Abstract
Adipocyte differentiation requires the coordinated activities of several nuclear transcription factors. Recently, mitochondria biogenesis was reported to occur during adipocyte differentiation and following treatment with thiazolidinediones in vitro and in vivo. Crif1 is a translational factor for mitochondrial DNA (mtDNA) and is important for transcription of the mitochondrial oxidative phosphorylation (OXPHOS) complex. To investigate the role of OXPHOS in adipogenesis, we analyzed adipocyte differentiation following disruption of Crif1 in vitro and in vivo. The adipose-specific Crif1 knockout mouse had a lower body weight and less fat mass than wild-type mice. Furthermore, adipocytes were smaller and had a dysplastic morphology in the adipose-specific Crif1 knockout mouse. 3T3-L1 adipocytes or adipose-derived stem cells (ADSCs) that lacked Crif1 expressed lower levels of mtDNA-encoded OXPHOS subunits, and adipocyte differentiation was disrupted. Rosiglitazone treatment did not induce adipogenesis or mitochondria biogenesis in Crif1 knockout ADSCs. These results show that mitochondrial OXPHOS and Crif1 are required for rosiglitazone- and hormone-induced adipogenesis.
Keywords: adipogenesis, CRIF1, mitochondrial oxidative phosphorylation
INTRODUCTION
White adipocyte differentiation involves a complex and highly orchestrated program of gene expression that is driven by synergistic interactions between transcription factors including the peroxisome proliferator activated receptor gamma (PPARγ), CCAAT/enhancer family [CCAAT/enhancer binding protein α, β, δ (C/EBPα, β, δ)], and sterol regulatory element binding protein-1c (SREBP-1c) (Farmer, 2006). These transcription factors regulate several intracellular pathways, processes, and secretory factors that influence adipogenesis. Understanding the processes responsible for adipogenesis is important because adipocyte dysfunction occurs in various chronic metabolic diseases. Improving adipocyte function by modulating adipogenic processes may help manage such diseases.
Marked mitochondrial biogenesis occurs during adipocyte differentiation (Newton et al., 2011; Wilson-Fritch et al., 2003). The concentration of mitochondrial proteins is increased 20–30-fold in differentiated 3T3-L1 adipocytes in comparison to the concentration in preadipocytes (Newton et al., 2011). Treatment with chemical inhibitors of respiratory chain function, such as rotenone, suppresses adipogenesis by modulating the expression of the key transcription factors C/EBPα, PPARγ, and SREBP-1c (Lu et al., 2010). The expression of PPARγ coactivator-1 (PGC-1), which is a key regulator of adipogenesis, correlates with mitochondrial biogenesis and adipogenesis (Gao et al., 2011). Consequently, mitochondria have been proposed to play an important role in adipogenesis programming and the progression of metabolic diseases. However, how sequential changes in the levels of mitochondrial proteins affect adipocyte differentiation and which genes are involved in mitochondrial development remain poorly understood.
Most cellular energy is released by OXPHOS in the electron transport chain in the inner mitochondrial membrane. The OXPHOS system in the mammalian respiratory chain is composed of at least 89 polypeptides that are encoded by nuclear DNA (nDNA) or mitochondria DNA (mtDNA) (Calvo and Mootha, 2010; Wallace, 2007). The human mtDNA genome encodes only 13 OXPHOS proteins, but these proteins provide an essential framework for the assembly of OXPHOS complexes in the inner mitochondrial membrane (Shoubridge, 2001). The mitochondrial ribosome is responsible for intra-mitochondrial synthesis of mtDNA-encoded OXPHOS polypeptides. Depletion of mitochondrial transcription factor A (TFAM) in 3T3-L1 cells impairs translocation of glucose transporter 4, down-stream of Akt, while adipogenic gene expression and lipid accumulation are not affected. However, mitochondria-related studies involving the knockdown of genes or inactivation of the OXPHOS complex have used cultured cell lines and have not been performed in vivo.
To investigate the role of mitochondrial OXPHOS in adipocyte differentiation, we generated adipose-specific Crif1 loss-of-function transgenic mouse. A primary mitochondrial function, which is important for the translation of the 13 subunits that constitute the mitochondrial OXPHOS complexes I, III, IV, and V in vitro and in vivo, was disrupted in this mouse model.
MATERIALS AND METHODS
Mice
Floxed Crif1 (Crif1flox/flox) mice were generated as previously described (Kwon et al., 2008). Adiponectin-Cre transgenic mice (C57BL/6J) were kindly provided by Dr. Evan D. Rosen. Mice were bred in our facility and housed at a constant temperature (22°C), with 50–60% humidity and a daily 12 h light/12 h dark cycle. All experiments were conducted using male mice housed in individual cages, and body weights were checked at 4 weeks of age. All mouse experiments were performed in the animal facility according to institutional guidelines, and the experimental protocols were approved by the institutional review board of the Korean Research Institute of Biotechnology and Bioscience, and Chungnam National University.
Histological and morphometric analysis
White adipose tissue (WAT) was fixed in 10% neutralized formalin, washed, and then embedded in paraffin. Paraffin tissue sections of 5 μm were deparaffinized, rehydrated, and stained with hematoxylin and eosin. For whole-mount fluorescence staining, epididymal WAT was fixed in 10% neutralized formalin. Formalin was removed with 20% sucrose, and WAT was then incubated with an anti-PECAM antibody (Millipore; MAB1398Z) and incubated for 10 min at room temperature with DAPI and BODIPY.
