Abstract
Lung cancer is a leading cause of cancer-related mortality across the world and tobacco smoking is the major risk factor. The Wnt signaling pathway is known to be involved in smoke-induced tumorigenesis in the lung. Promoter hypermethylation of Wnt inhibitory factor 1 (Wif1) has become a common event in a number of human tumors. Using a methylation-specific PCR, hypermethylation of the Wif1 gene promoter was evaluated in 139 primary non-small cell lung cancers (NSCLCs) and its correlation with clinicopathological and prognostic parameters was evaluated. Methylation of Wif1 was observed in 47.5% and 20.9% of neoplastic and adjacent normal lung tissues, respectively. Its methylation rate tended to be higher in stage I than stages II–IIIA. Results of Kaplan-Meier analysis showed no significant difference in overall survival according to Wif1 methylation status. However, Wif1 methylation showed an association with unfavorable prognosis of adenocarcinoma (AC) patients with EGFR mutation. According to our current findings, Wif1 promoter methylation is an early, frequent event as an epigenetic field manner and could be considered as a useful prognostic marker for AC patients with EGFR mutation. Further investigation into the therapeutic potential of this finding is warranted.
Keywords: EGFR, hypermethylation, MSP, NSCLC, prognosis, Wif1
INTRODUCTION
Lung cancer continues to be a major public health problem worldwide. Late diagnosis and lack of effective treatments for lung cancer are associated with unsatisfactory prognosis and high mortality (Jemal et al., 2010). In addition, lung cancer is a heterogeneous group of diseases made up of entities characterized by distinctive clinical, pathological, morphologic, and genetic features (Minna et al., 2002). However, the molecular basis of these variations in behavior and epidemiology is not well known. Of interest, growing evidence has demonstrated a remarkable difference in genetic and epigenetic factors leading to this neoplasm between East Asian and Western populations (Gazdar et al., 2004; Toyooka et al., 2003). In Korea, prevalence of male smoking is among the highest in the world, and mortality rates from smoking-related cancers, particularly lung cancer, are escalating (Jee et al., 2004). Therefore, advances in understanding of the molecular pathogenesis of lung cancer are critical to conquering lung cancer. Transcriptional silencing of genes by CpG island (CGI) methylation is now recognized as a critical component in initiation and progression of lung cancer (Kerr et al., 2007; Kim et al., 2007; Na et al., 2010; Toyooka et al., 2001). In addition, variation in methylation status has been associated with tobacco smoking (Liu et al., 2010).
Dysregulation of Wnt signaling is common in several types of cancer, including colon, lung, breast, and prostate (Mazieres et al., 2005; Paul et al., 2008). In particular, mutations of key Wnt signaling genes, such as APC, β-catenin, or Axin appear to be rare in lung cancer (Mazieres et al., 2005). The Wnt pathway is highly regulated and very complex, with different Wnt inhibitors, which consistently inhibit or facilitate Wnt signaling depending on molecular, cellular, and tissue context (Rubin et al., 2006). The Wnt pathway has been associated with normal lung development and homeostasis as well as chronic lung diseases such as fibrosis and asthma (Konigshoff et al., 2010; Van et al., 2008). However, there are contradictory findings indicating that the Wnt pathway is activated or down-regulated by cigarette smoke in bronchial epithelial cells (Lemjabbar-Alaoui et al., 2006; Wang et al., 2011). Wnt inhibitory factor 1 (Wif1), an extracellular antagonist that acts by binding to Wnt ligands, is a frequent target for epigenetic silencing in various human cancers (Ying et al., 2009). Of particular interest, Wif1 promoter hypermethylation is common in Western patients with lung cancer; however, it is rare in Japanese cases (Licchesi et al., 2008; Mazieres et al., 2004; Tang et al., 2006; Yoshino et al., 2009), which have been conducted in small numbers to draw definitive conclusions. We have recently found that DKK1 methylation is common in NSCLC and is a good prognostic indicator for a certain group of patients (Na et al., 2012). Herein, in order to further understand the molecular pathogenesis and clinical relevance of the Wnt pathway in lung cancer, we performed methylation-specific PCR (MSP) in order to determine the methylation status of the Wif1 gene promoter in Korean patients with NSCLC and investigated the relationship between methylation status and clinicopathological factors.
