Abstract
MicroRNAs (miRNAs) are important regulators of gene expression and play crucial roles in many biological processes including apoptosis, differentiation, development, and tumorigenesis. Recent estimates suggest that more than 50% of human protein coding genes may be regulated by miRNAs and that each miRNA may bind to 300–400 target genes. Approximately 1, 000 human miRNAs have been identified so far with each having up to hundreds of unique target mRNAs. However, the targets for a majority of these miRNAs have not been identified due to the lack of large-scale experimental detection techniques. Experimental detection of miRNA target sites is a costly and time-consuming process, even though identification of miRNA targets is critical to unraveling their functions in various biological processes. To identify miRNA targets, we developed miRTar Hunter, a novel computational approach for predicting target sites regardless of the presence or absence of a seed match or evolutionary sequence conservation. Our approach is based on a dynamic programming algorithm that incorporates more sequence-specific features and reflects the properties of various types of target sites that determine diverse aspects of complementarities between miRNAs and their targets. We evaluated the performance of our algorithm on 532 known human miRNA:target pairs and 59 experimentally-verified negative miRNA:target pairs, and also compared our method with three popular programs for 481 miRNA:target pairs. miRTar Hunter outperformed three popular existing algorithms in terms of recall and precision, indicating that our unique scheme to quantify the determinants of complementary sites is effective at detecting miRNA targets. miRTar Hunter is now available at http://203.230.194.162/~kbkim.
Keywords: computational approach, dynamic programming, miRNA, miRTar Hunter, seed region
INTRODUCTION
Eukaryotic small noncoding RNAs of approximately 21–25 nucleotides function as one of the central regulators of the expression of an extensive repertoire of genes involved in a remarkably wide range of biological processes-ranging from development, differentiation, and cell growth and proliferation to apoptosis and tumorigenesis (Chan et al., 2005; Esquela-Karscher and Slack, 2006). These noncoding RNAs belong to at least two general classes: microRNAs (miRNAs) and short interfering RNAs (siRNAs). miRNAs regulate the expression of protein coding genes at the post-transcriptional level by basepairing with transcripts of their target genes, subsequently leading to translational repression (Bartel, 2004; Esquela-Kersher and Slack, 2006), mRNA cleavage (Llave et al., 2002; Tang et al., 2003; Yekta et al., 2004), or miRNA-induced degradation (Bagga et al., 2005; Lim et al., 2005; Wu et al., 2006). However, these miRNAs have also been reported to activate target genes (Li et al., 2006). They are transcribed as long primary transcripts (pri-miRNAs) in the nucleus and processed into characteristic stem-loop precursor miRNAs (pre-miRNAs) by the enzyme Drosha. The pre-miRNAs are then transported into the cytoplasm, where they are transformed into small, single-stranded miRNAs with the aid of Dicer (Cullen, 2004). One strand of the mature miRNA is incorporated into the RNA-induced silencing complex (RISC) and binds preferentially to the 3′-untranslated region (3′-UTR) of the target mRNA through complementary base-pairing. The 5′-end sequence of the miRNA, which is 6–11 nucleotides in length, is called the “seed”. This sequence interacts with the target in an energetically favorable manner and is the determinant of target repression. Mutations in the seed region of a miRNA sequence can lead to inactive interactions (Doench and Sharp, 2004). The miRNA-mRNA interaction controls the expression level of a target gene by a number of mechanisms, including inhibition of translational initiation, inhibition of elongation, and induction of deadenylation, which decreases mRNA stability and increases the rate of mRNA degradation. miRNAs are involved in diverse regulatory pathways, as well as in disease development and progression, and have been used as diagnostic and prognostic markers and to evaluate treatment response (Dweep et al., 2011).
