).
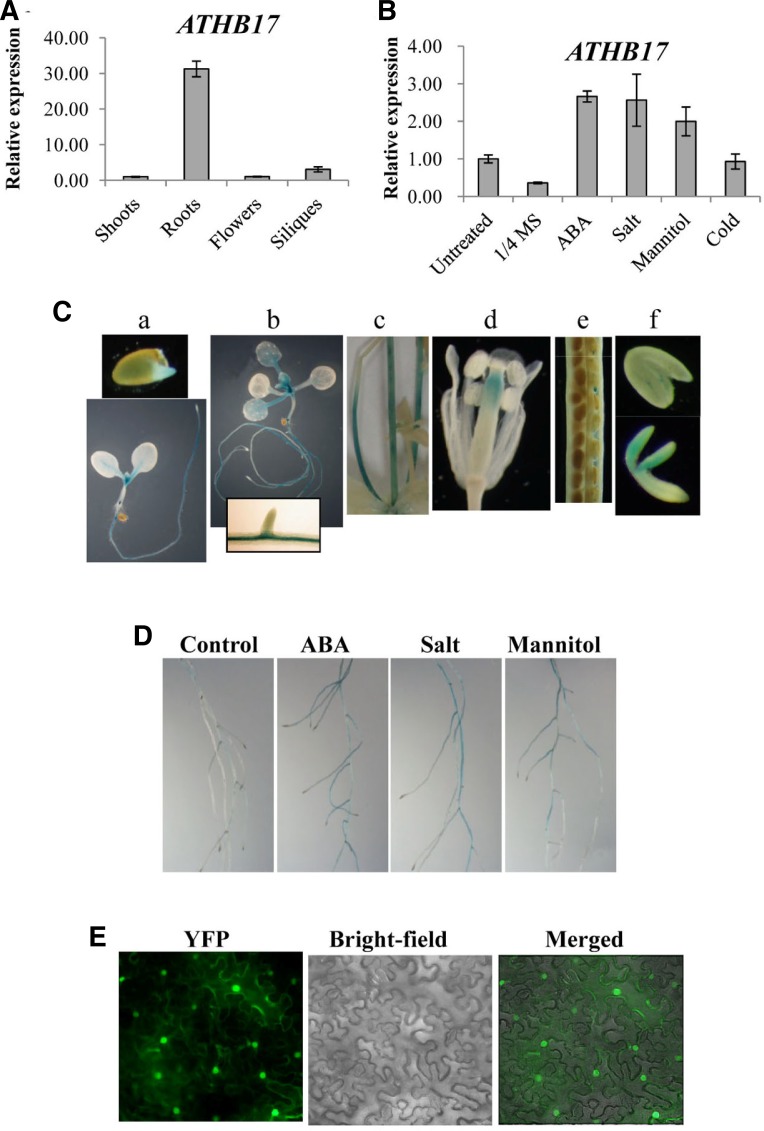
Fig. 6.
Expression patterns of ATHB17. (A) Tissue-specific expression patterns of ATHB17 were determined by Real-time PCR. The values represent the relative expression levels compared with that of the young seedling shoots. (B) Induction patterns of ATHB17. Induction levels of ATHB17 by ABA (100 μM, 4h), Salt (250 mM NaCl, 4 h), mannitol (600 mM, 4 h), and cold (4°C, 24 h) were determined by Real-time PCR. (C) ATHB17 promoter activity was determined by histochemical GUS staining. a, one-day-(top) and five-day-old (bottom) seedlings. b, 10-day-old seedlings. The inset in the bottom shows part of primary root. c, inflorescence stem. d, flower. e, silique. f, mature (top) and immature (bottom) embryos. GUS staining was carried out for 24 h. (D) ATHB17 promoter activity in 7-day-old seedling roots. Roots were treated with 1/2 MS (control) or with 1/2 MS medium containing ABA (100 μM), Salt (250 mM NaCl), or mannitol (600 mM) for 4 h before staining. GUS staining was for 1.5 h (E) Subcellular localization of ATHB17. Tobacco leaves were infiltrated with Agrobacterium harboring an ATHB17-YFP fusion construct and were observed with a fluorescence microscope 40 h after the infiltration. Fluorescence (YFP), bright-field, and merged images are shown.