MATERIALS AND METHODS
Patients and tissue samples
Tumor and corresponding non-malignant lung tissue specimens (n = 139) were provided by the National Biobank of Korea - Kyungpook National University Hospital (KNUH), which is supported by the Ministry of Health, Welfare and Family Affairs. All materials derived from the National Biobank of Korea - KNUH were obtained under institutional review board approved protocols. A summary of the clinicopathological characteristics of the patients which had previously been examined in another study (Lee et al., 2012) is provided in Table 1.
Table 1.
Correlation between Wif1 methylation and the clinicopathological features of NSCLCs
| Feature | Wif1 Methylation (%) | P |
|---|---|---|
| All subjects (n = 139) | 66 (47.5) | |
| Age (years) | ||
| ≤ 63 (n = 67) | 30 (44.8) | 0.328 |
| > 63 (n = 72) | 36 (50.0) | |
| Gender | ||
| Male (n = 101) | 48 (47.5) | 0.570 |
| Female (n = 38) | 18 (47.4) | |
| Smoking status | ||
| Ever (n = 104) | 51 (49.0) | 0.331 |
| Never (n = 35) | 15 (41.9) | |
| Histologic types | ||
| Squamous cell carcinoma (n = 60) | 30 (50.0) | 0.364 |
| Adenocarcinoma (n = 79) | 36 (45.6) | |
| EGFR mutations in Adenocarcinoma | ||
| Absent (n = 46) | 24 (52.2) | 0.122 |
| Present (n = 33) | 12 (36.4) | |
| TP53 mutations* | ||
| Absent (n = 54) | 25 (46.3) | 0.470 |
| Present (n = 49) | 24 (49.0) | |
| Pathologic stages | ||
| Stage I (n = 85) | 45 (52.9) | 0.068 |
| Stage II–IIIA (n = 54) | 21 (38.9) |
The mutation of TP53 was studied in 103 of the 139 NSCLCs.
Cell culture and 5-aza-2′-deoxycytidine (5-AzadC) treatment
A normal human bronchial epithelial cell (NHBE) and 4 human NSCLC cell lines (A549, H460, H1703, and H2009) were obtained from the American Type Culture Collection (ATCC, USA). All cells were propagated with the instructions from the ATCC. 5-AzadC treatment was performed as described previously (Lee et al., 2012).
Total RNA isolation and reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from the cultured cells using TRIzol (Invitrogen, Australia) according to the manufacture’s instructions. The structural integrity of the total RNA was confirmed by electrophoresis on 1.2% agarose-formaldehyde gels. Residual genomic DNA was digested with RNase-free DNase (Invitrogen). First strand cDNA was reverse-transcribed from 2 μg of total RNA in a total volume of 20 μl using oligo (dT) and a SuperScript preamplification kit (Invitrogen). The resulting cDNA was amplified by forward (5′-CCGAAATGGAGGCTTTTGTA-3′) and reverse (5′-TGGTTGAGCAGTTTGCTTTG-3′) primers. Amplified products were separated on 2% agarose gels, visualized using ethidium bromide, and photographed.
Genomic DNA isolation and methylation analysis
Genomic DNA was extracted using a QIAamp DNA Mini Kit (QIAGEN, USA). Bisulfite modification of geno-mic DNA was carried out by using an EZ DNA methylation kit (Zymo Research Co, USA) according to the manufacturer’s protocol. Bisulfite-treated genomic DNA was amplified using either a methylation-specific or an unmethylation-specific primer set; for the methylated reaction 5′-GGGCGTTTTATTGGGCGTAT-3′ (forward) and 5′-AAACCAACAATCAACGAAAC-3′ (reverse), and for the unmethylated reaction 5′-GGGTGTTTTATTGGG TGTAT-3′ (forward) and 5′-AAACCAACAATCAACAAAAC-3′ (reverse). All PCR amplifications were performed using reagents supplied in a GeneAmp DNA Amplification Kit with AmpliTaq Gold as the polymerase (PE Applied Biosystems, USA) on PTC-100 (MJ Research, USA). CpGenome™ Universal methylated and unmethylated DNA (Chemicon, USA) was used as a positive control for the methylated and unmethylated genes, respectively. Negative control samples without DNA were included for each set of PCR. PCR products were analyzed on 2% agarose gel, stained with ethidium bromide, and visualized under UV light. Each MSP was repeated at least once in order to confirm the results.