Cell culture and Oil red O staining
3T3-L1 preadipocytes were purchased from ATCC. Methylisobutylxanthine (IBMX), dexamethasone and insulin were obtained from Sigma-Aldrich. 3T3-L1 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% bovine calf serum (Gibco BRL). Lipofectamine® RNAiMAX reagent (Invitrogen) was used to transfect 3T3-L1 fibroblasts with Crif1 siRNA (GGA GUG CUC GCU UCC AGG AAC UAU U). Cells were cultured for 2 days, after which cells were differentiated by culturing in DMEM containing IBMX (0.5 mM), dexamethasone (1 μM), insulin (10 μg/ml), and 10% fetal bovine serum for a further 2 days. This medium was then replaced with DMEM containing insulin (10 μg/ml), which was exchanged every 2 days (Hemati et al., 1997). ADSCs were cultured from the Crif1flox/flox mouse, as previously described (Estes et al., 2010). After treatment with null or Cre recombinase adenoviruses, ADSCs were differentiated into adipocytes by culturing in M199 medium (Gibco BRL) containing IBMX (0.5 mM), dexamethasone (1 μM), insulin (10 μg/ml), and rosiglitazone (0.5 μM) and 10% fetal bovine serum. Differentiated 3T3-L1 adipocytes or ADSCs were fixed with 4% paraformaldehyde for 30 min at room temperature and then washed with 1x PBS. Staining was performed with freshly prepared 0.2% Oil red O solution for 30 min followed by three washes with water. For quantification, Oil red O was dissolved in 200 μl isopropanol and the absorbance at 540 nm was measured by ELISA.
Crif1-HA adenovirus
Crif1 DNA was amplified by PCR from mouse genomic DNA extracted from C57BL/6 mouse liver, and HA was fused at the C-terminal of Crif1. The Crif1-HA adenovirus was generated using the pAdEasy system as previously described (He et al., 1998).
Western blotting
Western blot analysis was performed using 12% acrylamide SDS-PAGE gels. Primary antibodies against OXPHOS complex subunits (ND6, NDUFA9, SDHA, UQCRC2, and ATP5A1) were purchased from Invitrogen. An anti-COX4 antibody (#4844), an anti-VDAC antibody (#4866) and an anti-PPARγ antibody (#2435) were purchased from Cell Signaling. An anti-ND1 antibody (sc-65237) and an anti-Crif1 antibody (sc-134882) were purchased from Santa Cruz Biotechnology. Secondary antibodies (goat anti-mouse and goat anti-rabbit) were obtained from Cell Signaling.
Blue native PAGE (BN-PAGE)
BN-PAGE and mitochondrial isolation was performed as previously described (Kim et al., 2012). Mitochondrial isolation was performed by suspending pellets of fully differentiated ADSCs in buffer B (210 mM mannitol, 70 mM sucrose, 1 mM EGTA, and 5 mM HEPES, pH 7.2). The mitochondrial fraction was used in the Native PAGE™ Novex® Bis-Tris Gel system (Invitrogen) with 0.5% n-dodecyl-β-D-maltoside to determine the content of the OXPHOS complex. After electrophoresis, the gel was transferred to a PVDF membrane, which was then incubated with an anti-OXPHOS antibody mixture (Invitrogen).
Oxygen consumption rate (OCR)
OCR was measured using an XF-24 analyzer (Seahorse Bioscience). ADSCs were plated on an XF-24 plate, treated with null or Cre recombinase adenovirus, and incubated in differentiation media. The day before OCR measurement, the sensor cartridge was calibrated with calibration buffer (Seahorse Bioscience) at 37°C. Fully differentiated ADSCs were washed and cultured in M199 (Gibco BRL) lacking sodium bicarbonate at 37°C. OCR was automatically calculated and recorded by the sensor cartridge and Seahorse XF-24 software.
Real-time PCR and Northern blot
