Abstract
In plants, transgenes with inverted repeats are used to induce efficient RNA silencing, which is also frequently induced by highly transcribed sense transgenes. RNA silencing induced by sense transgenes is dependent on RNA-dependent RNA polymerase 6 (RDR6), which converts single-stranded (ss) RNA into double-stranded (ds) RNA. By contrast, it has been proposed that RNA silencing induced by self-complementary hairpin RNA (hpRNA) does not require RDR6, because the hpRNA can directly fold back on itself to form dsRNA. However, it is unclear whether RDR6 plays a role in hpRNA-induced RNA silencing by amplifying dsRNA to spread RNA silencing within the plant. To address the efficiency of hpRNA-induced RNA silencing in the presence or absence of RDR6, Wild type (WT, Col-0) and rdr6-11 Arabidopsis thaliana lines expressing green fluorescent protein (GFP) were generated and transformed with a GFP-RNA interference (RNAi) construct. Whereas most GFP-RNAi-transformed WT lines exhibited almost complete silencing of GFP expression in the T1 generation, various levels of GFP expression remained among the GFP-RNAi-transformed rdr6-11 lines. Homozygous expression of GFP-RNAi in the T3 generation was not sufficient to induce complete GFP silencing in several rdr6-11 lines. Our results indicate that RDR6 is required for efficient hpRNA-induced RNA silencing in plants.
Keywords: double-stranded RNA, gene silencing, hairpin RNA, RDR6, RNA interference, single-stranded RNA
INTRODUCTION
Gene silencing is a mechanism that employs small RNAs and protein effectors to interfere with the expression of homologous genes at the transcriptional and post-transcriptional levels (Voinnet, 2008). Transcriptional gene silencing (TGS), which takes place through repression of transcription, is often associated with methylation of the corresponding homologous promoter (Neuhuber et al., 1994; Park et al., 1996). Post-transcriptional gene silencing (PTGS), occurring through sequence-specific degradation of target mRNAs, is characterized by accumulation of small interfering RNA (siRNA) and methylation of target gene sequences (Depicker and Montagu, 1997; Vaucheret et al., 2001). Gene silencing was first discovered in plants and is highly conserved among multicellular eukaryotes. It initiates with the formation of dsRNA, which is subsequently processed into siRNA. siRNAs are produced from dsRNAs by an RNase III-like enzyme called DICER, which has dsRNA-binding, RNA helicase and P-element-induced wimpy testes (PIWI)-Argonaute (AGO)-Zwille (PAZ) domains (Bernstein et al., 2001). Plants possess four DICER homologs: DCL1, DCL2, DCL3, and DCL4. DCL1 is responsible for producing various sizes of microRNAs (small RNAs encoded in the genome), whereas DCL2, DCL3, and DCL4 produce 22, 24, and 21 nucleotide (nt) siRNAs, respectively (Bartel, 2004; Dunoyer et al., 2005; Xie et al., 2004). The 24 nt siRNAs are reported to guide RNA-directed DNA methylation (RdDM) in plants (Wassenegger et al., 1994), whereas the 21 and 22 nt siRNAs are known to trigger cognate mRNA degradation and secondary siRNA biogenesis, respectively (Chen et al., 2010; Hamilton et al., 2002).
The guide strand of siRNA (usually the antisense strand) associates with AGO proteins to form the core of the RNA silencing effector complexes, which provide RNA-induced silencing complexes (RISCs) with substrate specificity for RNA cleavage (Brodersen and Voinnet, 2006). AGO proteins are characterized by PAZ, middle (Mid) and PIWI domains. The PAZ and Mid domains are thought to be involved in binding the 3′ and 5′ ends of small RNAs, respectively, whereas the PIWI domain, which is highly homologous to that of RNase H, is implicated in the cleavage of target RNAs (Hutvagner and Simard, 2008; Song et al., 2004). The Arabidopsis genome encodes 10 putative AGOs. AGO1, AGO7 and AGO10 bind to 21–22 nt siRNAs to guide PTGS, whereas AGO4, AGO6 and AGO9 bind to 24 nt siRNAs to mediate TGS (Morel et al., 2002; Vaucheret, 2008).
The initial response in gene silencing is amplified through the synthesis of ‘secondary’ siRNAs by RNA-dependent RNA polymerase (RDR) using the dicer-produced ‘primary’ siRNAs as templates (Baulcombe, 2007). The dsRNA source of primary siRNAs can be either exogenous or endogenous. Six RDR homologs have been identified in Arabidopsis. Three of them (RDR3a, RDR3b, and RDR3c) share a typical DFDGD amino acid motif in their catalytic domains; however, the function of this motif remains unclear. RDR1, RDR2, and RDR6 share the C-terminal canonical catalytic DLDGD motif and have orthologs in many plant species (Wassenegger and Krczal, 2006). In TGS, transposon sequences amplified by RDR2 are processed into 24 nt siRNA by DCL3 and subsequently incorporated into AGO4 or AGO6 (Chen et al., 2010; Hamilton et al., 2002). In PTGS, transgenes, virus RNAs or unwanted RNAs amplified by RDR6 are processed into 21 nt siRNA by DCL4 and subsequently incorporated into AGO1 (Hamilton et al., 2002; Qu et al., 2008; Xie et al., 2005). The amplification of aberrant RNAs by RDR6 in either a primer-dependent or a primer-independent manner enables the production of secondary siRNAs corresponding to sequences outside of primary targeted regions of a transcript (Himber et al., 2003; Moissiard et al., 2007). The RDR6-dependent dsRNA amplification, called transitivity, is crucial for the systemic spread of silencing and an efficient antiviral defense response in plants (Himber et al., 2003; Klahre et al., 2002; Voinnet, 2005).