Statistical analysis
The relationship between the methylation status and the clinicopathological characteristics was analyzed using a chi-square test or Fisher’s exact test for categorical variables. A P value of less than 0.05 was considered statistically significant. A logistic regression test was performed in order to estimate the relationship between methylation and the covariates of age, gender, exposure to tobacco smoke, and histology. The overall survival (OS) of NSCLC patients according to methylation status of the Wif1 gene was compared using the Kaplan-Meier method and the log-rank test. Hazard ratios (HR) and 95% confidence interval (CI) were estimated using a multivariate Cox proportional hazard model, with adjustment for age (≤ 63 versus > 63 years), gender (male versus female), smoking status (never-versus ever-smoker), and pathologic stage (I versus II–IIIA). The homogeneity test was performed for comparison of the difference between HR of different groups. The Statistical Analysis System for Windows, version 9.1 (SAS Institute, USA) was used in performance of all analyses.
RESULTS AND DISCUSSION
Methylation status of the Wif1 gene in tissues and inverse correlation with its mRNA expression
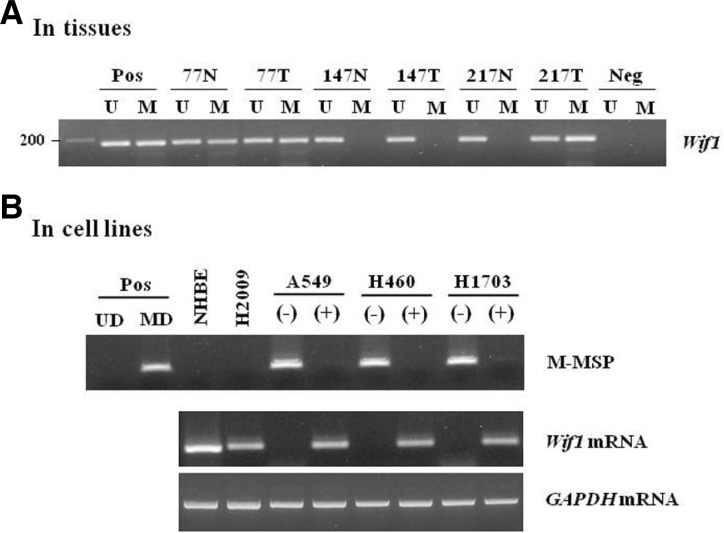
The methylation status of the human Wif1 gene was analyzed in 139 primary NSCLCs and matched nonmalignant lung tissues using the MSP method. Methylated and unmethylated allele-specific primers yielded a single band of the expected size; representative examples of the MSP analysis are shown in Fig. 1A. Unmethylated bands were detected in most of the nonmalignant and malignant tissues, thus confirming the integrity of the DNA in these samples. Bisulfite-sequencing of the representative PCR products confirmed their methylation status and showed that all cytosines at non-CpG sites were converted to thymine (data not shown), ruling out the possibility of incomplete bisulfite conversion. Wif1 hypermethylation was detected in 47.5% (66/139) of cancerous tissues and in 20.9% (29/139) of adjacent normal tissues (P < 0.0001), suggesting that its methyllation may be not an intrinsic, developmentally programmed event, but a tumor-associated de novo event. Our data is similar to recent finding that Wif1 are methylated in 31.1% of normal adjacent specimens and 68.9% of AC samples (Licchesi et al., 2008), although there was a difference of methylation prevalence due to MSP assay sensitivity (conventional vs nested MSP). Notably, Wif1 methylation was observed in a subset of nonmalignant lung tissues of the patients. Methylation of nonmalignant lung tissues concurred predominantly with that in the corresponding cancer tissues (data not shown), therefore, it is likely that the presence of Wif1 methylation in histologically nonmalignant tissues may be due to unavoidable contamination of nonmalignant cells with methylated cancer cells. Alternatively, it is possible that even phenotypically normal lung tissues can already harbor epigenetic alterations because the entire field of the lung was exposed to carcinogenic insult, such as cigarette smoking. Frequent occurrence of aberrant promoter methylation in histologically normal-appearing lung tissues has been reported, representing a field defect of widespread epigenetic change in lung tissues (Belinsky et al., 2002; Guo et al., 2004). Considering that methylation of the Wif1 gene is nearly as frequent in normal lung parenchyma as in low grade-atypical adenomatous hyperplasia (Licchesi et al., 2008), our findings suggest that Wif1 promoter hypermethylation may be a common and tumor-associated event as a field-cancerization manner.