To further investigate the temporal and spatial expression patterns of ATHB17, we generated transgenic plants harboring an ATHB17 promoter-GUS construct. The promoter activity was then determined by histochemical GUS staining. During the young seedling stage, the ATHB17 promoter was most active in roots, especially in the vascular tissues (Fig. 6C, a–c). In the aerial parts of seedlings, the ATHB17 promoter was most strong in the meristem region, and its weak activity was also observed in the vascular tissues of petioles (Fig. 6C, b). The ATHB17 promoter was also active in the inflorescence stem, lower part of style, funiculi, and embryos (Fig. 6C, c–f). Consistent with the RT-PCR result, treatment of seedlings with ABA, salt or mannitol enhanced the promoter activity in roots (Fig. 6D).
To determine the subcellular localization of ATHB17 protein, an ATHB17-EYFP fusion construct was prepared and introduced into tobacco leaf cells by Agroinfiltration (Witte et al., 2004). The localization of ATHB17 was then determined by observing YFP signal under microscope. As shown in Fig. 6E, the YFP signal was detected in the nucleus, indicating that ATHB17 was localized in the nucleus.
DISCUSSION
Our results indicate that ATHB17 is a positive regulator of ABA response. The original tagged line, in which the ATHB17 expression level is higher than the normal level, and the ATHB17 OX lines were hypersensitive to ABA, whereas its KO lines were partially insensitive to ABA. ABA inhibits both seed germination and postgermination seedling growth (Finkelstein et al., 2002). In our experiments to determine the developmental stage-dependence of ABA sensitivity, we observed ABA-hypersenitive response during cotyledon expansion/greening and root formation stage, but we did not observe clear phenotypes in our germination and root elongation assays (data not shown). Thus, it appears that ATHB17 affects a subset of ABA response, i.e., the ABA response during a limited developmental window of seedling establishment stage. Another aspect of ABA-assoiated phenotypes is drought tolerance. Consistent with the enhanced ABA sensitivity, the activation-tagged and the OX lines were drought-tolerant, whereas ATHB17 KO lines were more susceptible to water deficiency. Similarly, our results shown in Fig. 5 indicate that ATHB17 is a positive regulator of salt response.
Our expression analysis indicates that ATHB17 expression is generally higher in the root than in other tissues (Fig. 6A) and induced by ABA and abiotic stresses such as high salt and high osmolarity (Fig. 6B). Histochemical GUS assay further revealed that the ATHB17 promoter was more active in roots than in shoots and that it is strongly expressed in the shoot meristem region of seedling. The expression pattern is consistent with the finding that ATHB17 is involved in shoot growth regulation and responses to drought and salt.
ATHB17 belongs to the HD-Zip family of transcription factors, and our result in Fig. 6E indicates that it is localized in the nucleus. At present, we do not know what the target genes of ATHB17 are. However, several observations suggest that ATHB17 may require additional factor(s) for its full activity. First, our transcriptional assay shows that it possesses little transcriptional activity (data not shown). Second, we determined the expression levels of a number of ABA-responsive genes to identify ATHB17 target genes, but did not observe distinct changes in their expression levels in the ATHB17 OX lines (data not shown). Third, the phenotypes of the ATHB17 OX lines are not strong, implying that it alone may not be sufficient to alter the entire spectrum of ABA response.
As mentioned earlier, several ATHB proteins are known to be involved in ABA response. ATHB6, which was isolated as an ABI1-interacting protein, negatively regulate the ABA response during germination and stomatal closure (Himmelbach et al., 2002; Lechner et al., 2011). ATHB7 and ATHB12 are positive regulators of ABA inhibition of root and stem growth (Olsson et al., 2004). Recently, it has been demonstrated that ATHB12 inhibits stem growth by reducing GA20 oxidase (Son et al., 2010). Overexpression of another HD-Zip protein, ATHB5, enances ABA sensitivity during the germination and postgermination root growth stage, although its knockout plants do not exhibit any phenotypes (Johannesson et al., 2003). Expression of these HD-Zip protein genes are induced by ABA and, in the case of ATHB12, its tissue-specific expression patterns are somewhat similar to those of ATHB17 in vegetative tissues but not in reproductive tissues (Olsson et al., 2004). Our study showed that ATHB17 expression is also induced by ABA and that ATHB17 is involved in ABA-mediated growth regulation. However, details of tissue-specific expression patterns of ATHB17 (Fig. 6) differ from those of other HD-Zip proteins, and ATHB17 functions during a very narrow window of seedling development, i.e., the postgermination seedling establishment stage. Additionally, ATHB17 enhances drought tolerance and salt sensitivity, which was not reported for the HD-Zips mentioned above.
