Abstract
Symbiotic nodule formation on legume roots is characterized with a series of developmental reprograming in root tissues, including extensive proliferation of cortical cells. We examined a possible involvement of the target of rapamycin (TOR) pathway, a central regulator of cell growth and proliferation in animals and yeasts, during soybean nodule development. Our results show that transcription of both GmTOR and its key downstream effector, GmS6K1, are activated during nodulation, which is paralleled with higher kinase activities of these gene products as well. RNAi-mediated knockdown of GmS6K1 impaired the nodule development with severely reduced nodule weight and numbers. In addition, expression of a few nodulins including leghemoglobin was also decreased, and consequently nitrogen fixation was found to be reduced by half. Proteomic analysis of the GmS6K1-RNAi nodules identified glutamine synthetase (GS), an essential enzyme for nitrogen assimilation in nodules, as one of the proteins that are significantly down regulated. These results appear to provide solid evidence for a functional link between GmS6K1 and nodule development.
Keywords: kinase, nodulation, RNAi, TOR
INTRODUCTION
Leguminous plants form root nodule, a specialized tissue for nitrogen fixation and assimilation through symbiotic interaction with rhizobia. Intermolecular signaling between two symbiotic partners promotes the entry of rhizobia into root cells and the formation of nodule primordia by cell proliferation in root cortex (Oldroyd and Downie, 2008; Oldroyd et al., 2011). This stage of nodule development is characterized by vast synthesis of membrane lipids to enclose endosymbiotic bacteria and synthesis of proteins including nodule-specific proteins (nodulins) such as leghemoglobins (Ott et al., 2005), nodulin-24 (Cheon et al., 1994), nodulin-26 (Masalkar et al., 2010) and so on. Successful symbiosis results in the nitrogen fixation and assimilation into ureides in tropical legumes such as soybean.
In eukaryotes, the target of rapamycin (TOR) pathway has been extensively studied to be a central regulator of cell growth, incorporating various signals including nutrients, stress, energy status, hormones, and growth signals (Laplante and Sabatini, 2012; Ma and Blenis, 2009). TOR kinase is an atypical serine/threonine protein kinase, and is regulated by tuberous sclerosis (TSC)1/2 and the Ras homolog enriched in brain (Rheb) GTPase which transmit diverse upstream signals. TOR phosphorylates several proteins including S6 Kinase 1 (S6K1) and 4E-BP1, thereby promoting protein synthesis, ribosomal biogenesis and lipogenesis. S6K-deficient mice exhibited a small size phenotype (Pende et al., 2004), similarly to Drosophila deficient in S6K (Montagne et al., 1999), suggesting S6K1 as a regulator of cell size. T-DNA insertional mutants of TOR of Arabidopsis resulted in premature arrest of embryo development (Menand et al., 2002). Arabidopsis S6K (AtS6K1) was shown to be regulated by Raptor1, a TOR regulatory protein, upon osmotic stress (Mahfouz et al., 2006). Recently, AtS6K1 was suggested to regulate cell division possibly by association with the retinoblastoma-related 1 (RBR1)-E2FB complex (Henriques et al., 2010; Shin et al., 2012).
Toward better understanding of the coordinated nodule development in legumes, it is critical to understand how cell proliferation and growth in roots is initiated and controlled upon rhizobial infection. In this study, we detected the expression of TOR and S6K1, essential genes for TOR pathway, in root nodules and their biochemical activities as well. RNAi-mediated inhibition of S6K1 resulted in the impaired nitrogen fixation as well as poor nodule development. A few proteins such as glutamate synthetase, a key enzyme to nitrogen assimilatory, were identified to be reduced in S6K1-RNAi nodules by proteomic analysis. These data indicate that TOR signaling may play an important role possibly in early stage of nodule development.
MATERIALS AND METHODS
Plant materials and growth
Soybean (Glycine max cv. Sinpaldal 2) was grown in a growth chamber at 28°C with a photoperoid of 16 h light/8 h dark. For nodulation, three-day-old seedlings grown on moist, absorbent paper were inoculated with rhizobia (Bradyrhizobium japonicum USDA110), transferred to sterilized vermiculites, and further grown for a month.
Real-time RT-PCR
cDNAs were synthesized from total RNA with M-MLV Reverse Transcriptase (Promega). Real-time RT-PCR was performed using SYBR Green PCR Master Mix (Takara) and a Rotor-Gene 3000 (Corbett Research) detection system.