Fig. 1.
Hyperethylation of Wif1 in NSCLC patients (A) and correlation of its methylation with silencing of in cell lines (B). (A) Promoter methylation of the Wif1 gene was analyzed using MSP. CpGenome™ Universal methylated DNA (MD) or unmethylated DNA (UD) was used as a positive control for the methylated or unmethylated products, respectively. Water was used as a negative control. N, nonmalignant tissue; T, tumor tissues; U and M, amplified product with primers that recognize the unmethylated or methylated sequences. (B) Expression of Wif1 mRNA and its methylation status were determined in 5 cell lines by RT-PCR and MSP, res-pectively. Amplified product was visible at 188-bp for Wif1. Amplification of GAPDH was used as an internal loading control. The symbol (−) indicated vehicle alone treatment, whereas (+) indicated the 5-AzadC treatment for 3 days.
To determine whether promoter methylation was associated with transcriptional silencing of the Wif1 gene, the mRNA level and methylation status were investigated in NSCLC cell lines using RT-PCR and MSP analysis. Wif1 mRNA was found in NHBE and H2009 cells lacking methylated alleles, while its expression was hardly detected in the cell lines (A549, H460, and H1703) with methylated ones (Fig. 1B). Moreover, treatment of DNA demethylating agent 5-AzadC dramatically restored Wif1 mRNA expression in these cell lines. These results are agreement with previous finding by Mazieres et al. (2004), indicating an inverse correlation between Wif1 promoter methylation and its mRNA expression.
Correlation between methylation status and clinicopathological characteristics
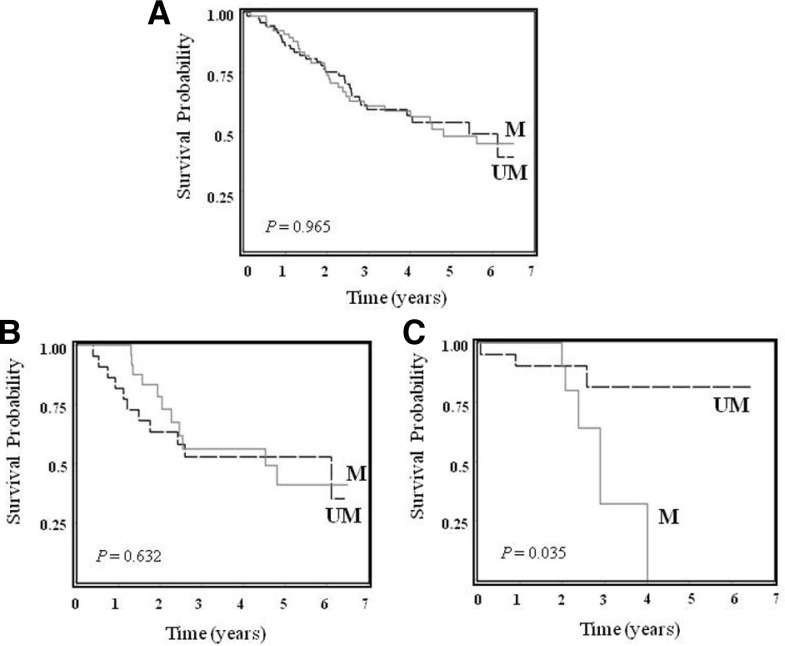
Wif1 methylation was more frequent in patients with stage I disease than those with stages II–IIIA with a borderline significance (P = 0.068). However, no correlations with other clinicopathological profiles, such as age, sex, smoking status, tumor histology, EGFR and TP53 mutations were found (Table 1). In addition, no significant difference in overall survival (OS) of total patients was observed according to Wif1 methylation status (Log-Rank P [PL-R] = 0.965, Fig. 2A). Of particular interest, when ACs were categorized according to EGFR mutation, Wif1 methylation showed an association with a worse OS in AC patients with EGFR mutation (P [PL-R] = 0.035, Figs. 2B and 2C). Multivariate analysis showed that Wif1 methylation exhibited a trend toward worse OS in AC patients with EGFR mutation (adjusted HR = 3.85, 95% CI = 0.79–18.72, P = 0.075) (Table 2), being comparable with a recent observation that in ACs with EGFR mutation group, patients with methylated Wnt antagonist SFRP5 has a significantly shorter progression free survival than those with unmethylated SFRP5 (Zhu et al., 2012). Crosstalk between Wnt and EGFR signaling in cancers has been well documented (Hu et al., 2010). Wnt binds to Frizzled receptor, leading to transactivation