Although thousands of miRNAs have been identified in animals, plants, and viruses, the targets for the majority of these miRNAs have not been identified due to the lack of large-scale experimental detection techniques. The identification of miRNA targets is critical to unraveling their functions in various biological processes. Many computational approaches have been developed in recent years to predict miRNA targets, for example, miR and a (Betel et al., 2008), TargetScan (Friedman et al., 2009), RNAHybrid (Kruger and Rehmsmeier, 2006), PicTar (Krek et al., 2005), and DIANA-microT (Maragkakis et al., 2009). To predict miRNA targets, these computational approaches typically evaluate three factors: (1) perfect or near-perfect complementarity between the miRNAs and their targets, (2) the thermodynamic stability, i.e. the free energy of the miRNA: target duplex, (3) conservation of the miRNA: target duplex across species. Imperfect complementarity between miRNAs and their targets in animals makes target prediction much harder than in plants, whose miRNAs almost always bind their targets with near-perfect complementarity (Doran and Strauss, 2007; Jones-Rhoades and Bartel, 2004). Many existing methods for animals make extensive use of the seed region, but a substantial number of miRNA:target pairs do not have recognizable seed regions. Most existing methods also use patterns of evolutionary conservation to find conserved targets. However, this type of information does not help to identify species-specific targets. In addition, for more than a decade, attempts to study the interactions of miRNAs with their targets have focused on the 3′-UTR region of mRNAs. However, recent studies of miRNA-target interactions have revealed that miRNAs may regulate gene expression by targeting promoters as well as protein coding regions (CDSs). Thus, it is of paramount importance to develop a new approach that can identify putative miRNA target sites not only in the 3′-UTR region, but also in other regions (promoter, 5′-UTR, and CDS) of a gene. In this context, focusing on human miRNAs, we have developed miRTar Hunter, a novel computational approach for predicting target sites regardless of the presence or absence of seed matches and across-species patterns of conservation. This approach is based on a dynamic programming algorithm that incorporates sequence-specific features and reflects the properties of various types of target sites that determine diverse aspects of complementarities between miRNAs and their targets. We evaluated the performance of our algorithm on 532 known human miRNA:target pairs and 59 experimentally-verified negative miRNA:target pairs. We also compared our method with three popular programs using a dataset of 481 predicted miRNA:target pairs. Performance evaluation revealed that miRTar Hunter outperformed the three popular existing algorithms in terms of recall and precision, indicating that our unique scheme to quantify the determinants of complementary sites is effective at detecting miRNA-target pairs.
MATERIALS AND METHODS
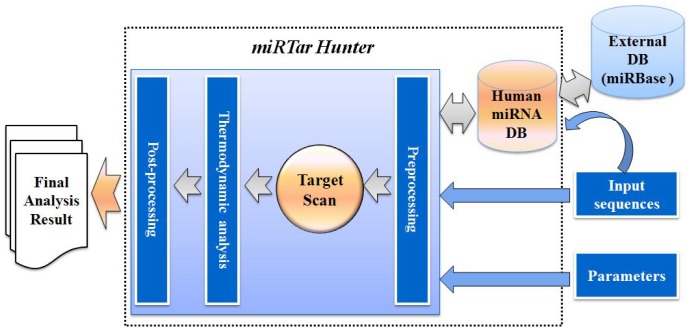
Overall configuration of miRTar Hunter
miRTar Hunter is a web-based system that runs on an Apache web server with a Linux operating system. It has a three-tier architecture composed of a client, an application server, and a back-end database. A schematic overview of the miRTar Hunter prediction is shown in Fig. 1; the pipeline allows researchers to analyze all possible scenarios to infer the regulatory relationships between miRNAs and targets. The client is a user-friendly web interface for the application server. The application server consists of preprocessing, target scanning, free energy calculation, and post-processing modules. The back-end database is a secondary database that contains a collection of the human miRNAs derived from miRBase database (Kozomara and Griffiths-Jones, 2011). After the parameters/options are specified and miRNAs and mRNAs are input into the system, input data and parameters/options are preprocessed, targets are predicted using a differentially-weighted Smith-Waterman algorithm, the free energy of possible interactions detected by the target scan module are calculated, and the final analysis results are then sorted, ranked, and displayed.
Fig. 1.
Schematic overview of miRTar Hunter. prediction pipeline. miRTar Hunter has a three-tier architecture consisting of a web interface, application server, and back-end database. The back-end database contains a collection of human miRNAs extracted from the miRBase database. The application server consists of a preprocessing module for input data and parameters, a target scan module that searches for complementarity matches between miRNAs and mRNA using a differentially-weighted Smith-Waterman algorithm, a free energy calculation module to calculate the free energy of possible interactions detected by target scan module, and a post-processing module to sort, rank, and display the final analysis results.