Total RNA was isolated using Trizol (Invitrogen), in accordance with manufacturer’s instructions. Complementary DNA (cDNA) was prepared with M-MLV Reverse Transcriptase and oligo-dT primers (Invitrogen). Real-time PCR was performed using cDNA, QuantiTect™ SYBR® Green PCR Master Mix (QIAGEN), and specific primers. Primer sequences were as follows: NDUFA9 F (5′-ACTGT GTTTG GGGCT ACAGG-3′), NDUFA9 R (5′-GATTG ATGAC CACGT TGCTG-3′), SDHA F (5′-ACACA GACCT GGTGG AGACC-3′), SDHA R (5′-GCACA GTCAG CCTCA TTCAA-3′), UQCRC2 F (5′-ATCAA AAGGG GCAAC AACAC-3′), UQCRC2 R (5′-CACTC AGGAA GCCCT CTGAC-3′), COX4 F (5′-TTGGC AAGAG AGCCA TTTCT-3′), COX4 R (5′-GCCCA CAACT GTCTT CCATT-3′), ATP5A1 F (5′-AGGCC TATCC TGGTG ATGTG-3′), ATP5A1 R (5′-CTTCA TGGTA CCTGC CACCT-3′), ND1 F (5′-ACGCA AAATC TTAGG GTACA-3′), ND1 R (5′-GAGTG ATAGG GTAGG TGCAA-3′), ND6 F (5′-CACCC AGCTA CTACC ATCAT-3′), ND6 R (5′-ATTGG GGGTG ATTAT AGAGG-3′), COX1 F (5′-ATTCG AGCAG AATTA GGTCA-3′), COX1 R (5′-CTCCG ATTAT TAGTG GGACA-3′), ATP6 F (5′-ACGAA AATCT ATTTG CCTCA-3′), ATP6 R (5′-TGGTG TGTGG ATTAG CATTA-3′), Crif1 F (5′-GAA CG CTGGG AGAAA ATTCA-3′), Crif1 R (5′-ATAGT TCCTG GAAGC GAGC A-3′), Ppargamma F (5′-ATCTT AACTG CCGGA TCCAC-3′), Ppargamma R (5′-TGGTG ATTTG TCCGT TGTCT-3′), Adiponectin F (5′-TCTCC TGTTC CTCTT AATCC TGCC-3′), Adiponectin R (5′-CATCT CCTTT CTCTC CCTTC TCTCC-3′), CD36 F (5′-TGCTG GAGCT GTTAT TGGTG-3′), CD36 R (5′-TGGGT TTTGC ACATC AAAGA-3′), Fasn F (5′-CCCTT GATGA AGAGG GATCA-3′), Fasn R (5′-ACTCC ACAGG TGGGA ACAAG-3′), C/ebpa F (5′-TGGAC AAGAA CAGCA ACGAG-3′), C/ebpa R (5′-TCACT GGTCA ACTCC AGCAC-3′), C/ebpb F (5′-CAAGC TGAGC GACGA GTACA-3′), C/ebpb R (5′-AGCTG CTCCA CCTTC TTCTG-3′), C/ebpd F (5′-ATCGA CTTCA GCGCC TACAT-3′), and C/ebpd R (5′-GCTTT GTGGT TGCTG TTGAA-3′). Relative expressions were calculated using Rotor-Gene™ 6000 realtime rotary analyzer software (Version 1.7, Corbett Life Science).
RESULTS
Knockout of Crif1 in adipocytes perturbs adipose tissue development
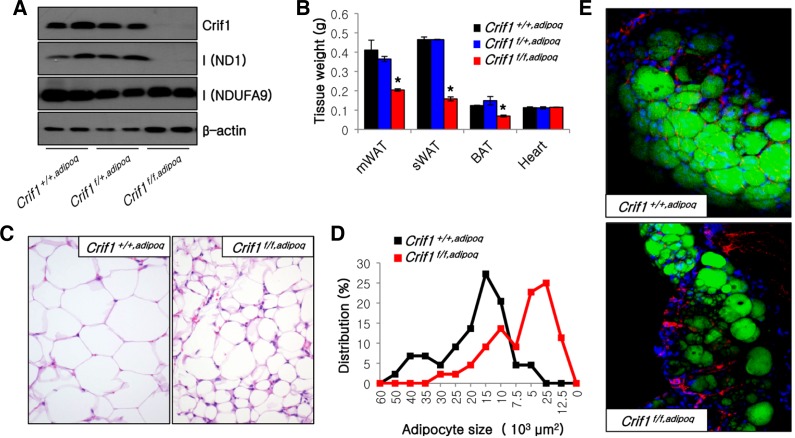
The mitochondrial protein Crif1 associates with the large subunit of the mitochondrial ribosome and plays a critical role in the intra-mitochondrial production and insertion of 13 OXPHOS subunits encoded by mtDNA (Kim et al., 2012). Crif1 is ubiquitously expressed in WAT and brown adipose tissue (BAT) (data not shown). To identify the function of Crif1 and mitochondrial OXPHOS in adipocytes, we generated adipocyte-specific Crif1 knockout mice by crossing floxed Crif1 mice (Kwon et al., 2008) with adiponectin-Cre transgenic mice. The epididymal WAT (eWAT) of Crif1f/f,adipoq mice exhibited marked depletion of Crif1 and ND1, a mtDNA-encoded OXPHOS complex I subunit, and a reduction in the level of NDUFA9, a nDNA-encoded OXPHOS complex I subunit (Fig. 1A). Crif1f/f,adipoq mice demonstrated a marked decrease in the weight of mesenteric WAT (mWAT), subcutaneous WAT (sWAT), and BAT (Fig. 1B). Hematoxylin and eosin staining of adipose tissues indicated that the adipocytes of Crif1f/f,adipoq mice were relatively smaller and irregularly size in comparison to those of Crif1+/+,adipoq mice (Fig. 1C). Distribution analysis of adipocyte size showed that the peak frequency was approximately 2.5 × 103 μm2 in Crif1f/f,adipoq mice and 15 × 103 μm2 in Crif1+/+,adipoq mice (Fig. 1D). BODIPY staining was reduced in the eWAT of Crif1f/f,adipoq mice, whereas nuclei (DAPI) and endothelial cell (anti-PECAM antibody) staining was unchanged. This indicates that mitochondrial OXPHOS dysfunction in these mice may perturb the development of adipose tissue (Fig. 1E). Collectively, data from the adipocytespecific Crif1 knockout mouse suggests that loss of Crif1 disrupts adipose tissue development in vivo.
Fig. 1.
Perturbation of adipose tissue development in Crif1f/f,adipoq mice. (A) Western blot analysis of eWAT probing for Crif1 and the OXPHOS complex I subunits NDUFA9 and ND1, which are encoded by nDNA and mtDNA, respectively. The β-actin was used as an internal control. (B) Tissue weight of mice (n = 10 for each group; means and s.d.s are shown; *P < 0.05). mWAT, mesenteric WAT; sWAT, subcutaneous WAT; BAT, brown adipose tissue. (C) Hematoxylin and eosin staining of eWAT. Magnification, × 200. (D) Distribution of adipocyte sizes in eWAT as shown in Fig. 1C. (E) Immunofluorescence image of eWAT stained with BODIPY (green), DAPI (blue) and an anti-PECAM antibody (red). Magnification, × 200.