Table 1.
List of primer sequences
| Primers | Primer sequences |
|---|---|
| FP1 | 5′-TGA AGG AAG TGG CGG AGG AAG AGA-3′ |
| RP1 | 5′-GCT CTT GCG GCG GAA ACG TTT-3′ |
| FP2 | 5′-TCT AGA AAT TTA CTT TGG TTA TGT CTT TCT-3′ |
| RP2 | 5′-GAG CTC CAT ACT CAA ATA ATT AAG ATA CAC-3′ |
| FP3 | 5′-AGC GGT CGA CAA TGA TTA AGA GAC AAG ACA GAA-3′ |
| RP3 | 5′-CAA GGA TCC TAA GGA GAA AAT ATT AAG ATG CGT T-3′ |
| FP4 | 5′-CAT CAG GAA GGA CTT GTA CGG-3′ |
| RP4 | 5′-GAT GGA CCT GAC TCG TCA TAC-3′ |
| FP5 | 5′-AAG GAG CTC ATG ATA AAA CTA CTA TTT ACG TAC ATA TG-3′ |
| RP5 | 5′-AAG CCC GGG ACG ATC ACG CTC TTG CGG-3′ |
Acknowledgments
This work was supported in part by grants from the Rural Development Administration, Republic of Korea [the Next Generation BioGreen 21 Program (TAGC grant PJ008198 to S.Y.K.)] and the Ministry of Education, Science and Technology (MEST) [the Mid-career Research Program (grant no. 2011-0015455) and the Basic Science Research Program (grant no. 2010-0023686) through NRF]. The authors are grateful to the Kumho Life Science Laboratory of Chonnam National University for providing equipments and plant growth facilities.
REFERENCES
- Aoyama T., Dong C.H., Wu Y., Carabelli M., Sessa G., Ruberti I., Morelli G., Chua N.H. Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco. Plant Cell. 1995;7:1773–1785. doi: 10.1105/tpc.7.11.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F.D., Manavella P.A., Dezar C.A., Chan R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007;12:419–426. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Barrero J.M., Millar A.A., Griffiths J., Czechowski T., Scheible W.R., Udvardi M., Reid J.B., Ross J.J., Jacobsen J.V., Gubler F. Gene expression profiling identifies two reguatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds. Plant J. 2010;61:611–622. doi: 10.1111/j.1365-313X.2009.04088.x. [DOI] [PubMed] [Google Scholar]
- Bechtold N., Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Ciarbelli A.R., Ciolfi A., Salvucci S., Ruzza V., Possenti M., Carabelli M., Fruscalzo A., Sessa G., Morelli G., Ruberti I. The Arabidopsis Homeodomain-leucine Zipper II gene family: diversity and redundancy. Plant Mol. Biol. 2008;68:465–478. doi: 10.1007/s11103-008-9383-8. [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R., Gampala S.S., Rock C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14(Suppl):S15–45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J.C., Hrmova M., Lopato S., Langridge P. Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol. 2011;190:823–837. doi: 10.1111/j.1469-8137.2011.03733.x. [DOI] [PubMed] [Google Scholar]
- Henriksson E., Olsson A.S.B., Johannesson H., Johansson H., Hanson J., Engstrom P., Soderman E. Homeoomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 2005;139:509–518. doi: 10.1104/pp.105.063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A., Hoffmann T., Leube M., Hohener B., Grill E. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 2002;21:3029–3038. doi: 10.1093/emboj/cdf316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;20:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesson H., Wang Y., Hanson J., Engstrom P. The Arabidopsis thaliana homeobox gene ATHB5 is a potential regulator of abscisic acid responsiveness in developing seelings. Plant Mol. Biol. 2003;51:719–729. doi: 10.1023/a:1022567625228. [DOI] [PubMed] [Google Scholar]