Target scan algorithm
The target prediction component of the miRTar Hunter utilizes the Smith-Waterman algorithm (Smith and Waterman, 1981), which is a well-known dynamic programming algorithm that is a core algorithm in many applications. This algorithm performs local sequence alignment to determine regions of similarity between two DNA or protein sequences. We modified it appositely and created the so-called “differentially-weighted Smith-Waterman” algorithm; this algorithm is optimized to search for maximal local complementarity alignments between a miRNA sequence and all possible positions in the input mRNA sequence. The algorithm consists of three steps: (1) initialize row 0 and column 0, (2) calculate the similarity matrix score, (3) trace back the similarity matrix to search for the optimal local alignment, using dynamic programming. The second step consumes the largest part of the total calculation time. The Smith-Waterman algorithm can be summarized as follows. For two sequences S and T, the length of S is n, |S|=n; the length of T is m, |T|=m; V(i, j) is the optimal alignment score of two subsequence S[1]...S[i] and T[1]...[j], and V(i, j) is calculated using Formulas 1 and 2 as shown below:
Initialization:
| (1) |
Recursion relation:
| (2) |
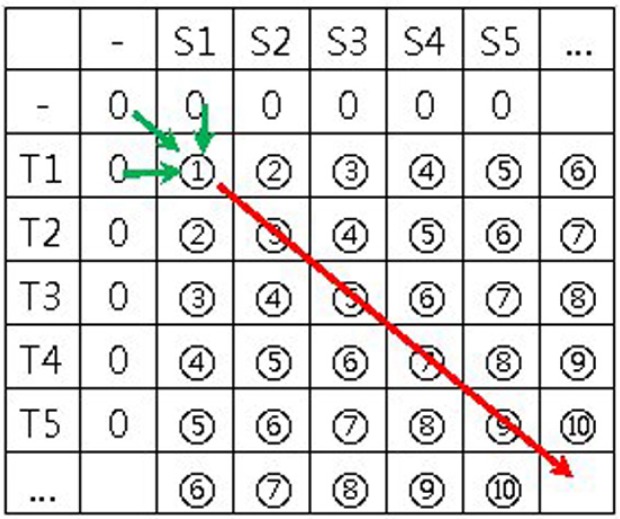
In these formulas, “−” represents a null character or gap; V(i, 0) represents the results obtained from comparing each character in S with a gap in T; V(0, j) is the result obtained from comparing each character in T with a gap in S; and (S[i], T[j]) is the value of the substitution matrix. When calculating the similarity matrix, the score of any matrix element V(i, j) always depends on the score of three other elements: i) the up-left neighbor element V(i−1, j−1), ii) the left neighbor V(i, j−1), iii) the up neighbor V(i−1, j). Therefore, the sequence of the similarity matrix is calculated as shown in Fig. 2. The calculation progresses from the top-left element to the bottom-right element in the direction shown by the arrow in Fig. 2.
Fig. 2.
Similarity matrix to calculate sequence and data dependency. While calculating the similarity matrix, the score of any matrix element always depends on the score of three other elements: the upleft neighbor element, the left neighbor, and the up neighbor. The similarity matrix is calculated from the top-left element to the bottom-right element in the direction shown by the arrow.