RNA interference (RNAi) using self-complementary hpRNA is an effective tool to elicit dsRNA-mediated gene silencing in plants (Mansoor et al., 2006; Wesley et al., 2001). The usefulness of RNAi technology in plants has been validated through the generation of knockdown mutants with novel traits, which are stably inherited by their progeny (Chuang and Meyerowitz, 2000; Davuluri et al., 2005). It has been proposed that RDR6 is dispensable for hpRNA-induced gene silencing (Beclin et al., 2002). A significant amount of primary siRNA may be produced through the synthesis of self-complementary hairpin RNA directly by RNAi in WT and plants with loss of function of the RDR6 gene. However, the synthesis of secondary siRNAs by RDR6 could also affect the efficiency of RNAi. To address this, we carefully compared the efficiency of RNAi in the presence or absence of the RDR6 gene in Arabidopsis thaliana. GFP-expressing WT and rdr6-11 plants were generated, and a construct harboring GFP-RNAi was used to induce gene silencing in the WT and rdr6-11 plants.
MATERIALS AND METHODS
Plant materials and growth conditions
For growth under sterile conditions, Arabidopsis thaliana seeds were surface sterilized (5 min in 70% ethanol, followed by a 15-min incubation in 5% [v/v] sodium hypochlorite and a five-fold rinse in sterile distilled water) and sown on Murashige and Skoog (MS) medium (Duchefa) supplemented with 3% sucrose, pH 5.7, and 0.25% (w/v) phytagel in Petri dishes. Arabidopsis thaliana plants were grown in a growth chamber under a long-day regime [16 h fluorescent light (120 μmol · m−2 · s−1) at 23°C and 60% relative humidity/8 h dark at 18°C and 75% relative humidity] after a 4-d vernalization period (4°C in dim light). Arabidopsis thaliana seedlings were rescued from the plates 14 days after germination, transferred to soil and grown under the same conditions.
Genetic analysis
A genomic DNA region of rdr6-11 was amplified by PCR using primer pair P1 (5′-TACTGTCCCTGGCGATCTCT-3′) and P2 (5′-CCACCTCACACGTTCCTCTT-3′), followed by digestion with Taq1 to verify the point mutation (C to T) and homozygosity (Peragine et al., 2004). P3 (5′-CTGTGTCAATGTCGGATCTCTCGAT-3′ and LBSalk (5′-GCGTGGACCGCTTGCTGCAACT-3′) were used to verify T-DNA insertion in rdr6-16, and LBSail (5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTC-3′) and P2 were used to verify T-DNA insertion in rdr6-17.
Expression of GFP in WT and rdr6-11 backgrounds
The pCAMBIA1302 vector carrying genes for the selectable marker hygromycin phosphotransferase (HPT) and green fluorescent protein (GFP) driven by the CaMV 35S promoter was used to generate GFP-expressing plants (Hajdukiewicz et al., 1994). pCAMBIA1302 vector was introduced into Agrobacterium tumefaciens strain GV3101 via electroporation, and the resulting strain was used to transform Arabidopsis plants by the floral dip method (Clough and Bent, 1998). Transgenic seeds were selected on MS plates containing 50 μg/ml hygromycin. Transgenic lines whose T2 progeny phenotypes indicated likely single-copy T-DNA insertions (segregating 3:1 for hygromycin resistance) were selected for further testing. The cut-off for the probability of a line to contain a single-copy T-DNA insertion was p < 0.05, as determined by the Chi-square test: [(observed resistant plants −0.75)2 / (0.75)] + [(observed sensitive plants −0.25)2 / (0.25)]. Progeny sets with Chi-square values less than 3.841 were retained for further study. Expression of GFP in WT and rdr6-11 backgrounds was assessed by immunoblot analysis using a GFP monoclonal antibody (Abcam). Representative lines expressing similar amounts of GFP were selected for further experiments.
Construction of GFP-RNAi and Arabidopsis transformation
To create the GFP-RNAi vector, the HPT selectable marker gene in the pCAMBIA1302 vector was replaced with the PPT (phosphinothricin) resistance gene. A 198 nt intron was inserted between the SpeI and BglII sites upstream of the GFP gene and a full-length GFP was inserted at the BglII and NcoI sites in the antisense orientation upstream of the intron. The resulting GFP-RNAi construct was transferred into Agrobacterium tumefaciens strain GV3101 and used to silence GFP expression in both rdr6-11 and WT backgrounds. Transgenic plants carrying the GFP-RNAi were selected on MS media containing 7.5 μg/ml phosphinothricin. T1 plants resistant to phosphinothricin were transferred to soil when their first true leaves appeared. GFP expression in independent T1 lines was assessed by immunoblot analysis using a GFP monoclonal antibody. Plants displaying continued GFP expression were selected and allowed to produce T2 seeds. A single insertion of T-DNA and homozygosity were confirmed as described above.
Protein extraction and immunoblot analysis
Total soluble protein was extracted from rosette leaves of 30-d-old Arabidopsis plants. Approximately 0.1 g of leaf tissue was collected in a microcentrifuge tube and ground into a fine powder in liquid nitrogen with a prechilled tissue homogenizer. Fine powder was mixed with 100 μl of pre-chilled extraction buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride (PMSF) and kept on ice for 10 min. The soluble proteins were separated by centrifugation at 15, 000 × g for 15 min at 4°C. Protein concentration was determined by spectrophotometry according to the Bradford method (Bradford, 1976). For immunoblot analysis, 20 μg soluble protein was separated by SDS-PAGE and transferred to a Protran nitrocellulose membrane (Whatman GmbH) using an XCell II (Invitrogen) electrotransfer unit. The membrane was stained with Ponceau S solution (Sigma) to examine protein loading and transfer before proceeding with the detection. The membrane was blocked with 5% skim milk (w/v) in TTBS buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 0.05% Tween 20 overnight at 4°C. The membrane was incubated with monoclonal GFP antibody in fresh blocking solution for 6 h and washed 3 times with TTBS buffer for 15 min. The membrane was subsequently incubated with a goat anti-mouse IgG HRP-conjugate secondary antibody (Agrisera) at a 1:5, 000 dilution for 6 h. Washing was carried out 3 times with TTBS buffer every 15 min. Antibody reactivity was visualized with a chemiluminescent peroxidase substrate (Bionote).