Kinase assay
Equal amount of soluble proteins (200 μg) were incubated with 5 to 30 μl of the GST-fusion protein substrate resin at 4°C for 30 min by gentle rotation. A substrate-mediated kinase pull down assay was performed according to the method described in Shin et al. (2012).
Generation of transgenic root nodules
To make a GmS6K1-RNAi construct, a 250 bp-fragment of GmS6K1 was amplified by PCR with PrimeSTAR Taq DNA polymerase (Takara) using the following primers: primers for GmS6K1-sense, 5′-TATGCTCGAGAGGTTATGCGGAAGGACAA-3′ and 5′-CATTGGTACCGAGATGGGAAACTGCACAA-3′; primers for GmS6K1-antisense, 5′-GCTATCTAGATTATG CGGAAGGACAAGA-3′ and 5′-TTGAAGCTTGATGGGAAACT GCACAAAC-3′. The amplified fragments were inserted into HindIII/XbaI and XhoI/KpnI sites of pKANNIBAL (Wesley et al., 2001). The GmS6K1-RNAi construct was transferred into the binary plasmid pCAMBIA1304 which contained 35S-GUS, and the resulting plasmid was introduced into Agrobacterium rhizogenes (K599) by the freeze-thaw method. Transgenic root nodules containing the GmS6K1-RNAi construct were produced as previously described (Lee et al., 2005).
Acetylene reduction assay
Ethylenes produced per g (fresh weight) of nodules were determined as previously described (Oh et al., 2001).
Two-dimensional electrophoresis (2-DE) analysis
Harvested nodules (800 μg) were dissolved in 350 μl rehydration buffer [7 M urea, 2 M thiourea, 2.5% (w/v) DTT, 4% (w/v) CHAPS and 2% IPG buffer (pH 4–7)] and then loaded onto an immobiline™ dry strip (pH 4–7, 18 cm, GE Healthcare) in a rehydration tray. The IEF was performed on a Multiphor III (GE Healthcare) at 20°C for total of 57 kVh. The 2-D separation was performed on 8–16% (v/v) linear gradient SDS-polyacrylamide gels and then imaged using a GS-710 imaging calibrated densitometer (Bio-Rad).
Identification of proteins by LC-MS/MS
Protein spots were excised from the stained 2-DE gels, destained with destaining solution (25 mM ammonium bicarbonate, 50% acetonitrile), digested with sequencing grade trypsin (Promega), desalted using a GELoader tip (Eppendorf) packed with 1.5 μg of Poros 20 R2 resin (PerSpectiveBiosystems), and applied on the C18 RP-HPLC column (75 um × 150 mm). Agilent 1100 Series LC system (Agilent Technologies) was used to separate tryptic peptides, which were eluted with 0–40% acetonitrile gradient for 60 min. The eluant was analyzed by Finnigan LCQ Deca (ThermoQuest) equipped with a nano-electrospray ion source.
RESULTS AND DISCUSSION
High expression of TOR signaling components during soybean nodulation
As nodule development in legumes is accompanied with extensive cortical cell proliferation and cell expansion in response to rhizobial signaling and infection into root tissues, we explored possibility of TOR signaling pathway contributing to soybean nodule development, in a similar fashion to what has been observed in animals and yeast, coordinating cellular growth and metabolism after integrating various external environmental cues (Laplante and Sabatini, 2012). Blast searches of the soybean genome database, Phytozome (http://www.phytozome.net/soybean.php), for conserved sequences of TOR kinase and its key downstream effectors ribosomal S6 kinase (S6K1) and ribosomal protein S6 (RPS6) identified several soybean orthologs; 2 paralogs of GmTOR kinase, which we tentatively named GmTORa and GmTORb, 3 paralogs of GmS6K1 as GmS6K1a to -c, and 6 paralogs of GmRPS6 as GmRPS6a to -f (Supplementary Figs. 1–3). Real time RT-PCR showed that both GmTORs and GmS6K1s were induced in 7-day-old nodules by about 6 and 8 folds, respectively, though our primer design did not allow to distinguish which particular paralogs were upregulated (Fig. 1). By contrast, expression of GmRPS6s remained relatively constant, at slightly higher level than in root tissue, throughout the entire stages of nodule development. Thus far, the only other occasion where an active TOR transcription was detected was in Arabidopsis meristem (Menand et al., 2002). While it is not known if TOR kinase is involved in controlling cell proliferation in plants as well, let alone how it is being orchestrated, this strong induction of GmTORs and GmS6K1s obtained in our result suggests that function of TOR pathway may be required for soybean root nodules development although no information is available at present on how the cell proliferation during nodulation is connected to cellular signaling such as TOR pathway. S6K1 has been known as a central regulator of cell and body size in animals (Arsham and Neufeld, 2006) and with recent reports of S6K1 being involved in the control of cell size in plants (Henriques et al., 2010; Montagne et al., 1999), it is highly likely that GmS6K1s play a critical role in nodule growth.