The target scan algorithm of miRTar Hunter is very similar to the Smith-Waterman algorithm. However, instead of building alignments based on matching nucleotides (A-A or U-U, for example), the scores are based on the complementarity of nucleotides (A=U or = =). The complementarity ≡ scoring matrix used for this analysis also allows ==U ‘wobble’ pairs, which is important for the accurate detection of RNA:RNA duplexes (Wuchty et al., 1999). Table 1 shows the parameters used for complementarity alignments. A score of +7 is assigned to =:= pairs, +5 to A:T pairs, +1 to =:U wobble pairs, and −3 to all other non-complementary pairs. The algorithm uses affine gap penalties (linear in the length of a gap after an initial opening penalty) for gap opening (−8) and gap extension (−2). In addition, following observation of known target sites, complementarity scores (positive and negative values) at the 5′ seed region (positions 1 to 11 counting from the 5′ end of miRNA) and 3′ region (positions 12 to 15) are multiplied by scaling factors of 4 and 2, respectively, so as to reflect the types of miRNA target sites (observed 5′-3′ asymmetry), including 7mer-A1 sites, 7mer-m8 sites, 8mer sites, 6mer sites, offset 6mer sites, 3′-supplementary sites, and 3′-compensatory sites. Seven of these types of target sites were clearly defined by Bartel’s group (Bartel, 2009). The scaling factor is an adjustable parameter that can be optimized as new experimental information becomes available regarding the types of target sites or seed regions. Furthermore, we applied four empirical rules to ensure that the proposed miRNA:mRNA duplexes followed experimentally determined patterns: no mismatches at positions 2 to 4 (counting from the 5′ end); fewer than five mismatches between positions 3–12; at least one mismatch between positions 9 and L-5 (where L is the total alignment length); and fewer than two mismatches in the last five positions of the alignment. Using these parameters, the target scan algorithm optimizes the complementarity alignment score between a miRNA sequence and a mRNA sequence, sums over all aligned positions, and creates a ranking of all non-overlapping complementarity alignments in decreasing order of complementarity alignment score down to some threshold value (default value 100). The detection of suboptimal alignments follows heuristics previously used in sequence alignment (Schneider and Sander, 1996; Waterman and Eggert, 1987). The key extension of the miRTar Hunter algorithm compared to the Smith-Waterman sequence alignment algorithm is the addition of weighted scores for certain positions and regions in the complementarity alignments according to the four empirical rules and seven types of target sites described above.
Table 1.
Parameters used for complementarity sequence alignment of miRNAs and mRNAs
| Parameters | Symbol | Score | Examples | |||
|---|---|---|---|---|---|---|
| Complemetarity | | | 7 | C | G |
or | G | C |
|
| 5 | A | U |
or | U | A |
|||
| Wobble | : | 1 | G : U |
or | U : G |
|
| Non-Complementarity | −3 | A G |
G A |
|||
| Gap | Opening | – | −8 | A G |
A – |
G G |
| Extension | −2 | A – |
C – |
G – |
||
| Weight | 5′ region | 4 | ||||
| 3′ region | 2 | |||||
The various parameters were designed based on known mechanisms of miRNA target recognition and knowledge of diverse types of target sites.
To analyze the time complexity of our algorithm, we analyzed each individual part of the algorithm. To initialize the matrix, the scores of row 0 and column 0 need to be input. This has a time complexity of O(m+n). To fill the matrix, each cell of the matrix is traversed and a constant number of operations is performed in each cell, and thus the time complexity of this part is O(mn). In the traceback, the algorithm requires the maximum cell be found, and this must be done by traversing the entire matrix, making the time complexity for the traceback O(mn). It is also possible to keep track of the largest cell during the matrix filling segment of the algorithm, although this will not change the overall complexity. Thus the total time complexity of our algorithm is O(m+n) + O(mn) + O(mn) = O(mn). Because this algorithm fills a single matrix of size mn and stores at most n positions for the traceback, the total space complexity of this algorithm is O(mn) + O(n) = O(mn).
Free energy calculation
To estimate the thermodynamic properties of the predicted miRNA:mRNA duplex from each complementarity alignment, miRTar Hunter uses the minimum free energy folding routine (MFE Fold), one of folding routines from the Vienna 1.3 RNA secondary structure programming library (RNAlib) (Hofacker, 2003). The calculation of minimum free energy structures is based on dynamic programming algorithm originally developed by Zuker and Stiegler (1981). The expanded thermodynamic parameters are more computationally intensive than the initial target scan, but allow potential hybridization sites to be scored according to their respective folding energies. The miRNA sequence and mRNA sequence from a complementarity alignment are joined into a single sequence with an eight base sequence linker containing artificial ‘X’ bases that cannot base pair. This strand-linker-strand configuration assumes the phase space entropy of strand-strand association is constant for all miRNA-target complementarities (Hofacker, 2003; Schneider and Sander, 1996). The minimum energy of this structure with the last complementary base pair (from the initial sequence alignment) constrained is then calculated using MFE Fold and checked against a threshold value. After target scanning and free energy calculation, predicted target sites for each miRNA are classified into types of target sites and sorted first according to complementarity alignment score and then according to free energy.