Mitochondrial biogenesis is critical for adipocyte differentiation
It has been previously reported that mitochondrial biogenesis is associated with adipocyte differentiation (Luo et al., 2008; Wilson-Fritch et al., 2003). In this study, we studied mitochondrial biogenesis during adipogenesis of 3T3-L1 cells. The mtDNA copy number was measured using real-time PCR. The mtDNA copy number started to increase on day 1 and peaked at day 3 of 3T3-L1 adipocyte differentiation (Fig. 2A). To determine whether this increase in mtDNA copy number was accompanied by an increase in the levels of the OXPHOS complex proteins, Western blotting of various OXPHOS subunits was performed on whole cell lysates obtained from 3T3-L1 adipocytes (Fig. 2B). The levels of various subunits of OXPHOS complexes I, II, III, and IV were induced on the first day of differentiation. The OXPHOS complex V subunit, ATP5A1, was expressed at a relatively high level in all samples (Fig. 2B).
Fig. 2.
Mitochondrial biogenesis during 3T3-L1 adipocyte differentiation. (A) The relative amount of mtDNA was analyzed by real-time PCR during 3T3-L1 adipocyte differentiation. (n = 4; means and s.d.s are shown; *P < 0.05). Ctr, control; R, Rosiglitazone; EtBr, ethidium bromide. (B) Western blot analysis of OXPHOS subunits encoded by nDNA (NDUFA9, SDHA, SDHB, UQCRC2, COX4, and ATP5A1) and mtDNA (ND6) during 3T3-L1 adipocyte differentiation. (C) BN-PAGE analysis of assembled OXPHOS complexes in isolated mitochondria during 3T3-L1 adipocyte differentiation. (D) Western blot analysis of Crif1, VDAC and OXPHOS subunits encoded by nDNA (NDUFA9, SDHA, UQCRC2, COX4, and ATP5A1) and mtDNA (ND1) in isolated mitochondria during 3T3-L1 adipocyte differentiation.
BN-PAGE analysis showed that the level of assembled OXPHOS complexes increased during adipocyte differentiation of 3T3-L1 cells (Fig. 2C). To normalize the expression levels of OXPHOS subunits, Western blotting of various OXPHOS subunits was performed on mitochondria isolated from differentiating 3T3-L1 cells. The expression of OXPHOS subunits in these isolated mitochondria increased as differentiation progressed. Crif1 was expressed in undifferentiated 3T3-L1 cells and its expression steadily increased during differentiation (Fig. 2D). These data indicate that mitochondrial biogenesis, which correlates with an increase in the mtDNA copy number and increased expression of OXPHOS complex subunits, is linked with adipocyte differentiation of 3T3-L1 cells.
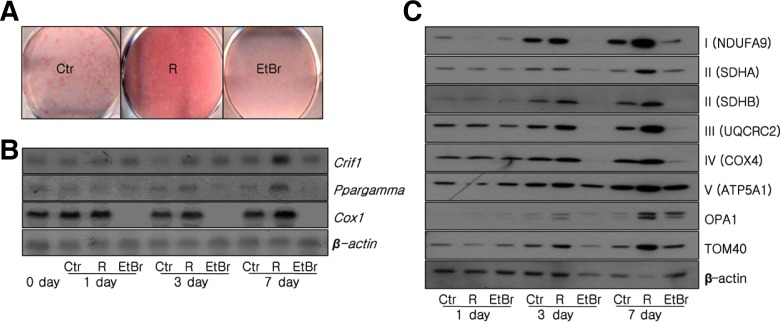
Adipose mitochondria function is suppressed in the insulin resistant state and the reversal of insulin resistance by treatment with PPAmγ agonists, such as rosiglitazone, is associated with an increase in adipose mitochondrial biogenesis (Sears et al., 2009; Wilson-Fritch et al., 2003; 2004). Treatment with a low dose of rosiglitazone over a prolonged period stimulated the differentiation of 3T3-L1 cells. Rosiglitazone treatment increased the intensity of Oil red O staining, whereas staining was reduced when cells were treated with ethidium bromide (EtBr), which depletes mtDNA (Fig. 3A). Treatment with rosiglitazone promoted differentiation of 3T3-L1 cells and induced mmNA expression of Crif1 and Cox1, a subunit of OXPHOS complex IV (Fig. 3B). Western blotting revealed that rosiglitazone treatment increased the expression of subunits of OXPHOS complexes I, II, III, IV, and V, whereas treatment with EtBr did not (Fig. 3C). These data indicate that PPAmγ-induced adipogenesis is linked with the biogenesis of mitochondrial OXPHOS complexes.
Fig. 3.
Modulation of mitochondrial OXPHOS following treatment with rosiglitazone or EtBr. 3T3-L1 cells were treated with 1 μM rosiglitazone or 100 ng/ml EtBr. (A) Oil red O staining at day 7 of 3T3-L1 adipocyte differentiation. (B) Northern blot analysis of Crif1, Ppargamma, Cox1 and β-actin at days 0, 1, 3, 7 of 3T3-L1 adipocyte differentiation. (C) Western blot analysis of the OXPHOS complex subunits NDUFA9, SDHA, SDHB, UQCRC2, COX4 and ATP5A1, as well as OPA1, TOM40, and β-actin, at days 1, 3 and 7 of 3T3-L1 adipocyte differentiation.