Fig. 1.
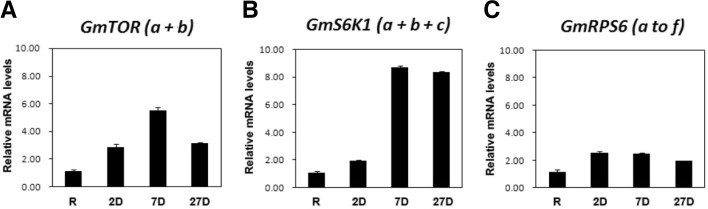
Expression of TOR, S6K1, and RPS6 during nodulation. Expression of GmTOR (a + b) (A), GmS6K1 (a + b + c) (B) and GmRPS6 (a to f) (C), respectively, in soy-bean tissues including root (R), 2-day-old nodules (2D), 7-day-old nodules (7D) and 27-day-old nodules (27D) was examined. Relative transcript levels were measured by real time RT-PCR, with ubiquitin as control. Data are representative of three independent experiments. Bars represent standard deviations (n = 3).
Increasing S6K1 activity during nodule development
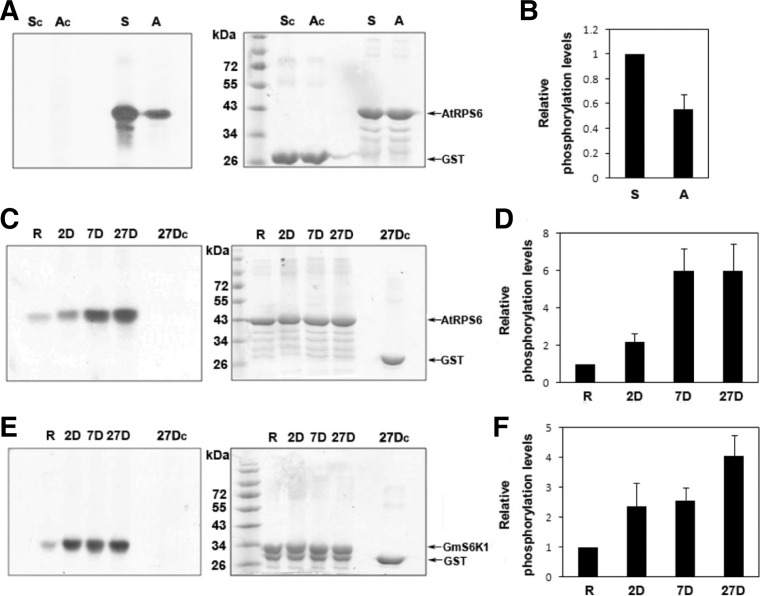
In order to make certain that the elevated expression of GmTORs and GmS6K1s during nodulation actually corresponds to higher activities of these kinases, the substrate-mediated kinase pull down (Shin et al., 2012) was employed to detect their kinase activities at three different stages of the nodule development from which the transcription analyses of GmTORs and GmS6K1s were performed. A carboxy-terminal region of RPS6 has been shown to be phosphorylated by S6K1 in animal cells (Krieg et al., 1988; Radimerski et al., 2000) as well as in plant (Williams et al., 2003). In addition, the GFP-S6K1 was shown to be able to phosphorylate a carboxy-terminal region of Arabidopsis RPS6 (a.a. 150–249; AtRPS6-CT) (Mahfouz et al., 2006). The AtRPS6- CT was used as a substrate for the pull down and kinase reaction of the GmS6K1s because the overall amino acids sequence of AtRPS6 is more than 95% identical to those of GmRPS6s with highly conserved C-terminal phosphorylation motif shared among them (Supplementary Fig. 3). It has been demonstrated that phosphorylation of the carboxy-terminal region of S6K1 by mTOR1 is essential for activation of S6K1 (Cheatham et al., 1995; Weng et al., 1995). The carboxy-terminal region of AtS6K1 (a.a. 392–465; AtS6K1-CT) also contains the conserved phosphorylation motifs although their phosphorylation has not been experimentally confirmed until very recently (Shin et al., 2012). In this study, GmS6K1-CT homologous to AtS6K1-CT was used to examine the level of GmS6K1 phosphorylation, which possibly reflects the status of GmS6K1 activation by GmTOR. Total protein extract from actively nitrogen-fixing nodules (27- day-old nodules) was subjected to the substrate-mediated kinase pull down with glutathione-S-transferase (GST) fusion protein of AtRPS6-CT (GST-AtRPS6-CT) followed by kinase assay in vitro, and showed a strong kinase activity specifically phosphorylating the AtRPS6, which was about twice as strong as that obtained with Arabidopsis leaf extracts (Figs. 2A and 2B). This kinase activity was largely reflective of the expression profile of GmS6K1s (Fig. 1), thus it was only about 2-fold higher than that of the roots in 2-day-old nodules but exhibited marked increase (approx. 