Data collection
As stated above, we created a secondary database for human miRNAs obtained from the miRBase database. The secondary database contains 1, 448 human miRNA entries hyperlinked to miRBase through the miRNA name and identifier number. Human miRNAs can be retrieved from this database by their names, accession numbers, and mature sequences. The retrieved miRNAs can be used directly as input data for target prediction analysis.
To assess the performance of miRTar Hunter, we collected 481 predicted miRNA:mRNA target pairs from the miRNAMap database (Hsu et al., 2008), 532 experimentally-verified positive miRNA:target pairs from the TarBase database (Papadopoulos et al., 2009), and 59 experimentally-verified negative miRNA: target pairs from the web site (http://www.isical.ac.in/~bioinfo_miu/download20.htm) of the Machine Intelligence Unit of the Indian Statistical Institute. The miRNA:target pair of the web site consists of a miRNA name and the GenBank accession number of a target gene. Mature sequences of the miRNAs in the pairs were retrieved from miRBase by their names and transcript sequences of the target genes in the pairs were retrieved from GenBank by their accession numbers. The miRNA:target pair of the TarBase database consists of a mature miRNA sequence and the name of a target gene. In this case, the transcript sequence of a target gene was retrieved from GENE database in National Center for Biotechnology and Information (NCBI) by its gene name. The experimentally-verified positive and negative <mature miRNA sequence, target transcript sequence> pair data were employed to evaluate the performance of our method in terms of recall and precision. The target mRNA sequences from miRNAMap has been identified by three representative computational tools, namely miRanda, TargetScan, and RNAHybrid. We used predicted target data to indirectly compare our method with the three popular programs.
RESULTS
User-friendly Web Interface and diverse options/parameters for customized analysis
miRTar Hunter is implemented in C, PHP (Personal Hypertext Preprocessor), and SQL (Structured Query Language) languages on a MySQL database management system and a Linux operating system. Figure 3 shows the first page of the miRTar Hunter web interface, which is composed of five main sections: “Enter microRNAs”, “Sequence Submission”, “Core Algorithm Options”, “Alignment Options”, and “Types of microRNA Target Sites”. There are three different options for user input of microRNA(s): “Enter microRNAs” is for entering either the microRNAs in multi-FASTA format directly into a textbox or for uploading a file of microRNA(s) or retrieving microRNA(s) from the human miRNA database. The “Sequence Submission” box allows users to input mRNA sequences in FASTA format directly into a textbox or to upload a file containing the input mRNA. The “Core Algorithm Options” has four parameters: complementarity score threshold, free energy threshold, the weight for the 5′ region (or seed region), and the weight for the 3′ region. The gap opening penalty and gap extension penalty can be altered in the “Alignment Options” section. All options and parameters provided can be specified by the user to be tailored to his/her intent. The “Types of microRNA Target Sites” section allows the user to select multiple items among 7mer-A1 sites, 7mer-m8 sites, 8mer sites, 6mer sites, offset 6mer sites, 3′-supplementary sites, and 3′-compensatory sites. The web interface was designed to allow users to perform customized analyses.
Fig. 3.
User-friendly web interface of miRTar Hunter. This web interface has five main sections: “Enter microRNAs”, “Sequence Submission”, “Core Algorithm Options”, “Alignment Options”, and “Types of microRNA Target Sites”. The web interface is designed to allow customization of various options/parameters for tailored analyses.
Performance evaluation of miRTar Hunter
A variety of quantitative measures have been proposed to characterize the accuracy of prediction methods in the bioinformatics field. A common way of the evaluating the performance of a prediction method is using recall and precision. These measures are named for their origin in information retrieval. Recall and precision are defined as
| (1) |
Fundamentally, accuracy is related to the degree of concordance between predicted and actual miRNA targets. The outcome of a prediction experiment results in a number of correctly predicted data from both classes, i.e., “true positives (TP)” and “true negatives” (TN). In the same sense, mispredicted data are called “false positives” (FP) and “false negatives” (FN). In the context of miRNA target prediction programs, recall is the proportion of actual miRNA targets that have been correctly predicted as miRNA targets and precision or confidence (as it is called in data mining) denotes the proportion of predicted targets that are actually targets.