of EGFR signaling by matrix metalloproteinase-mediated release of soluble EGFR ligands. Upon activation, EGFR could transactivate β-catenin through the receptor tyrosine kinase-PI3K/Akt pathway, and β-catenin might also form a heterodimer with EGFR, leading to activation of the EGFR pathway. A meta-analysis has demonstrated a frequent association of EGFR over-expression in NSCLC with poor prognosis, even in the presence of contradicting results (Nakamura et al., 2006). In addition, somatic mutations of the EGFR gene hyperactivate the downstream pro-survival pathways and consequently confer oncogenic shock (Sharm et al., 2007). Similarly, the EGFR kinase domain mutation is associated with overexpression of EGFR (Ohtsuka et al., 2006). Taken together, it is tempting to speculate that synchronous alterations of Wif1 and EGFR might act synergistically in enhancement of Wnt and EGFR signaling, tumor formation, and progression, contributing to a poor prognosis. However, further investigation with large numbers of patients is needed in order to confirm this clinical implication.
Fig. 2.
Kaplan-Meier survival curves of NSCLC patients according to Wif-1 methylation status. Kaplan-Meier survival curve for all patients (A), AC patients without EGFR mutations (B), and AC patients with EGFR mutations (C). P-values from log-rank test. UM denotes patients with unmethylation, while M indicates patients with methylation.
Table 2.
Association of Wif1 methylation with overall survival in NSCLC patients
| Variables | PL-R | Crude HR (95% CI) | P | PH | Adjusted HR (95% CI) | P | PH |
|---|---|---|---|---|---|---|---|
| Overall subjects (n = 139) | 0.965 | 0.96 (0.58–1.58) | 0.857 | 1.13 (0.68–1.89)‡ | 0.644 | ||
| EGFR mutations in AC | |||||||
| Absent (n = 46) | 0.632 | 0.82 (0.35–1.89) | 0.633 | 0.054 | 1.00 (0.43–2.35)¶ | 0.968 | 0.141 |
| Present (n = 33) | 0.035 | 4.21 (0.99–17.98) | 0.052 | 3.85 (0.79–18.72)¶ | 0.075 |
Hazard ratios (HRs), 95% confidence intervals (CIs), and their corresponding P-values were calculated using multivariate Cox proportional hazard models, adjusted for age, gender, smoking status, tumor histology, and pathologic stage
HRs, 95% CIs, and their corresponding P-values were calculated using multivariate Cox proportional hazard models, adjusted for age, gender, smoking status, and pathologic stage
Collectively, although the present study is limited by the small number of samples and the virtual lack of information on protein, it shows that Wif1 promoter is frequently methylated in NSCLCs to down-regulate its mRNA expression. In addition, Wif1 methylation is significantly associated with unfavorable prognosis of AC patients with EGFR mutation.
Acknowledgments
This work was supported by the Korea Research Foundation Grant funded by the Korean Government [The Ministry of Education & Human Resources Development (MOEHRD), Basic Research Promotion Fund] (2010-0010000).