7-fold) in both 7-day- and 27-day-old nodules (Figs. 2C and 2D). Although we cannot rule out the possibility of other unknown kinases being also pulled down and phosphorylating the AtRPS6, the assay performed with nodules expressing RNAi construct of GmS6K1 clearly demonstrated that GmS6K1s were at least the primary kinases phosphorylating AtRPS6 in our results (Figs. 3G and 3H). The kinase activity of GmTORs was also examined by using a GST-fused C-terminal region of GmS6K1s (GST-GmS6K1-CT) as a substrate in the same manner (Fig. 2E). Despite the fact that the specificity for GmS6K1 phosphorylation is not established yet, considering Arabidopsis S6K1 was shown to interact specifically with At-TOR (Mahfouz et al., 2006), the phosphorylation of GmS6K1-CT in our data is more than likely to represent portion of the GmTOR kinase activity. Under this premise, it appears that GmTORs also become active in 2-day-old nodules and maintain their activity as nodules develop further (Fig. 2C), even the GmTOR transcripts diminishes after 7-day post nodule development (Fig. 1), an observation consistent with the high level of GmS6K1 activity in mature nodules. Expression of GST-fusions in E. coli, at our hands, showed smaller sizes of proteins that appeared to be proteins with premature stops during protein synthesis or nonspecific proteins. But the topmost bands were always the proteins of our interest as they were identified in Western blots. These results indicate that GmTORs and GmS6K1s indeed become active upon nodule initiation and may contribute to nodule formation in soybean. Both kinases seem to become activated in early stages of nodules such as 2-day-old nodules in which nodule organogenesis including cortical cell division occurs (Oldroyd et al., 2011).
Fig. 2.
Kinase activities of GmS6K1 and GmTOR during nodule development. Phosphorylations of AtRPS6-CT-GST (A-C) or GST-GmS6K1-CT (E) according to the substrate-mediated kinase pull-down method (Shin et al., 2012) were shown. GST fusion protein substrates (AtRPS6-CT or GmS6K1-CT) were allowed to incubate with proteins from soybean tissues indicated or Arabidopsis leaf extracts; S for soybean nodules, A for Arabidopsis leaf extracts, R for root, 2D for 2-day-old nodules, 7D for 7-day-old nodules, and 27D for 27-day-old nodules. SC, AC and 27DC: the negative control kinase reaction with 200 μg of soybean 27-day-old nodule proteins (SC) or Arabidopsis leaf extracts (AC) or 27-day-old nodules (27DC) input using GST protein as substrate. Relative phosphorylation level was shown as the means and standard deviations (SD) of the means of three independent experiments. (A, C, and E) Left panels; autoradiographs of the kinase reactions with cellular proteins indicated. Right panels; Coomassie-stained gels of the reaction PAGE from which the autoradiograph shown in left panel was generated. (B, D, and F) Quantification of the relative kinase activities shown in (A, C, and E). Kinase activity was normalized to the activity with soybean nodules (B) or roots (D, F) and was defined as “1.” Three replicates were conducted. Error bars indicate standard error.