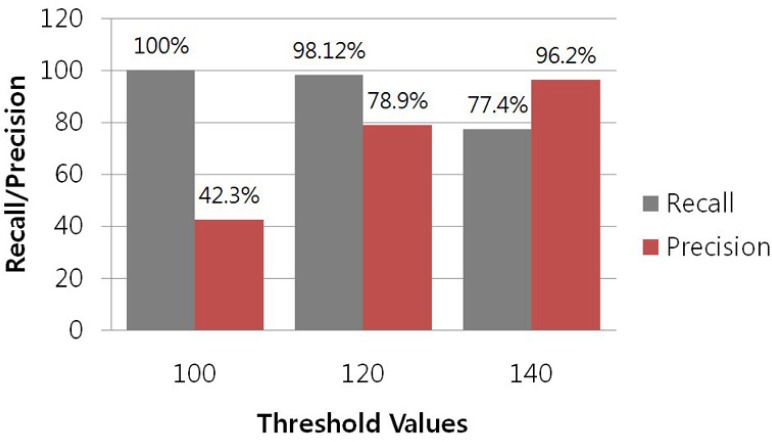
As stated above, to assess the predictive accuracy of our target scan algorithm in terms of recall and precision, we used 532 experimentally-verified positive miRNA:target pairs from the TarBase and 59 experimentally-verified negative miRNA: target pairs from the web site (http://www.isical.ac.in/~bioinfo_miu/download20.htm) of the Machine Intelligence Unit of the Indian Statistical Institute. The results of the performance test are shown in Fig. 4. Our target scan algorithm displayed much higher recall than the programs TargetScan, miRanda and RNAHybrid. Sethupathy and colleagues evaluated mammalian target prediction programs (Sethupathy et al., 2006) based on experimentally-supported miRNA-target interactions provided by TarBase, and estimated the performance of each program by determining the program’s recall. They reported that most programs had a relatively low recall of about 60–65%. In contrast, our method showed a relatively high recall of 100%, 98.12%, and 77.4% for threshold values of 100, 120, and 140, respectively. However, false positive data are major problems in miRNA target predictions. Similar to the existing programs, our method had a low precision of 42.3% for a threshold value of 100. However, it showed relatively high precisions of 78.8% and 96.2% for threshold values of 120 and 140, respectively. It is difficult to avoid a high rate of false positives in miRNA target prediction in animals because of the partial complementarity of miRNAs and their targets in animals. We also indirectly compared the performance of miRTar Hunter with that of Target-Scan, miRanda, and RNAHybrid. For this indirect comparison, we selected 481 predicted miRNA:target pairs from the miRNAMap database (Hsu et al., 2008) that had been predicted by Target-Scan, miRanda, and RNAHybrid. These pairs were input into miRTar Hunter, and the complementarity alignment score for each pair was calculated. Figure 5 shows the distribution of complementarity alignment scores, which were all above the threshold value of 100. This result indicates that miRTar Hunter can detect any target sites predicted by the three popular programs with a threshold value of 100. A direct comparison of our algorithm with existing algorithms is challenging, because the different published methods are based on different principles, making it difficult to an accurate comparison. In addition, many methods are not available for download for independent testing on a common dataset, whereas the datasets used by these methods are highly diverse, which is why we only indirectly compared these methods.
Fig. 4.
Performance of the target scan algorithm implemented in miRTar Hunter based on 532 experimentally-verified positive miRNA: target pairs and 59 experimentally-verified negative miRNA:target pairs. Our method had sensitivities of 100%, 98.12%, and 77.4% for threshold values of 100, 120, and 140, respectively, which is much higher than that of other existing programs. In previous work, most existing programs had almost the same recall of about 60–65% (Sethupathy et al., 2006). Similar to other programs, our method had a low precision of 42.3% for the threshold value of 100. However, it had relatively high precisions of 78.8% and 96.2% for the threshold values of 120 and 140, respectively.
Fig. 5.