REFERENCES
- Belinsky SA, Palmisano WA, Gilliland ED, Crooks LA, Divine KK, Winters SA, Grimes MJ, Harms HJ, Tellez CS, Smith TM, et al. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002;62:2370–2377. [PubMed] [Google Scholar]
- Gazdar AF, Shigematsu H, Herz J. Mutations and addiction to EGFR: the Achilles ‘heal’ of lung cancers? Trends Mol Med. 2004;10:481–486. doi: 10.1016/j.molmed.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Guo M, House MG, Hooker C, Han Y, Heath E, Gabrielson E, Yang SC, Baylin SB, Herman JG, Brock MV. Promoter hypermethylation of resected bronchial margins: a field defect of changes? Clin Cancer Res. 2004;10:5131–5136. doi: 10.1158/1078-0432.CCR-03-0763. [DOI] [PubMed] [Google Scholar]
- Hu T, Li C. Convergence between Wnt-β-catenin and EGFR signaling in cancer. Mol Cancer. 2010;9:236–242. doi: 10.1186/1476-4598-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee SH, Samet JM, Ohrr H, Kim JH, Kim IS. Smoking and cancer risk in Korean men and women. Cancer Cases Control. 2004;15:341–348. doi: 10.1023/B:CACO.0000027481.48153.97. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Kerr KM, Galler JS, Hagen JA, Laird PW, Laird-Offringa IA. The role of DNA methylation in the development and progression in lung adenocarcinoma. Disease Marker. 2007;23:5–30. doi: 10.1155/2007/985474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Cha SI, Lee JH, Lee YM, Choi JE, Kim MJ, Lim JS, Lee EB, Kim CH, Park TI, et al. Aberrant DNA methylation profiles of non-small cell lung cancers in a Korean population. Lung Cancer. 2007;58:1–6. doi: 10.1016/j.lungcan.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Konigshoff M, Eickelberg O. WNT signaling in lung disease. Am J Respir Cell Mol Biol. 2010;42:21–31. doi: 10.1165/rcmb.2008-0485TR. [DOI] [PubMed] [Google Scholar]
- Lee SM, Park JY, Kim DS. Methylation of TMEFF2 gene in tissue and serum DNA from patients with non-small cell lung cancer. Mol Cells. 2012;34:171–176. doi: 10.1007/s10059-012-0083-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemjabbar-Alaoui H, Dasari V, Sidhu S, Mengistab A, Finkbeiner W, Gallup M, Basbaum C. Wnt and hedgehog are critical mediators of cigarette smoke-induced lung cancer. PLoS One. 2006;1:1–11. doi: 10.1371/journal.pone.0000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licchesi JDF, Westra WH, Hooker CM, Machida EO, Baylin SB, Herman JG. Epignetic alteration of Wnt pathway antagonists in progressive glandular neoplasia of the lung. Carcinogenesis. 2008;29:895–904. doi: 10.1093/carcin/bgn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Killian JK, Yang M, Walker RL, Hong JA, Zhang M, Davis S, Zhang Y, Hussain M, Xi S, et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene. 2010;29:3650–3664. doi: 10.1038/onc.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazieres J, He B, You L, Xu Z, Lee AY, Mikami I, Reguart N, Rosell R, McCormick F, Jablons DM. Wnt inhibitory factor is silenced by promoter hypermethylation in human lung cancer. Cancer Res. 2004;64:717–720. doi: 10.1158/0008-5472.CAN-04-1389. [DOI] [PubMed] [Google Scholar]
- Mazieres J, He B, You L, Xu Z, Jablons DM. Wnt signaling in lung cancer. Cancer Lett. 2005;222:1–10. doi: 10.1016/j.canlet.2004.08.040. [DOI] [PubMed] [Google Scholar]
- Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1:49–52. doi: 10.1016/s1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- Na YK, Lee SM, Hong HS, Kim JB, Park JY, Kim DS. Hypermethylation of growth arrest DNA-damage inducible gene 45 in non-small cell lung cancer and its relationship with clinicopathologic features. Mol Cells. 2010;30:89–92. doi: 10.1007/s10059-010-0092-1. [DOI] [PubMed] [Google Scholar]