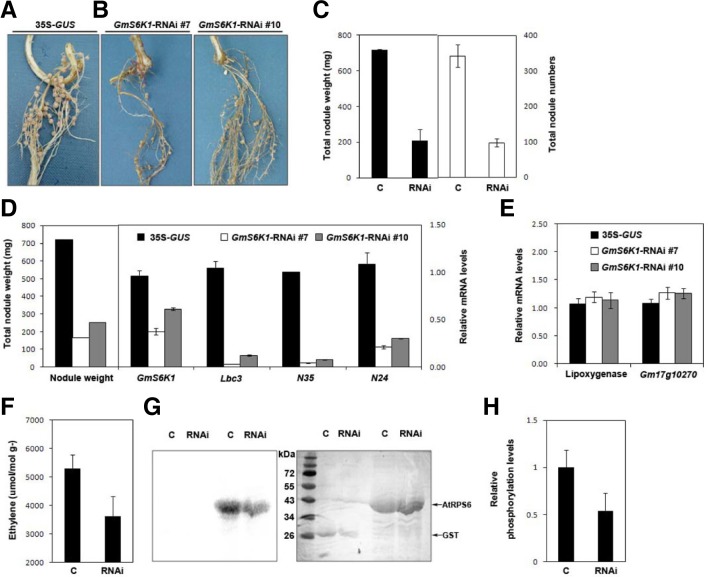
Fig. 3.
Effects of the expression of an RNAi construct against GmS6K1 on nodule development. (A, B) Nodules formed on soybean hairy roots containing an empty vector containing 35S-GUS (A, control) or the GmS6K1-RNAi cloned in the vector (B). GUS-positive (transgenic) hairy roots with 35S-GUS or 35S-GUS-GmS6K1-RNAi were inoculated with B. japonicum (USDA110). GmS6K1-RNAi plant #7 showed severely impaired nodulation while GmS6K1-RNAi plant #10 had moderately affected nodules. These experiments were repeated three times and representative results are shown. In each experiment, 8 to 13 plants per construct were used. (C) Total nodule weights (mg) and total nodule numbers of control (C, 35S-GUS) and GmS6K1-RNAi (RNAi) plants were observed. (D) Relative transcript levels were examined for representative plants of different nodule weights. Transcript levels of GmS6K1, leghemoglobin (Lbc3), nodulin-35 (N35), nodulin-24 (N24), lipoxygenase and Gm17g10270 were measured by real time RT-PCR in controls (35S-GUS), GmS6K1-RNAi #7 and GmS6K1-RNAi #10 nodules. Ubiquitin was used as control. (F) Nitrogenase activities were determined by the acetylene reduction assay. Data are averaged from three independent experiments. (G) Kinase activity of GmS6K1 in nodules expressing 35S-GUS (C) or the GmS6K1-RNAi (RNAi). Left panel, autoradiograph of the kinase reactions with cellular proteins indicated. Right panel, Coomassie-stained gel of the reaction PAGE from which the autoradiograph shown in left panel was generated. (H) Quantification of the relative kinase activities shown in (G). Kinase activity was normalized to the activity with control nodules. Data were presented as the average and standard deviations (SD) of the means of three independent experiments.
The effects of phytohormones, especially auxin and cytokinin, have been reported to affect nodule organogenesis (Lohar et al., 2004; Oldroyd et al., 2011), and the activity of plant S6K1 was also found to be positively regulated by these hormones (Turck et al., 2004). Therefore, it seems probable that control of nodulation by these phytohormones is mediated through the activation of TOR signaling components. In order to explore the connection between phytohormones and TOR signaling in root nodules, 27-day-old soybean nodules were incubated with auxin or cytokinin, and their extracts were assayed for GmS6K1 activity in the same manner as described above. Both auxin and cytokinin treatment increased slightly the GmS6K1 activity in dose-dependent manner (Supplementary Fig. 4). Despite the magnitude of change in GmS6K1 activity was rather smaller than expected, perhaps due to a certain levels of endogenous auxin and cytokinin already present in root nodules, which would have rendered the effect of the additional exogenous hormones on the GmS6K1 activity become marginal, the data nonetheless demonstrates positive effects of both auxin and cytokinin on the activity of GmS6K1 in developing nodule, supporting a possible link between these hormones and the TOR signaling pathway in regulating nodulation. For clear dissection of the relationship between TOR signaling and nodule organogenesis, however, more extensive examination of these hormonal effects needs to be carried out, including studies using inhibitors of hormone transport or mutants of hormone biosynthesis, precise implementation of which in nodule system is rather limited at present technical level. Regardless, our data obtained from these analyses of kinase activities suggest that the functional TOR pathway in soybean may contribute to nodule development.