The distribution of complementarity alignment scores of miRNA-target pairs predicted by miRanda, TargetScan, and RNAHybrid. The predicted data were extracted from the miRNAMap database. Our method yielded complementarity alignment scores above the threshold value of 100 for the predicted data. This result indicates that miRTar Hunter can detect any target sites predicted by the three popular programs with the threshold value set to 100.
DISCUSSION
Experimental identification of miRNA target sites is a costly and time-consuming process. While recent estimates suggest that more than 50% of human protein coding genes may be regulated by miRNAs and that each miRNA may bind to hundreds of target genes, the latest release of the TarBase database contains information on only 995 human in vivo miRNA-target gene interactions involving 103 distinct miRNAs and 825 distinct genes, a far cry from the actual extent of miRNA targeting (Bartel, 2009; Papadopoulos et al., 2009). Computational prediction of miRNA-target gene interactions is a valuable tool for guiding wet-lab experiments, and it remains the only option for systematic genome-wide analysis. It is also a challenging task because of the difficulty of distinguishing true miRNA-mRNA interactions from the noisy background of millions of possible miRNA-mRNA combinations and, more generally, because the basic mechanisms of miRNA target recognition remain largely unknown. In recent years, several target prediction algorithms based on different principles have been developed (Bartel, 2009). However, the two recurring parameters used by the available methods are the existence of a seed match and evolutionary conservation of the target site across multiple species. Utilization of these powerful constraints in prediction algorithms leads to more reliable detection of functional duplexes, but at the same time, limits our ability to identify biologically relevant miRNA target sites that do not fulfill these requirements. By definition, organism-specific or simply poorly conserved sites cannot be predicted at all if a conservation filter is applied. It has also been suggested that the seed match requirement may be too stringent, and that several non-canonical types of target sites exist that cannot be detected by seed match based methods. Furthermore, many potential miRNA-target interactions that do involve conserved seed regions may be non-functional in a physiological context.
In light of the above, we sought to develop a computational technique free from both the seed requirement and the conservation filter and thus capable of predicting species-specific and non-canonical as well as canonical target sites. While miRTar Hunter does not require a perfect seed match, it weights the seed region and the non-seed region differentially according to various types of target sites. However, our algorithm does have flaws that can be improved upon. If multiple miRNAs target the same site on a transcript, only the miRNA with the highest complementarity alignment score and lowest energy score is reported for that site. This is a possible source of false negatives because different miRNAs are expressed at different times in cell development. It is possible that multiple miRNAs can bind to overlapping sites, but because they are never expressed at the same time in the cell, they do not usually compete for binding. Furthermore, even though we modified the Smith-Waterman alignment algorithm specifically for miRNA/mRNA sequence comparisons, it can be argued that the Smith-Waterman alignment was originally designed to compare evolutionarily-related sequences, and that miRNAs and their mRNA targets do not fall into this category. Finally, the parameter values and penalties assigned throughout our algorithm are arbitrary and should be modified as greater knowledge regarding the biology of miRNA targeting accumulates. Ultimately, we intend to further refine the rules for miRNA-target interactions as more and more miRNA interactions are characterized and validated.
Acknowledgments
This work was supported by a grant from the Korea Centers for Disease Control and Prevention.
REFERENCES
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 mirnas results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource targets and expression. Nucleic Acids Res. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kenneth SK. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran J, Strauss WM. Bioinformatic trends for the determination of miRNA-target interactions in mammals. DNA Cell Biol. 2007;26:353–350. doi: 10.1089/dna.2006.0546. [DOI] [PubMed] [Google Scholar]
- Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:1–11. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu P, Wong YH, Chen YH, Chen GH, Huang HD. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. 2008;36:D165–169. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein E, MacMenamin P, Piedade ID, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Sander C. The HSSP database of protein structure-sequence alignments. Nucleic Acids Res. 1996;24:201–205. doi: 10.1093/nar/24.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalizan microRNA targets. Nat. Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman MS, Eggert M. A new algorithm for best subsequence alignments with application to tRNA-rRNA comparisons. J Mol Biol. 1987;197:723–728. doi: 10.1016/0022-2836(87)90478-5. [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchty S, Fontana W, Hofacker IL, Schuster P. Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers. 1999;49:145–165. doi: 10.1002/(SICI)1097-0282(199902)49:2<145::AID-BIP4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamic and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]