Quantitative analysis of GFP silencing using confocal microscopy
Fluorescence images of GFP were obtained using a FluoView FV1000 confocal microscope (Olympus). Excitation and emission spectra in the GFP channel were at 488 nm and 510–540 nm, respectively. Roots of 14-d-old plants grown in MS media were used for imaging. The images were recorded with equal exposure time under non-saturated conditions.
RNA gel blot analysis
Total RNA was extracted from Arabidopsis leaves using the Nucleospin RNA II kit (Macherey-Nagel) according to the manufacturer’s instructions. Approximately 0.1 g of Arabidopsis leaf tissue was collected in a microcentrifuge tube and ground to a powder in liquid nitrogen. Total RNA (20 μg per lane) was denatured, resolved on a 1.2% agarose/formaldehyde gel and transferred onto a Hybond N+ membrane (Amersham Biosciences). RNA was immobilized by UV cross-linking (1, 000 μJ/cm2) at 254 nm. Membranes were prehybridized in hybridization buffer containing 1% BSA, 1 mM EDTA, 50 mM Na2HPO4 (pH 7.2), and 1% SDS for at least 2 h at 50°C, followed by overnight hybridization with a 32P-labeled probe complementary to the full-length GFP gene in hybridization buffer at 65°C. Membranes were washed two times in non-stringent wash solution (2× SSC, 0.1% SDS) at 60°C for 15 min, followed by one wash in stringent wash solution (0.2× SSC, 0.1% SDS) at 60°C for 15 min, and exposed to X-ray film at −70°C for 2 days.
Quantitative and semi-quantitative RT-PCR
First-strand cDNA was synthesized in a volume of 20 μl containing 1 μg total RNA, 50 pmol of oligo (dT) 18 primer, 10 pmol of dNTP, 10 units of ReverTra Ace (Toyobo) and first-strand synthesis buffer (Toyobo) according to the manufacturer’s protocol. Expression of GFP-RNAi was measured by quantitative and semi-quantitative PCR using primer pair G1 (5′-TGCCTTTCATTCTTTGTCTCC-3′) and G2 (5′-CCCAAGTGACATTTACAGCAA-3′). The synthesized cDNAs were diluted 50 times with sterilized water and used as a template for quantitative real-time PCR. Real-time quantitative reverse transcription (qRT)-PCR was performed in a Bio-Rad C1000™ thermal cycler according to manufacturer’s protocol. Semi-quantitative RT-PCR was performed with the same primer pair using Ex-Taq polymerase (Takara) and the PCR products were visualized on an ethidium bromide-containing 2% agarose gel.
siRNA gel blot analysis
For GFP-siRNA analysis, low molecular weight RNA was extracted using the mirPremier microRNA Kit (Sigma) following the manufacturer’s protocol. Three μg RNA was load onto a 15% polyacrylamide gel containing (0.1× Tris-boric acid-EDTA (TBE), 8 M urea, 15% polyacrylamide and run until the bromophenol blue dye reached the bottom of the gel. The separated RNA was transferred to a Hybond N+ membrane. The membrane was exposed to UV-radiation to crosslink RNA to the membrane. The membrane was prehybridized in ULTRAhyb-Oligo Hybridization buffer for 2 h at 40°C, followed by overnight hybridization with a 32P-labeled probe comprising full-length GFP at same temperature. After overnight hybridization, the membrane was washed at 40°C for 15 min with 2× SSC, 0.05% SDS, followed by washing in 0.1× SSC, 0.1% SDS. The membrane was exposed to a phosphor imaging screen for 24 h. The screen was read using a phosphor imaging system (Cyclone Plus Phosphor Imager, Perkin Elmer).
RESULTS
rdr6 mutants exhibit developmental phenotypes
BLAST searches of the T-DNA Express database (http://signal.salk.edu/cgi-bin/tdnaexpress) using the Arabidopsis RDR6 cDNA (NM_114810) sequence as a query revealed that the RDR6 gene composed of three exons is present as a single copy in the Arabidopsis genome at the At3g49500 locus. To examine whether or not the function of RDR6 is relevant to hpRNA-induced gene silencing, mutant alleles with loss-of-function mutations in RDR6 were selected by searching the T-DNA Express database. Arabidopsis mutant lines (Salk_110377 and Sail_617_H07) containing a T-DNA insertion in the first or second exon of RDR6 were obtained from the ABRC (http://www.arabidopsis.org). Homozygous mutant plants were selected based on kanamycin or basta resistance and designated as rdr6-16 and rdr6-17, respectively (Fig. 1A). T-DNA insertions in RDR6 were analyzed using a gene-specific primer (P3) and T-DNA left border primers (LBSalk and LBSail). rdr6-11, a loss-of-function mutant that has a single nucleotide substitution (C805T from the first ATG codon in the second exon) leading to a premature stop codon (R269stop) in RDR6 (Peragine et al., 2004), was isolated by PCR-based restriction mapping and used in this study as well (Fig. 1B). Compared with WT, rdr6-11, rdr6-16 and rdr6-17 displayed a precocious juvenile-to-adult transition, and dark-green, elongated and curled-down leaf phenotypes (Fig. 1C). To determine whether the phenotypes in rdr6-11, rdr6-16 and rdr6-17 plants could be explained by homozygosity of recessive mutations, the mutants were backcrossed to WT (Col-0) plants. None of the F1 plants exhibited the aforementioned phenotypes, indicating that the mutations in the rdr6-11, rdr6-16 and rdr6-17 plants are recessive. The ratio of plants with normal leaves to those with curled-down leaves was 3:1 in the F2 generation, indicating that the phenotypes in rdr6-11, rdr6-16 and rdr6-17 were each caused by a single mutation (Table 1).