Impaired nodulation caused by RNAi-mediated inhibition of GmS6K1s expression
The functional requirement of GmS6K1 in soybean nodule development was further evaluated by generating transgenic nodules expressing RNAi construct of GmS6K1. An N-terminal region of 250-bp sequence conserved in all three GmS6K1 paralogs (Supplementary Fig. 2, dashed arrows) was chosen for the interference and introduced into Agrobacterium rhizogenes to form transgenic hairy roots of soybean (Lee et al., 2005) after being cloned downstream of a nodule-specific leghemoglobin (Lbc3) promoter. Significantly impaired nodulation was observed in GmS6K1-RNAi roots with apparent reduction in both weight and number of nodules (Fig. 3). Two soybean mRNA sequences having the highest homology with GmS6K1s - Gm17g10270 and lipoxygenase gene -were selected as nonspecific off-target controls in this experiment. While the transcript levels of these off-targets remained unchanged (Fig. 3E), expression of GmS6K1s showed substantial decrease in all transgenic GmS6K1s-RNAi nodules (Fig. 3D), confirming specific inhibition of only GmS6K1s in these plants. Moreover, the degree of defect in nodulation in the GmS6K1s-RNAi roots was proportional to the repression level of the GmS6K1 expression such that transgenic plant #7 with strongly impaired nodules showed one-third of GmS6K1s expression compared with control, while plant #10 with moderately affected nodules had two-thirds of GmS6K1s expression (Fig. 3D), suggesting a consistent relationship between nodule development and GmS6K1s expression. In addition, GmS6K1s-RNAi root nodules exhibited reduced expressions of essential nodule-specific genes; leghemoglobin c3 (Lbc3) which functions as oxygen carrier in nodules, nodulin 24 which is a symbiosome membrane protein, and nodulin 35, a subunit of uricase. Moreover, nitrogen-fixing ability was significantly affected in GmS6K1s-RNAi root nodules as examined by acetylene reduction assay (Fig. 3F). Twenty seven-day-old nodules on GmS6K1s-RNAi hairy roots showed about a half-level kinase activity of GmS6K1 compared with those on 35S-GUS control (Figs. 3G and 3H). All the data from GmS6K1s-RNAi root nodules suggest strongly that GmS6K1s are essential for nodule formation.
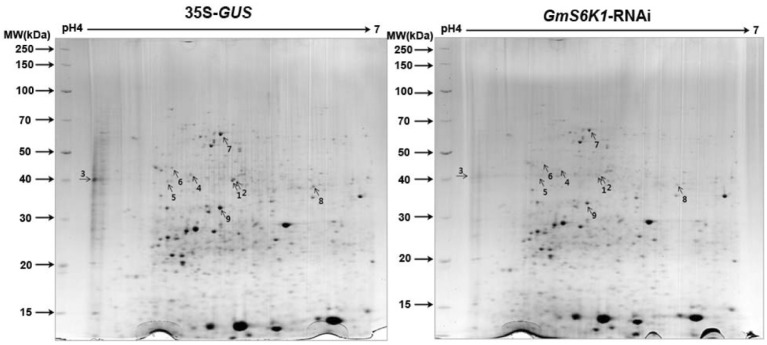
In order to obtain more insight into how GmS6K1s affect nodule development, GmS6K1s-RNAi root nodules were analyzed by a proteomic approach. Proteins from GmS6K1s-RNAi nodules were compared with those from control nodules by two-dimensional differential gel electrophoresis (Fig. 4). Down-regulated proteins (> 2 fold) were selected and identified by Liquid Chromatography-Mass Spectrometry (LC-MS) analysis (Table 1). Among them, 6 spots contained identical amino acid sequences to known soybean proteins (two glutamine synthetases-β1 and β2, heat shock protein 90-2, and glyceraldehyde- 3-phosphate dehydrogenase), while others to non-soybean proteins (Supplementary Fig. 5). For three spots, we could not find the corresponding soybean proteins, but proteins from other species. With DNA sequences corresponding to the identified peptide sequences from the LC-MS analysis, we blasted against the soybean genome database (http://www.phytozome.net/soybean) and found two soybean proteins which had very high homologies to the identified proteins (92% to spots # 6 and 80% to spot #7) and were annotated with the same functions, and a soybean protein with 69% homology to spot #9 (Supplementary Fig. 6). There may be other soybean paralogs which match completely with amino acid sequences from the LC-MS analysis but not yet included in the present soybean protein sequence database.