- Na YK, Lee SM, Kim DS, Park JY. Promoter methylation of Wnt antagonist DKK1 gene and prognostic value in Korean patients with non-small cell lung cancers. Cancer Biomarkers. 2012;12:73–79. doi: 10.3233/CBM-2012-00295. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Kawasaki N, Taguchi M, Kabasawa K. Survival impact of epidermal growth factor receptor overexpression in patients with non-small cell lung cancer: a meta-analysis. Thorax. 2006;61:140–145. doi: 10.1136/thx.2005.042275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka K, Ohnishi H, Furuyashiki G, Nogami H, Koshiishi Y, Ooide A, Matsushima S, Watanabe T, Goya T. Clinico-pathological and biological significance of tyrosine kinase domain gene mutations and overexpression of epidermal growth factor receptor for lung adenocarcinoma. J Thorac Oncol. 2006;8:787–795. [PubMed] [Google Scholar]
- Paul S, Dey A. Wnt signaling and cancer development: therapeutic implication. Neoplasma. 2008;55:165–176. [PubMed] [Google Scholar]
- Rubin JS, Barshishat-Kupper M, Feroze-Merzoug F, Xi ZF. Secreted WNT antagonists as tumor suppressors: pro and con. Front Biosci. 2006;11:2093–105. doi: 10.2741/1952. [DOI] [PubMed] [Google Scholar]
- Sharm SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- Tang M, Torres-Lanzas J, Lopez-Rios F, Esteller M, Sanchez-Cespedes M. Wnt signaling promoter hyperemthylation distinguishes lung primary adenocarcinomas from colorectal metastasis to the lung. Int. J Cancer. 2006;119:2603–2606. doi: 10.1002/ijc.22211. [DOI] [PubMed] [Google Scholar]
- Toyooka S, Toyook KO, Maruyama R, Virmani AK, Girard L, Miyajima K, Harada K, Ariyoshi Y, Takahashi T, Sugio K, et al. DNA methylation profiles of lung tumors. Mol Cancer Ther. 2001;1:61–67. [PubMed] [Google Scholar]
- Toyooka S, Maruyama R, Toyooka KO. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int. J Cancer. 2003;103:153–160. doi: 10.1002/ijc.10787. [DOI] [PubMed] [Google Scholar]
- Van Scoyk M, Randall J, Sergew A, Williams LM, Tennis M, Winn RA. Wnt signaling pathway and lung disease. Transl Res. 2008;151:175–180. doi: 10.1016/j.trsl.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Wang R, Ahmed J, Wang G, Hassan I, Strulovici-Barel Y, Hackett NR, Crystal RG. Down-regulation of the canonical Wnt β-catenin pathway in the airway epithelium of healthy smokers and smokers with COPD. PLoS One. 2011;6:e14793. doi: 10.1371/journal.pone.0014793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y, Tao Q. Epigenetic disruption of the WNT/β-catenin signaling pathway in human cancers. Epigenetics. 2009;4:307–312. doi: 10.4161/epi.4.5.9371. [DOI] [PubMed] [Google Scholar]
- Yoshino M, Suzuki M, Tian L, Moriya Y, Hoshino H, Okamoto T, Yoshida S, Shibuya K, Yoshino I. Promoter hypermethylation of the p16 and Wif-1 genes as an independent prognostic marker in stage IA non-small cell lung cancers. Int J Oncol. 2009;35:1201–1209. doi: 10.3892/ijo_00000437. [DOI] [PubMed] [Google Scholar]
- Zhu J, Wang Y, Duan J, Bai H, Wang Z, Wei L, Zhao J, Zhou M, Wang S, Yang L, et al. DNA methylation status of Wnt antagonist SFRP5 can predict the response to the EGFR-tyrosine kinase inhibitor therapy in non-small cell lung cancer. J Exp Clin Cancer Res. 2012;31:80–88. doi: 10.1186/1756-9966-31-80. [DOI] [PMC free article] [PubMed] [Google Scholar]