- Kang J., Choi H., Im M., Kim S.Y. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. 2002;14:343–357. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner E., Leonhardt N., Eisler H., Parmentier Y., Alioua M., Jacquet H., Leung J., Genschik P. MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev Cell. 2011;21:1116–1128. doi: 10.1016/j.devcel.2011.10.018. [DOI] [PubMed] [Google Scholar]
- Lee S., Cho D., Kang J., Kim S.Y. An ARIA-interacting AP2 domain protein is a novel component of ABA signaling. Mol Cells. 2009;27:409–416. doi: 10.1007/s10059-009-0058-3. [DOI] [PubMed] [Google Scholar]
- Liu Y.G., Mitsukawa N., Oosumi T., Whittier R.F. Efficient isolation and mapping of Arabidopsis-thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee K., Brocchieri L., Burglin T.R. A compreensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009;26:2775–2794. doi: 10.1093/molbev/msp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Katsumata H., Abe M., Yabe N., Komeda Y., Yamamoto K.T., Takahashi T. Characterization of the class IV homeodomain-leucine zipper gene family in Araidopsis. Plant Physiol. 2006;141:1363–1375. doi: 10.1104/pp.106.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A.S.B., Engstrom P., Soderman E. The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 2004;55:663–677. doi: 10.1007/s11103-004-1581-4. [DOI] [PubMed] [Google Scholar]
- Park M.Y., Kang J.Y., Kim S.Y. Overexpression of AtMYB52 confers ABA hypersensitivity and drought tolerance. Mol Cells. 2011;31:447–454. doi: 10.1007/s10059-011-0300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge M.J., Otsuga D., Alonso J.M., Ecker J.R., Drews G.N., Clark S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S., Ohgishi M., Goda H., Higuchi K., Shimada Y., Yoshida S., Koshiba T. The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J. 2002;32:1011–1022. doi: 10.1046/j.1365-313x.2002.01488.x. [DOI] [PubMed] [Google Scholar]
- Schena M., Davis R.W. Structure of homeoboxleucine zipper genes suggests a model for the evolution of gene families. Proc. Natl. Acad. Sci USA. 1994;91:8393–8397. doi: 10.1073/pnas.91.18.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M., Lloyd A.M., Davis R.W. The HAT4 gene of Arabidopsis encodes a developmental regulator. Genes Dev. 1993;7:367–379. doi: 10.1101/gad.7.3.367. [DOI] [PubMed] [Google Scholar]
- Son O., Hur Y.S., Kim Y.K., Lee H.J., Kim S., Kim M.R., Nam K.H., Lee M.S., Kim B.Y., Park J., et al. ATHB12, an ABA-inducible homeodomain-leucine zipper (HD-Zip) protein of Arabidopsis, negatively regulates the growth of the Inflorescence stem by decreasing the expression of a gibberellin 20-oxidase gene. Plant Cell Physiol. 2010;51:1537–1547. doi: 10.1093/pcp/pcq108. [DOI] [PubMed] [Google Scholar]
- Sorin C., Salla-Martret M., Bou-Torrent J., Roig-Villanova I., Martinez-Garcia J.F. ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J. 2009;59:266–277. doi: 10.1111/j.1365-313X.2009.03866.x. [DOI] [PubMed] [Google Scholar]
- Wang Y., Henriksson E., Soderman E., Henriksson K.N., Sundberg E., Engstrom P. The Arabidopsis homeobox gene, ATHB16, regulates leaf development and the sensitivity to photoperiod in Arabidopsis. Dev. Biol. 2003;264:228–239. doi: 10.1016/j.ydbio.2003.07.017. [DOI] [PubMed] [Google Scholar]
- Witte C.P., Noel L.D., Gielbert J., Parker J.E., Romeis T. Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol. Biol. 2004;55:135–147. doi: 10.1007/s11103-004-0501-y. [DOI] [PubMed] [Google Scholar]
- Xiong L., Schumaker K.S., Zhu J.-K. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14(Suppl):S165–183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]