Crif1 is induced during adipogenesis and is required for the differentiation of adipocytes
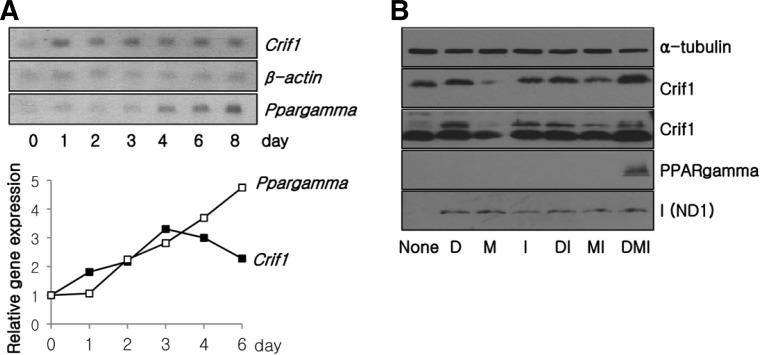
Expression of Crif1 mmNA was induced during the early phase of adipogenesis, in a similar manner to OXPHOS complex subunits (Figs. 2A and 4A). To explore the functional link between mitochondrial OXPHOS function and adipocyte differentiation in vitro, we monitored the expression of Crif1 and OXPHOS complex subunits in 3T3-L1 fibroblasts stimulated with various differentiation signals. Crif1 expression was increased by treatment with insulin and/or dexamethasone (Fig. 4B). Treatment with IBMX, dexamethasone and insulin increased the expression of the OXPHOS subunits, ND1, and the expression of PPAmgamma, a marker of differentiated adipocytes (Fig. 4B).
Fig. 4.
Crif1 expression during 3T3-L1 adipocyte differentiation. (A) Northern blot analysis of Crif1 and Ppargamma mRNA during 3T3-L1 adipocyte differentiation (upper). The graph shows the relative densities of Crif1/β-actin or Ppargamma/β-actin mRNA compared with the value at day 0 from the Northern blots. (B) Western blot analysis of 3T3-L1 fibroblasts treated with various combinations of hormones that induce adipocyte differentiation. D, dexamethasone; M, IBMX; I, insulin.
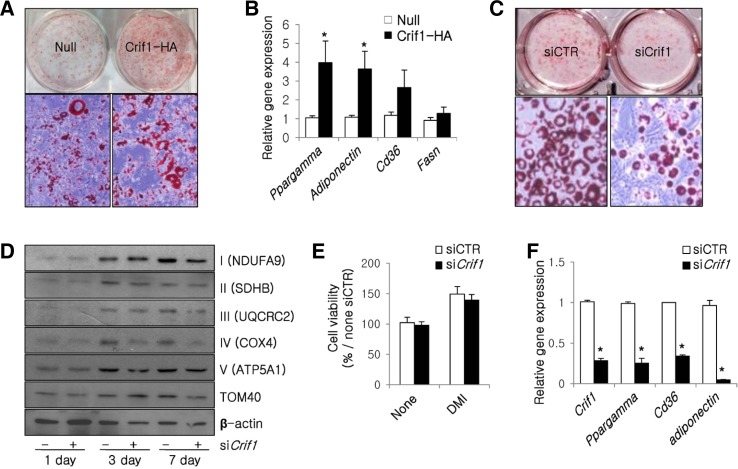
The induction of Crif1 expression with half the dose of dexamethasone promoted 3T3-L1 adipocyte differentiation as indicated by enhanced Oil red O staining and increased expression of Ppargamma, adiponectin, Cd36 and fatty acid synthase (Fasn) (Figs. 5A and 5B). By contrast, when Crif1 was depleted in 3T3-L1 cells using siRNA, the protein levels of the NDUFA9 and COX4, were decreased (Fig. 5D). The mRNA levels of these subunits were unchanged (data not shown). These results suggest that the depletion of Crif1 inhibits lipid accumulation and decreases the expression of OXPHOS complexes due to defects in mitochondrial translation (Figs. 5C and 5D). Furthermore, depletion of Crif1 reduced the maximal induction of Ppargamma, Cd36, and adiponectin (Fig. 5F), but did not affect cell viability (Fig. 5E). These observations indicate that Crif1 plays a crucial role in adipocyte differentiation by regulating the expression of adipogenic genes.
Fig. 5.
Crif1 controls adipogenesis by modulating the expression of various genes. (A) Oil red O staining of 3T3-L1 cells infected with adenoviruses. Null, null adenovirus; Crif1-HA, Ha-tagged Crif1 adenovirus. Magnification, × 100 (upper). (B) Real-time PCR analysis of Ppargamma, adiponectin, Cd36 and Fasn in 3T3-L1 cells at day 7 of differentiation (n = 6; means and s.d.s are shown; *P < 0.05). (C) Oil red O staining of 3T3-L1 cells treated with Crif1 siRNA. Magnification, × 100 (upper). (D) Western blot analysis of differentiating 3T3-L1 cells probing for OXPHOS complex subunits NDUFA9, SDHB, UQCRC2, COX4, and ATP5A1. TOM40 and β-actin were used as internal controls. (E) Viability of 3T3-L1 cells treated with Crif1 siRNA or control siRNA. Cells were cultured in the presence or absence of differentiation hormones. The relative cell viability was normalized against that of the control siRNA sample that was not treated with hormones (n = 6; means and s.d.s are shown; *P < 0.05). D, dexamethasone; M, IBMX; I, insulin. (F) Real-time PCR analysis of Crif1, Pparγ, Cd36 and adiponectin mRNA levels following treatment with Crif1 siRNA or control siRNA (n = 6; means and s.d.s are shown; *P < 0.05).