Fig. 1.
Identification of rdr6 mutants in Arabidopsis. (A) Schematic representation of the genomic structure of the RDR6 gene and its mutant alleles. Black boxes represent exons composing an open reading frame (ORF), solid lines between the boxes indicate introns, and grey boxes represent exons composing the five-and three-prime untranslated regions (5′ and 3′ UTRs). Locations of the mutation and T-DNA insertions are indicated with inverted triangles. Gene-specific primers (P1, P2 and P3) used in genotyping are shown with arrows. (B) Identification of homozygous rdr6-11. Genomic DNAs were amplified by PCR using the P1 and P2 primers and subjected to restriction digestion using Taq1. SM, size marker; WT, wild-type; He, heterozygote; Ho, homozygote. (C) Phenotypes of 30-d-old WT, rdr6-11, rdr6-16 and rdr6-17 plants grown in soil.
Table 1.
Segregation analysis of the F2 population for rdr6 mutant alleles
| Line | Total | Normal | Curled-down | χ2 |
|---|---|---|---|---|
| rdr6-11 | 98 | 72 | 26 | 0.122 |
| rdr6-16 | 105 | 80 | 25 | 0.079 |
| rdr6-17 | 97 | 79 | 18 | 2.147 |
rdr6-11, rdr6-16 and rdr6-17 mutants were backcrossed to Col-0 plants. The F2 populations were subjected to segregation analysis using the curled-down leaf phenotype as a marker. The total numbers of tested seedlings and the numbers of seedlings exhibiting normal and curled-down leaf phenotypes are listed. The hypothesis of a segregation ratio normal:curled-down = 3:1 was accepted in all F2 populations of the mutant alleles according to the Chi-square test with a 0.05 significance level.
hpRNA-induced gene silencing is efficient in the WT background
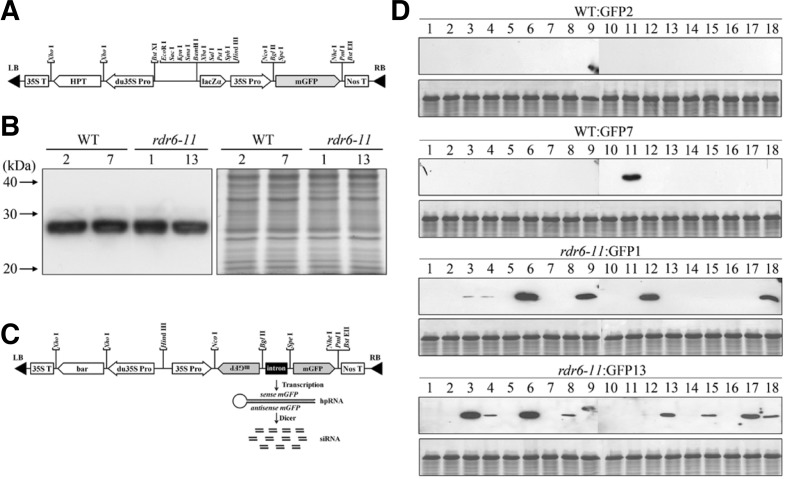
rdr6-11 was originally derived from a T-DNA insertion line, but the T-DNA was subsequently crossed out. Thus, the mutation is not associated with the T-DNA insertion. To avoid possible sequence similarity-dependent TGS caused by multiple T-DNA insertions, rdr6-11 was used as the representative rdr6 mutant allele in this work. To generate WT and rdr6-11 plants that constitutively express GFP, the pCAMBIA1302 vector (http://www.cambia.org/daisy/cambia/585.html) carrying the GFP gene under the control of the CaMV 35S promoter was used (Fig. 2A). Transgenic lines with a single copy of the T-DNA insertion were isolated in the T2 generation and homozygous transgenic plants were identified in the T3 generation based on the segregation ratios of hygromycin resistance. Expression of GFP in the T3 generation with WT and rdr6-11 backgrounds was evaluated by immunoblot analysis using a GFP antibody. Transgenic lines (WT:GFP2, 7 and rdr6-11:GFP1, 13) that exhibited similar GFP expression levels were used to examine the efficiency of hpRNA-based GFP knockdown in WT and rdr6-11 backgrounds (Fig. 2B).
Fig. 2.
Suppression of GFP expression by the introduction of GFP-RNAi in WT and rdr6-11 plants. (A) Schematic representation of the pCAMBIA1302 vector carrying GFP. (B) Expression of GFP in the T3 generation with WT and rdr6-11 backgrounds. Immunoblot analysis of GFP was conducted using a monoclonal GFP antibody (left panel). Coomassie Brilliant Blue (CBB) staining was used to confirm that an equal amount of protein was loaded in each lane (right panel). (C) Schematic representation of the GFP-RNAi construct. (D) Suppression of the GFP expression in the T1 generation of GFP-RNAi transformants with GFP-expressing WT and rdr6-11 backgrounds. Immunoblot analysis was conducted using a monoclonal GFP antibody. Suppression of the GFP expression in transformants with the WT backgrounds (WT:GFP2 and WT:GFP7) and the rdr6-11 backgrounds (rdr6-11:GFP1 and rdr6-11:GFP13) are shown in the two upper panels and two lower panels, respectively. Coomassie blue-stained gels are shown below each blot as loading controls.