Fig. 4.
Two-dimensional differential gel electrophoresis analysis of GmS6K1-RNAi nodules. Two-dimensional gel electrophoresis of 27-day-old GmS6K1-RNAi nodules was compared with control (35S-GUS). Arrows and spot numbers indicate proteins for further identification by LC-MS/MS.
Table 1.
Identification of proteins reduced in GmS6K1-RNAi nodules by LC-MS/MS
| Spot label | NCBI BLAST | Protein name | Mass | Fold change |
|---|---|---|---|---|
| 1 | gil10946357 | Cytosolic glutamine synthetase Gsbeta1 [Glycine max] | 38.9 | 3.9 |
| 2 | gil363807092 | Cytosolic glutamine synthetase beta2 [Glycine max] | 39.1 | 3.9 |
| 3 | gil10946357 | Cytosolic glutamine synthetase Gsbeta1 [Glycine max] | 38.9 | 6.4 |
| 4 | gil351725976 | Heat shock protein 90-2 [Glycine max] | 80.1 | 4.2 |
| 5 | gil351725976 | Heat shock protein 90-2 [Glycine max] | 80.1 | 9.1 |
| 6 | gil383100964 | Early-responsive to dehydration 2 [Arabidopsis halleri subsp. Halleri] | 68.8 | 2.6 |
| 7 | gil308807030 | Putative TCP-1/cpn60 chaperonin family protein (ISS) [Ostreococcus tauri] | 55.0 | 2.6 |
| 8 | gil351727206 | Glyceraldehyde-3-phosphate dehydrogenase [Glycine max] | 36.7 | 2.1 |
| 9 | gil111378664 | Putative auxin efflux carrier [Zea mays] | 65.1 | 2.0 |
Glutamine synthetase (GS) β always showed higher activity in cytoplasm of nodules than root and shoots as cytosolic GS is critical to assimilate ammonium resulting from N2 fixation into glutamate in nodules. The mechanism of repression of GS β in GmS6K1s-RNAi root nodules is unknown at present. In previous studies of yeasts treated with limiting nitrogen sources or rapamycin, GLN1 (a yeast ortholog of GS) was rather induced by activated GLN3 (Cardenas et al., 1999), but whether Sch9 (a yeast ortholog of S6K1) was not involved in the induction was not examined. It will be intriguing to find out if the activator of GS in plants such as a Dof factor (Reuda-Lopéz et al., 2008) would be functional in GmS6K1s-RNAi root nodules. Recently, GS was found to interact with the symbiosome membrane nodulin 26, a NH3 transporter in soybean nodule (Masalkar et al., 2010). The binding of GS to nodulin 26 was proposed to promote efficient assimilation of fixed nitrogen. Reduced amount of GS β1 in GmS6K1s-RNAi root nodules could cause inefficient nitrogen assimilation, resulting in reduced induction of essential nodulins (Fig. 3) and thus the overall poor development of root nodules. However, it is also possible that the reduced GS β1 level is just a result of poor nodule development, rather than a cause of the observed effects in GmS6K1s-RNAi soybean.
We have so far presented evidence that TOR pathway is essentially required for proper nodule development, enabling soybean to perform efficient nitrogen fixation and assimilation. During early nodulation stage, cytokinin affects nodule organogenesis, causing cortical cell division. It will be of interest how TOR pathway is activated during the early stage of nodulation.
Supplementary Material
Acknowledgments
This work was supported by the SRC Research Center for Women’s Diseases of Sookmyung Women’s University (2010) (to CIC).