Treatment with rosiglitazone fails to promote adipogenesis in Crif1 null ADSCs
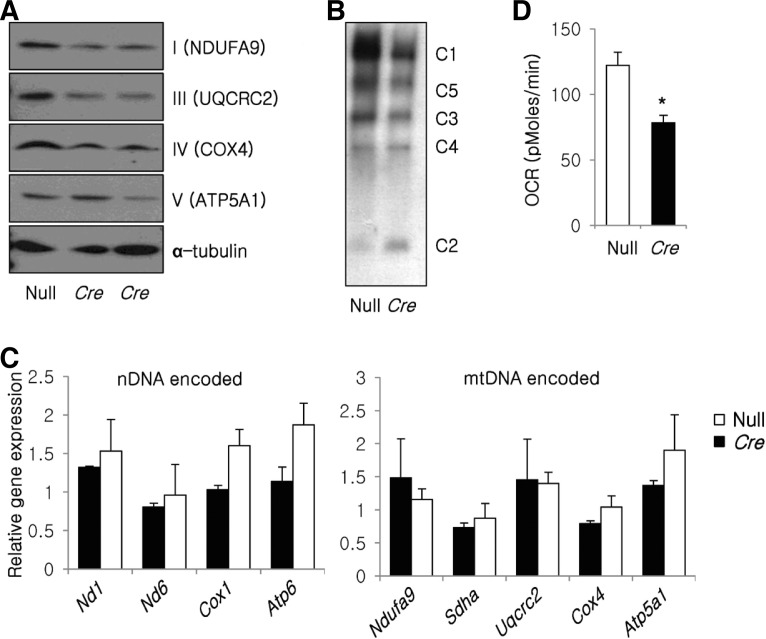
To identify the role of mitochondrial OXPHOS in rosiglitazone-induced adipogenesis, we studied the ability of rosiglitazone to promote adipogenesis in Crif1 null ADSCs. These cells were generated by infecting stromal vascular fraction cells isolated from Crif1flox/flox mice with an adenovirus expressing Cre recombinase. The translation of subunits of OXPHOS complex I (NDUFA9), III (UQCRC2) and IV (COX4) was perturbed in Crif1 null ADSCs (Fig. 6A), and the assembly of the OXPHOS complex was also disrupted (Fig. 6B). The mRNA levels of these subunits were unchanged (Fig. 6C). This defect in the assembly of the OXPHOS complex led to decreased oxygen consumption by mitochondria (Fig. 6D).
Fig. 6.
Mitochondrial dysfunction in Crif1 null ADSCs. Control and Crif1-deficient ADSCs were generated by infection with null or Cre recombinase-containing adenovirus, respectively. Null, control ADSCs; Cre, Crif1-deficince ADSCs. (A) Western blot analysis of the OXPHOS complex subunits NDUFA9, UQCRC2, COX4, and ATP5A1 in null and Crif1-deficient ADSCs. α-tubulin was used as an internal control. (B) BN-PAGE analysis of OXPHOS complexes in control and Crif1-deficient ADSCs. (C) mRNA levels of nDNA-encoded and mDNA-encoded OXPHOS subunits. (n = 4; means and s.d.s are shown; *P < 0.05). Nd1, Nd6 and Ndufa9 for OXPHOS complex I; Sdha for OXPHOS complex II; Uqcrc2 for OXPHOS complex III; Cox1 and Cox4 for OXPHOS complex IV; Atp6 and Atp5a1 for OXPHOS complex V. (D) Basal OCRs were measured using a Seahorse XF-24 flux analyzer (n = 8; means and s.d.s are shown; *P < 0.05).
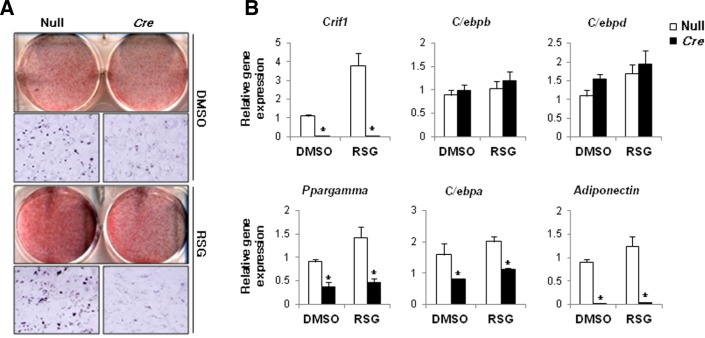
Treatment with rosiglitazone failed to promote adipogenesis in Crif1 null ADSCs. The defect in mitochondrial OXPHOS in Crif1 null ADSCs blocked rosiglitazone-induced lipid accumulation (Fig. 7A). As adipogenesis is controlled by the coordinated activities of many transcription factors, we monitored the expression levels of some of these transcription factors in ADSCs on days 1, 3, and 7 of the differentiation time course. The expression levels of C/ebpβ and C/ebpδ, which are involved in 3T3-L1 adipocyte differentiation and are induced at early time-points, were not perturbed in Crif1 null ADSCs. Furthermore, rosiglitazone treatment increased the expression of these transcription factors. However, the expression levels of Ppargamma and C/ebpα were decreased in untreated Crif1 null ADSCs and rosiglitazone treatment did not induce their expression (Fig. 7B). The expression of adiponectin was abolished in Crif1 null ADSCs, irrespective of whether they were treated with rosiglitazone. These data suggest that mitochondrial OXPHOS may play a role in the generation of endogenous PPARγ ligands.