A GFP-RNAi construct carrying the full-length open reading frame (ORF) of GFP in both sense and antisense orientations and a phosphinothricin (PPT) resistance gene was introduced into the genomes of GFP-expressing WT and rdr6-11 lines (Fig. 2C). Primary transformants (T1) of the GFP-RNAi construct were obtained by selection on PPT-containing media, and 18 independent T1 plants for each background were transferred to soil and grown for further analysis. To investigate the efficiency of hpRNA-induced GFP knockdown, the T1 GFP-RNAi plants were analyzed by immunoblot analysis (Fig. 2D). Among the 36 independently transformed GFP-RNAi lines with WT backgrounds, GFP expression was detected in only one line (WT-GFP7: GFP-RNAi11). Since only one GFP-RNAi line with the WT background displayed inefficient hpRNA-induced gene silencing, we wondered whether the continued expression of GFP in the WT-GFP7:GFP-RNAi11 line was due to aberrant GFP-RNAi expression. When the expression of GFP-RNAi in WT-GFP7:GFP-RNAi11 was analyzed by RT-PCR using a primer set for the intron between the antisense- and sense-GFP, no GFP-RNAi expression was observed, suggesting incomplete T-DNA integration by partial gene transfer (Supplementary Fig. S1). By contrast, GFP protein was detected in several GFP-RNAi lines in the rdr6-11 background. A lack of GFP-RNAi expression possibly caused by incomplete T-DNA integration was also observed in three (line 6 of rdr6-11:GFP1 and line 3 and 6 of rdr6-11:GFP13) of the 36 independently transformed GFP-RNAi lines with rdr6-11:GFP backgrounds (Supplementary Fig. S1). Together, these results suggest that hpRNA-induced gene silencing is efficient in the WT backgrounds but not in the rdr6-11 backgrounds.
Efficient hpRNA-induced gene silencing is abolished in rdr6-11
The different efficiencies of hpRNA-induced gene silencing in the WT and rdr6-11 backgrounds prompted us to undertake comparative analyses of GFP expression in the plants. Different expression levels of a given transgene in heterozygous versus homozygous transgenic plants is well known (Beaujean et al., 1998). In addition, the expression level of a transgene in independently transformed eukaryotic organisms is known to be dependent on the site of integration and the copy number of the transgene (Hobbs et al., 1993; Ni et al., 1995). The different levels of GFP silencing in the independent GFP-RNAi plants could be caused by different levels of GFP-RNAi expression, which in turn could be affected by copy number and position effects of T-DNA insertion. T-DNA copy number was assessed by chi-square analysis in the T2 generation of the GFP-RNAi lines. GFP-RNAi lines with single copies of the T-DNA insertion were selected for further analysis (Table 2). Homozygous plants were identified in the T3 generation based on the segregation ratios of hygromycin resistance and used to analyze the efficiency of hpRNA-induced gene silencing in the WT and rdr6-11 backgrounds.
Table 2.
Segregation analysis of T2 GFP-RNAi lines with GFP-expressing WT and rdr6-11 backgrounds
| Line | Total | Resistant | Sensitive | χ2 | Background |
|---|---|---|---|---|---|
| 1 | 65 | 50 | 15 | 0.096 | WT:GFP2 |
| 2 | 67 | 45 | 22 | 1.645 | WT:GFP2 |
| 3 | 73 | 57 | 16 | 0.277 | WT:GFP2 |
| 4 | 75 | 61 | 14 | 1.203 | WT:GFP2 |
| 5 | 75 | 61 | 14 | 1.604 | WT:GFP2 |
| 7 | 73 | 49 | 24 | 2.416 | WT:GFP2 |
| 10 | 73 | 55 | 18 | 0.005 | WT:GFP2 |
| 11 | 76 | 50 | 29 | 3.439 | WT:GFP2 |
| 12 | 68 | 45 | 23 | 2.824 | WT:GFP2 |
| 6 | 78 | 58 | 20 | 0.017 | rdr6-11:GFP1 |
| 9 | 61 | 52 | 19 | 1.230 | rdr6-11:GFP1 |
| 12 | 69 | 55 | 14 | 0.816 | rdr6-11:GFP1 |
| 18 | 47 | 28 | 19 | 5.965 | rdr6-11:GFP1 |
| 3 | 47 | 37 | 10 | 0.348 | rdr6-11:GFP13 |
| 4 | 78 | 58 | 20 | 0.017 | rdr6-11:GFP13 |
| 6 | 75 | 62 | 13 | 2.351 | rdr6-11:GFP13 |
| 8 | 61 | 42 | 19 | 1.229 | rdr6-11:GFP13 |
| 13 | 69 | 55 | 14 | 0.816 | rdr6-11:GFP13 |
| 15 | 47 | 37 | 10 | 0.347 | rdr6-11:GFP13 |
| 17 | 59 | 43 | 16 | 0.141 | rdr6-11:GFP13 |
| 18 | 72 | 55 | 17 | 0.074 | rdr6-11:GFP13 |
The total numbers of tested seedlings and the numbers of seedlings exhibiting resistance (resistant) or sensitivity (sensitive) to phosphinothricin are listed. The Chi-square test was used to assess the copy number of T-DNA insertions and a significance level of 0.05 was used.