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Arsham AM, Neufeld TP. Thinking globally and acting locally with TOR. Curr Opin Cell Biol. 2006;18:589–597. doi: 10.1016/j.ceb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatham L, Monfar M, Chou MM, Blenis J. Structural and functional analysis of pp70S6K. Proc. Natl. Acad. Sci. USA. 1995;92:11696–11700. doi: 10.1073/pnas.92.25.11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon C-I, Hong Z, Verma DP. Nodulin-24 follows a novel pathway for integration into the peribacteroid membrane in soybean root nodules. J Biol Chem. 1994;269:6598–6602. [PubMed] [Google Scholar]
- Henriques R, Magyar Z, Monardes A, Khan S, Zalejski C, Orellana J, Szabados L, de la Torre C, Koncz C, Bögre L. Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. EMBO J. 2010;29:2979–2993. doi: 10.1038/emboj.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg J, Hofsteenge J, Thomas G. Identification of the 40S ribosomal protein S6 phosphorylation sites induced by cycloheximide. J Biol Chem. 1988;263:11473–11477. [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Shin KH, Kim YK, Suh JY, Gu YY, Kim MR, Hur YS, Son O, Kim JS, Song E, et al. Induction of thioredoxin is required for nodule development to reduce reactive oxygen species levels in soybean roots. Plant Physiol. 2005;139:1881–1889. doi: 10.1104/pp.105.067884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohar DP, Schaff JE, Laskey JG, Kieber JJ, Bilyeu KD, Bird DM. Cytokinins play opposite roles in lateral root formation, and nematode and Rhizobial symbioses. Plant J. 2004;38:203–214. doi: 10.1111/j.1365-313X.2004.02038.x. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Mahfouz MM, Kim S, Delauney AJ, Verma DP. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell. 2006;18:477–490. doi: 10.1105/tpc.105.035931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masalkar P, Wallace IS, Hwang JH, Roberts DM. Interaction of cytosolic glutamine synthetase of soybean root nodules with the C-terminal domain of the symbiosome membrane nodulin 26 aquaglyceroporin. J Biol Chem. 2010;285:23880–23888. doi: 10.1074/jbc.M110.135657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA. 2002;99:6422–6427. doi: 10.1073/pnas.092141899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- Oh HS, Son O, Chun JY, Stacey G, Lee MS, Min KH, Song ES, Cheon C-I. The Bradyrhizobium japonicum hsfA gene exhibits a unique developmental expression pattern in cowpea nodules. Mol Plant Microbe Interact. 2001;14:1286–1292. doi: 10.1094/MPMI.2001.14.11.1286. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, Downie JA. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet. 2011;45:119–144. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- Ott T, van Dongen JT, Günther C, Krusell L, Desbrosses G, Vigeolas H, Bock V, Czechowski T, Geigenberger P, Udvardi MK. Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr Biol. 2005;15:531–535. doi: 10.1016/j.cub.2005.01.042. [DOI] [PubMed] [Google Scholar]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1-/-S6K2-/- mice exhibit perinatal lethality and rapamycin-sensitive5’- terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radimerski T, Mini T, Schneider U, Wettenhall REH, Thomas G, Jenö P. Identification of insulin-induced sites of ribosomal protein S6 phosphorylation in Drosophila melanogaster. Biochemistry. 2000;39:5766–5774. doi: 10.1021/bi9927484. [DOI] [PubMed] [Google Scholar]
- Reuda-Lopéz M, Crespillo R, Cánovas FM, Ávila C. Differential regulation of two glutamine synthetase genes by a single Dof transcription factor. Plant J. 2008;56:73–85. doi: 10.1111/j.1365-313X.2008.03573.x. [DOI] [PubMed] [Google Scholar]
- Shin YJ, Kim S, Du H, Choi S, Verma DP, Cheon C-I. Possible dual regulatory circuits involving AtS6K1 in the regulation of plant cell cycle and growth. Mol. Cells. 2012;33:487–496. doi: 10.1007/s10059-012-2275-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Zilbermann F, Kozma SC, Thomas G, Nagy F. Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis. Plant Physiol. 2004;134:1527–1535. doi: 10.1104/pp.103.035873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Q-P, Andrabi K, Kozlowski MT, Grove JR, Avruch J. Multiple independent inputs are required for activation of the p70 S6 kinase. Mol Cell Biol. 1995;15:2333–2340. doi: 10.1128/mcb.15.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Werner-Fraczek J, Chang I-F, Bailey-Serres J. Regulated phosphorylation of 40S ribosomal protein S6 in root tips of maize. Plant Physiol. 2003;132:2086–2097. doi: 10.1104/pp.103.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.