Fig. 7.
Crif1-deficient ADSCs do not differentiate in response to rosiglitazone. (A) Oil red O staining in fully differentiated control (Null) and Crif1-deficient (Cre) ADSCs. (B) Real-time PCR analysis of Crif1, C/ebpβ, C/ebpδ, Ppargamma, C/ebpα, and adiponectin at day 3 of ADSC differentiation induced with 1 μM rosiglitazone. Control cells were treated with DMSO for vesicle (n = 4, means and s.d.s are shown; n = 6; *P < 0.05).
In summary, these findings indicate that mitochondrial OXPHOS in adipose tissue is crucial for hormone- and rosiglitazoneinduced adipogenesis in vitro and in vivo.
DISCUSSION
In the present study, we showed that mitochondrial OXPHOS affects adipose tissue development in vivo and is required for adipogenesis in response to hormones and PPARγ ligands in vitro. The most important function of mitochondria is to produce energy; however, mitochondria may also modulate the expression of genes that control tissue specific functions. Comparative proteomic studies with the mitochondrial proteome have revealed that tissue specific remodeling controls mitochondrial function in metabolically active tissues (Bugger et al., 2009).
Mitochondria biogenesis during adipocyte differentiation in 3T3-L1 cells has been studied using proteomics and microarray analyses. Thiazolidinediones are PPARγ agonists that stimulate mitochondrial biogenesis and remodeling (Wilson-Fritch et al., 2003; 2004). To verify the role of mitochondria and mitochondrial biogenesis in adipocyte differentiation, a number of studies have targeted specific genes, such as TFAM (Shi et al., 2008) and apoptosis inducing factor (AIF) (Shen et al., 2011). TFAM is a critical regulator of the transcription of mtDNA-encoded OXPHOS subunits and AIF is also important for the function of the OXPHOS complex. The ablation of these genes in adipocytes perturbs or inhibits adipogenesis. We showed that Crif1 regulates the biogenesis of the OXPHOS complex by modulating the translation of mtDNA-encoded OXPHOS subunits. Therefore, homozygous and heterozygous ablation of Crif1 is an appropriate model to study the function of mitochondrial OXPHOS during adipose development in vivo. Interestingly, Crif1 expression transiently increased during adipocyte differentiation in vitro. However, Crif1 expression was not dependent on PPARγ expression and was not matched with induction of OXPHOS expression when adipocytes were treated with PPARγ agonists. These observations indicate that Crif1 expression, which is independent of PPARγ expression and PPARγ agonist stimulation, is linked with mitochondrial OXPHOS complex biogenesis and adipocyte differentiation. However, the regulatory pathways that control Crif1 expression in adipose tissue remain unknown.
In this study, we showed that loss of Crif1 impairs mitochondrial OXPHOS and adipocyte differentiation in vitro and in vivo. It remains unclear how the disruption of mitochondrial OXPHOS modulates gene expression to perturb adipocyte differentiation. It has been recently reported that reactive oxygen species generated by the mitochondrial OXPHOS complex promote the differentiation of adipocytes. We did not measure the level of reactive oxygen species in adipose tissue of Crif1 knockout mice. However, it is unlikely that changes in the levels of reactive oxygen species are responsible for the defects in the development of adipose tissue observed in Crif1 knockout mice. PPARγ and its co-activators control transcriptional networks that govern gene expression patterns in adipocytes and thereby regulate adipogenesis (Hwang et al., 2011). It is unclear how endogenous PPARγ ligands control adipogenesis. Mitochondrial OXPHOS regulates fatty acid metabolism, and it is possible that changes in the levels of fatty acid metabolites following mitochondrial OXPHOS dysfunction affect the levels or behaviors of PPARγ ligands. Defects in the expression or activity of endogenous PPARγ ligands may affect adipogenesis in Crif1 knockout mice.
Thiazolidinediones induce mitochondrial biogenesis and adipogenesis (Wilson-Fritch et al., 2003; 2004); however, these processes have not been reported to be dependent on each other. We found that rosiglitazone treatment did not promote adipogenesis in Crif1-deficient cells. Crif1 null ADSCs did not differentiate into fully matured adipocytes when treated with rosiglitazone. These results suggest that the mitochondrial OXPHOS capacity of adipose tissue may determine whether cells undergo adipogenesis in response to PPARγ agonists.
It is interesting to speculate whether the anti-diabetic effects of PPARγ stimulation are dependent on the mitochondrial OXPHOS capacity of adipose tissue in humans. A significant number of diabetic patients do not respond adequately to treatment with thiazolidinediones. Furthermore, the anti-diabetic effects of thiazolidinediones gradually decrease when patients are treated with these drugs for a prolonged period of time. Our study suggests that the mitochondrial OXPHOS capacity of adipose tissue in diabetic patients is an important factor for determining the anti-diabetic effects of thiazolidinediones.
Acknowledgments
We would like to thank Dr. Evan D. Rosen for providing us with the adiponectin-Cre transgenic mice. M.S. acknowledges financial support from the National Research Foundation on Mitochondria and Metabolic Diseases (NRF, 2010-0020527), MOE, and the Korean Healthcare Technology R&D project (A100588), MHW, Korea.