To determine the efficiency of hpRNA-induced gene silencing in the WT and rdr6-11 backgrounds, expression of GFP-RNAi was analyzed by quantitative and semi-quantitative RT-PCR in several homozygous T3 plants. Line 2 of the WT and 16 of rdr6-11 backgrounds in which no GFP expression was detected by immunoblot analysis showed the highest expression of GFP-RNAi among the lines of each background (Fig. 3). By contrast, relatively lower expression of GFP-RNAi was observed in the other lines of the rdr6-11 background, in which remaining GFP expression was detected by immunoblot analysis (Fig. 3). However, lower expression of GFP-RNAi was also observed in three lines of the WT background (3, 4 and 7) in which no GFP expression was detected by immunoblot analysis (Fig. 3). These results indicate that the efficiency of hpRNA-induced gene silencing is closely associated with the expression level of GFP-RNAi and higher in the WT backgrounds.
Fig. 3.
Expression of GFP-RNAi in the WT and rdr6-11 backgrounds. (A) Quantitative real-time PCR and (B) semi-quantitative RT-PCR were performed using a primer pair for an intron of the GFP-RNAi construct in the selected homozygous lines of the WT and rdr6-11 backgrounds. TUBULIN was used as an internal control for quantitative and semi-quantitative RT-PCR.
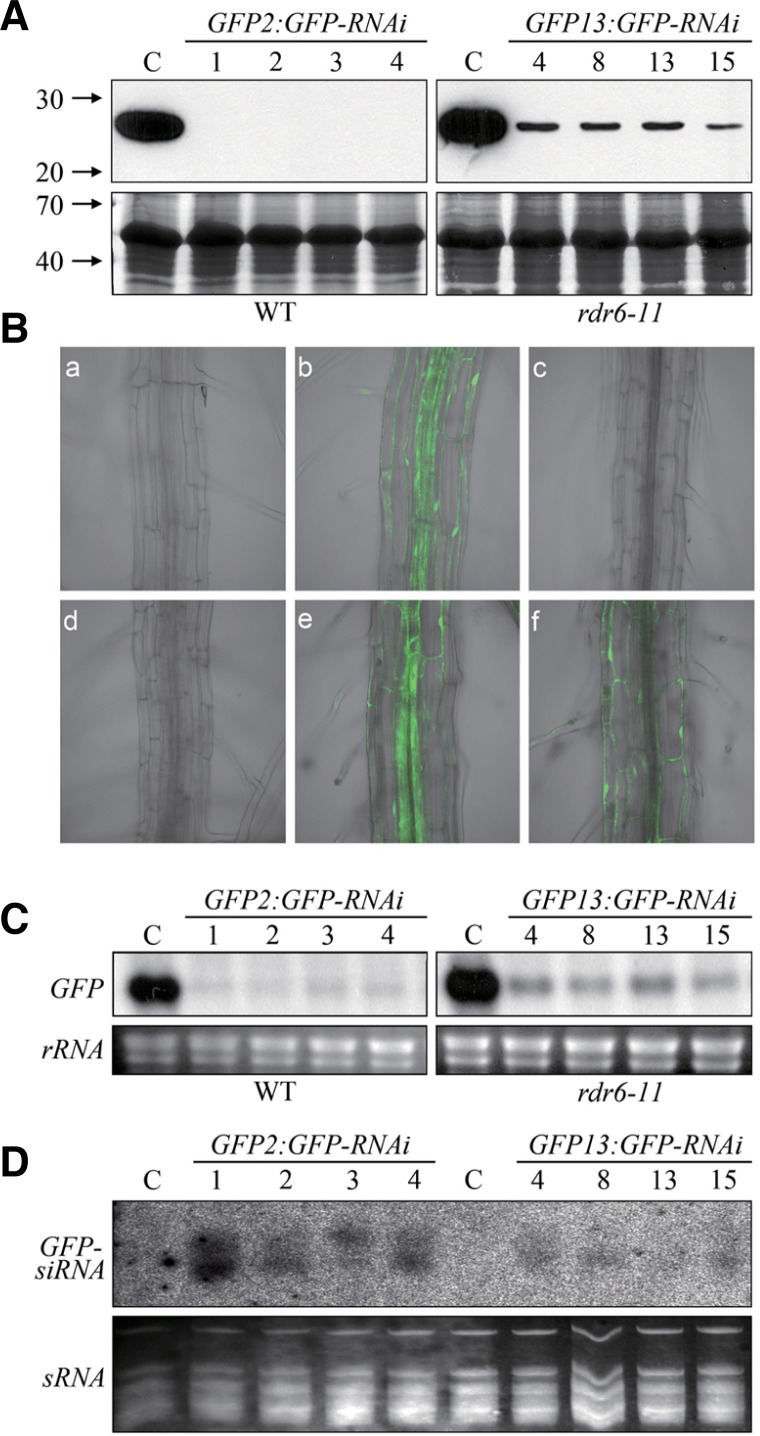
Several GFP-RNAi lines were used to analyze the efficiency of hpRNA-induced gene silencing and siRNA synthesis in the WT and rdr6-11 backgrounds. When the expression of GFP was analyzed by immunoblot analysis, GFP expression was not detected in four independent GFP-RNAi homozygous plants with the WT:GFP background; however, it was detectable in four independent GFP-RNAi homozygous plants with the rdr6-11:GFP background (Fig. 4A). To confirm the results by in vivo visualization of GFP expression, we also carried out confocal microscopy analysis using the homozygous GFP-RNAi plants in the WT:GFP and rdr6-11:GFP backgrounds (Fig. 4B). Whereas the fluorescence of GFP detected in the WT background was efficiently suppressed by the introduction of GFP-RNAi, weak fluorescence was still observed in the GFP-RNAi homozygous plant in the rdr6-11 background (Fig. 4B). These results suggest that RDR6 is involved in hpRNA-induced gene silencing, probably regulating the efficiency of RNA degradation. To examine degradation of GFP mRNA in the WT and rdr6-11 plants, northern blot analysis was carried out (Fig. 4C). Consistent with the results from immunoblot and confocal microscopy analyses, the amounts of remaining GFP mRNA were higher in the four independent homozygous GFP-RNAi plants with the rdr6-11:GFP background than in those of the four independent GFP-RNAi homozygous plants with the WT:GFP background (Fig. 4C). Finally, to address whether RDR6 is involved in the secondary siRNA synthesis, the amount of GFP-siRNAi in the WT and rdr6-11 backgrounds was directly measured by Northern blot analysis. Consistent with other results, the siRNAi gel blotting analysis showed that the amount of GFP-siRNAi is higher in the WT but lower in the rdr6-11 backgrounds (Fig. 4D). This result suggests that RDR6 is required for efficient gene silencing through the synthesis of secondary siRNAs in plants.
Fig. 4.
Analyses of the efficiency of gene silencing in WT and rdr6-11 plants. (A) Expression of GFP in homozygous T3 generation GFP-RNAi plants with GFP-expressing WT and rdr6-11 backgrounds. Immunoblot analysis was conducted using a monoclonal GFP antibody (upper panel). Coomassie Brilliant Blue (CBB) staining was used to confirm that an equal amount of protein was loaded in each lane (lower panel). GFP-expressing WT and rdr6-11 backgrounds were used as controls (C). (B) Confocal microscopy analysis of GFP expression in the homozygous T3 generation GFP-RNAi plants with GFP-expressing WT and rdr6-11 backgrounds. (a) WT, (b) WT:GFP2, (c) WT-GFP2:GFP-RNAi1, (d) rdr6-11, (e) rdr6-11:GFP13, (f) rdr6-11-GFP13:GFP-RNAi4. (C) Northern blot analysis of homozygous T3 generation GFP-RNAi plants with GFP-expressing WT and rdr6-11 backgrounds. Membranes were hybridized with a 32P-labeled full-length GFP cDNA probe. Ribosomal RNA (rRNA) is shown to confirm equal loading. (D) Northern blot analysis of GFP-siRNA in T3 GFP-RNAi plants. Membranes were hybridized with a 32P-labeled full-length GFP cDNA probe. Ethidium bromide-stained low molecular weight RNAs are shown as loading controls below the Northern blots. 30-d-old plants grown in soil were used for the immunoblot and Northern blot analyses. The 14-d-old plants grown in MS media were used for confocal microscopy analysis.
DISCUSSION
Developmental phenotypes such as a precocious juvenile-to-adult transition, and dark-green, elongated and curled-down leaf have been observed in rdr6-11, rdr6-16 and rdr6-17 (Fig. 1C). Auxin response factors ARF3 and ARF4 are known to regulate a subset of the traits associated with the vegetative-to-reproductive transition and leaf heteroblasty (Pekker et al., 2005). It has also been reported that the production of tasiR-ARF, a trans-acting siRNA that targets both ARF3 and ARF4, requires ZIP/AGO7, SGS3, DCL4 and RDR6 (Hunter et al., 2006). These results implicate RDR6 in epigenetic regulation of genes that are involved in transcriptional control of plant development.
In this study, we compared the efficiency of hpRNA-induced gene silencing in the presence or absence of RDR6 using GFP as a reporter gene. The GFP-RNAi construct was designed to carry a full-length ORF of GFP in both sense and antisense orientations under the control of the CaMV35S promoter. Thus, it can produce GFP siRNAs, which target GFP mRNA from the 5′ to 3′ end. Even though an RNAi knockdown technique was employed to induce gene silencing, almost complete silencing of GFP expression was observed by immunoblot analysis in most lines of the WT backgrounds excepting one aberrant T-DNA integration line (Fig. 2D). This result may be due to several factors such as efficient GFP-siRNA production from the long hairpin RNA containing full-length GFP sequence, TGS induced by the CaMV35S promoter, which is used for both GFP and GFP-RNAi expression, and/or by co-suppression effects of the GFP gene expression. The efficiency and frequency of transitive silencing of an endogenous gene depends on the length of its sequence identity with the primary target (Bleys et al., 2006). Gene silencing mediated by promoter homology occurs at the level of transcription (Park et al., 1996). Mutants of C. elegance that are defective in transposon silencing and RNAi are also resistant to co-suppression, indicating that RNAi and co-suppression are mediated by the same molecular machinery, possibly through the formation of dsRNA/RNA duplex as a precursor to generate siRNA (Ketting and Plasterk, 2000). Thus, the length of hairpin RNA, TGS mediated by promoter homology and co-suppression effects of the CaMV35S:GFP might all contribute to the strong silencing of GFP expression in the WT backgrounds. It is difficult to determine whether each mechanism contributes to the result in a cooperative or independent manner in this study. However, it appears that the three mechanisms commonly employ siRNA to initiate gene silencing.
RDR6 is thought to initiate RNA silencing by recognizing transgenic transcripts with aberrant features and converting them to dsRNAs, which are then processed into siRNAs (Bro Brodersen and Voinnet, 2006). The activity of RDRs is implicated in the facilitation of strong and persistent silencing that is initiated by small amounts of ‘trigger’ RNA, including transposon-derived and viral-derived RNA (Voinnet, 2008). Deep sequencing analysis revealed that the rdr1 and rdr6 mutants exhibit significantly reduced levels of small RNAs (Qi et al., 2009). The efficiency of gus silencing is correlated with the amount of secondary siRNA suggesting that the amplification of secondary siRNAs is required to exceed the threshold for efficient silencing (Bleys et al., 2006). Taken together, these results indicate that RDRs are required for the amplification of secondary siRNAs and efficient gene silencing. Amplification of secondary siRNA might be necessary to enhance the efficiency of RNA silencing, and RDR6 is likely required for this amplification step. Thus, we speculate that the amount of siRNA in the GFP-RNAi lines was sufficient to induce efficient gene silencing in the GFP-expressing WT backgrounds but not the rdr6-11 backgrounds (Fig. 5). Based on our results, we propose that RDR6 not only initiates RNA silencing by recognizing aberrant RNAs and converting them to dsRNAs but also promotes amplification of secondary siRNAs to enhance the efficiency of RNA silencing (Fig. 5).