REFERENCES
- Bugger H., Chen D., Riehle C., Soto J., Theobald H.A., Hu X.X., Ganesan B., Weimer B.C., Abel E.D. Tissue-specific remodeling of the mitochondrial proteome in type 1 diabetic akita mice. Diabetes. 2009;58:1986–1997. doi: 10.2337/db09-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S.E., Mootha V.K. The mitochondrial proteome and human disease. Annu. Rev. Genomics Hum. Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes B.T., Diekman B.O., Gimble J.M., Guilak F. Isolation of adipose-derived stem cells and their induction to a chondrogenic phenotype. Nat. Protoc. 2010;5:1294–1311. doi: 10.1038/nprot.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C.L., Liu G.L., Liu S., Chen X.H., Ji C.B., Zhang C.M., Xia Z.K., Guo X.R. Overexpression of PGC-1beta improves insulin sensitivity and mitochondrial function in 3T3-L1 adipocytes. Mol. Cell. Biochem. 2011;353:215–223. doi: 10.1007/s11010-011-0789-2. [DOI] [PubMed] [Google Scholar]
- He T.C., Zhou S., da Costa L.T., Yu J., Kinzler K.W., Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemati N., Ross S.E., Erickson R.L., Groblewski G.E., MacDougald O.A. Signaling pathways through which insulin regulates CCAAT/enhancer binding protein alpha (C/EBPalpha) phosphorylation and gene expression in 3T3-L1 adipocytes. Correlation with GLUT4 gene expression. J. Biol. Chem. 1997;272:25913–25919. doi: 10.1074/jbc.272.41.25913. [DOI] [PubMed] [Google Scholar]
- Hwang H.H., Moon P.G., Lee J.E., Kim J.G., Lee W., Ryu S.H., Baek M.C. Identification of the target proteins of rosiglitazone in 3T3-L1 adipocytes through proteomic analysis of cytosolic and secreted proteins. Mol Cells. 2011;31:239–246. doi: 10.1007/s10059-011-0026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.J., Kwon M.C., Ryu M.J., Chung H.K., Tadi S., Kim Y.K., Kim J.M., Lee S.H., Park J.H., Kweon G.R., et al. CRIF1 Is essential for the synthesis and insertion of oxidative phosphorylation polypeptides in the mammalian mitochondrial membrane. Cell Metab. 2012;16:274–283. doi: 10.1016/j.cmet.2012.06.012. [DOI] [PubMed] [Google Scholar]
- Kwon M.C., Koo B.K., Moon J.S., Kim Y.Y., Park K.C., Kim N.S., Kwon M.Y., Kong M.P., Yoon K.J., Im S.K., et al. Crif1 is a novel transcriptional coactivator of STAT3. EMBO J. 2008;27:642–653. doi: 10.1038/sj.emboj.7601986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R.H., Ji H., Chang Z.G., Su S.S., Yang G.S. Mitochondrial development and the influence of its dysfunction during rat adipocyte differentiation. Mol. Biol. Rep. 2010;37:2173–2182. doi: 10.1007/s11033-009-9695-z. [DOI] [PubMed] [Google Scholar]
- Luo G.F., Yu T.Y., Wen X.H., Li Y., Yang G.S. Alteration of mitochondrial oxidative capacity during porcine preadipocyte differentiation and in response to leptin. Mol. Cell. Biochem. 2008;307:83–91. doi: 10.1007/s11010-007-9587-2. [DOI] [PubMed] [Google Scholar]
- Newton B.W., Cologna S.M., Moya C., Russell D.H., Russell W.K., Jayaraman A. Proteomic analysis of 3T3-L1 adipocyte mitochondria during differentiation and enlargement. J. Proteome Res. 2011;10:4692–4702. doi: 10.1021/pr200491h. [DOI] [PubMed] [Google Scholar]
- Sears D.D., Hsiao G., Hsiao A., Yu J.G., Courtney C.H., Ofrecio J.M., Chapman J., Subramaniam S. Mechanisms of human insulin resistance and thiazolidinedione-mediated insulin sensitization. Proc. Natl. Acad. Sci USA. 2009;106:18745–18750. doi: 10.1073/pnas.0903032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S.M., Yu Y., Wu Z.X., Zheng Y., Chen G.Q., Wang L.S. Apoptosis-inducing factor is a target gene of C/EBP alpha and participates in adipocyte differentiation. FEBS Lett. 2011;585:2307–2312. doi: 10.1016/j.febslet.2011.05.061. [DOI] [PubMed] [Google Scholar]
- Shi X., Burkart A., Nicoloro S.M., Czech M.P., Straubhaar J., Corvera S. Paradoxical effect of mitochondrial respiratory chain impairment on insulin signaling and glucose transport in adipose cells. J. Biol. Chem. 2008;283:30658–30667. doi: 10.1074/jbc.M800510200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoubridge E.A. Nuclear genetic defects of oxidative phosphorylation. Hum. Mol. Genet. 2001;10:2277–2284. doi: 10.1093/hmg/10.20.2277. [DOI] [PubMed] [Google Scholar]
- Wallace D.C. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 2007;76:781–821. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- Wilson-Fritch L., Burkart A., Bell G., Mendelson K., Leszyk J., Nicoloro S., Czech M., Corvera S. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol. Cell. Biol. 2003;23:1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Fritch L., Nicoloro S., Chouinard M., Lazar M.A., Chui P.C., Leszyk J., Straubhaar J., Czech M.P., Corvera S. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J. Clin. Invest. 2004;114:1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]