Fig. 5.
Model for RDR6 function in RNAi-induced gene silencing. Since hpRNA can directly fold back to form dsRNA, RNA silencing induced by self-complementary hpRNA takes place even in the absence of RDR6. The resulting dsRNA is then sliced by Dicer, creating primary siRNA. This primary siRNA is incorporated into RISC and subsequently induces mRNA degradation, which can produce aberrant RNAs. In WT plants, RDR6 recognizes the aberrant RNAs and amplifies them to form dsRNA. Dicer then slices the dsRNA to produce secondary siRNA. In the rdr6 mutant plants, there is no secondary siRNA production because of the absence of RDR6. Thus, only primary siRNAs are present. As a result, rdr6 plants have a lower amount of siRNA, which is not sufficient to induce complete RNA silencing. It appears that RDR6 enhances hpRNA-induced gene silencing in plants via the production of secondary siRNAs using aberrant RNAs as templates
Supplementary Material
Acknowledgments
This research was supported by grants from the Next-Generation BioGreen Program (SSAC, PJ008109), the Rural Development Administration, the TDPAF (609004-5), the Ministry for Food, Agriculture, Forestry and Fisheries, and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012R1A1A2001074), Republic of Korea. R. Harmoko, W.I. Fanata, J.Y. Yoo, K.S. Ko were supported by scholarships from the Brain Korea 21 program.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baulcombe DC. Molecular biology. Amplified silencing. Science. 2007;315:199–200. doi: 10.1126/science.1138030. [DOI] [PubMed] [Google Scholar]
- Beaujean A, Sangwan RS, Hodges M, Sangwan-Norreel BS. Effect of ploidy and homozygosity on transgene expression in primary tobacco transformants and their androgenetic progenies. Mol Gen Genet. 1998;260:362–371. doi: 10.1007/s004380050905. [DOI] [PubMed] [Google Scholar]
- Beclin C, Boutet S, Waterhouse P, Vaucheret H. A branched pathway for transgene-induced RNA silencing in plants. Curr Biol. 2002;12:684–688. doi: 10.1016/s0960-9822(02)00792-3. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bleys A, Vermeersch L, Van Houdt H, Depicker A. The frequency and efficiency of endogene suppression by transitive silencing signals is influenced by the length of sequence homology. Plant Physiol. 2006;142:788–796. doi: 10.1104/pp.106.083956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA. 2010;107:15269–15274. doi: 10.1073/pnas.1001738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Meyerowitz EM. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Davuluri GR, van Tuinen A, Fraser PD, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J, et al. Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat Biotechnol. 2005;23:890–895. doi: 10.1038/nbt1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depicker A, Montagu MV. Post-transcriptional gene silencing in plants. Curr Opin Cell Biol. 1997;9:373–382. doi: 10.1016/s0955-0674(97)80010-5. [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs SL, Warkentin TD, DeLong CM. Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol Biol. 1993;21:17–26. doi: 10.1007/BF00039614. [DOI] [PubMed] [Google Scholar]
- Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig SR. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Plasterk RH. A genetic link between co-suppression and RNA interference in C. elegans. Nature. 2000;404:296–298. doi: 10.1038/35005113. [DOI] [PubMed] [Google Scholar]
- Klahre U, Crete P, Leuenberger SA, Iglesias VA, Meins F., Jr High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc. Natl. Acad. Sci. USA. 2002;99:11981–11986. doi: 10.1073/pnas.182204199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansoor S, Amin I, Hussain M, Zafar Y, Briddon RW. Engineering novel traits in plants through RNA interference. Trends Plant Sci. 2006;11:559–565. doi: 10.1016/j.tplants.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Moissiard G, Parizotto EA, Himber C, Voinnet O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA. 2007;13:1268–1278. doi: 10.1261/rna.541307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhuber F, Park YD, Matzke AJ, Matzke MA. Susceptibility of transgene loci to homology-dependent gene silencing. Mol Gen Genet. 1994;244:230–241. doi: 10.1007/BF00285450. [DOI] [PubMed] [Google Scholar]
- Ni M, Cui D, Einstein J, Narasimhulu S, Vergara CE, Gelvin SB. Strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase gene. Plant J. 1995;7:661–676. [Google Scholar]
- Park YD, Papp I, Moscone EA, Iglesias VA, Vaucheret H, Matzke AJ, Matzke MA. Gene silencing mediated by promoter homology occurs at the level of transcription and results in meiotically heritable alterations in methylation and gene activity. Plant J. 1996;9:183–194. doi: 10.1046/j.1365-313x.1996.09020183.x. [DOI] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Bao FS, Xie Z. Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS One. 2009;4:e4971. doi: 10.1371/journal.pone.0004971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris TJ. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. USA. 2008;105:14732–14737. doi: 10.1073/pnas.0805760105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Beclin C, Fagard M. Post-transcriptional gene silencing in plants. J Cell Sci. 2001;114:3083–3091. doi: 10.1242/jcs.114.17.3083. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 2008;13:317–328. doi: 10.1016/j.tplants.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Wassenegger M, Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11:142–151. doi: 10.1016/j.tplants.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.