Abstract
Homologous recombination occurs closely between homologous chromatids with highly ordered recombinosomes through RecA homologs and mediators. The present study demonstrates this relationship during the period of “partner choice” in yeast meiotic recombination. We have examined the formation of recombination intermediates in the absence or presence of Shu1, a member of the PCSS complex, which also includes Psy3, Csm2, and Shu2. DNA physical analysis indicates that Shu1 is essential for promoting the establishment of homolog bias during meiotic homologous recombination, and the partner choice is switched by Mek1 kinase activity. Furthermore, Shu1 promotes both crossover (CO) and non-crossover (NCO) pathways of meiotic recombination. The inactivation of Mek1 kinase allows for meiotic recombination to progress efficiently, but is lost in homolog bias where most double-strand breaks (DSBs) are repaired via stable intersister joint molecules. Moreover, the Srs2 helicase deletion cells in the budding yeast show slightly reduced COs and NCOs, and Shu1 promotes homolog bias independent of Srs2. Our findings reveal that Shu1 and Mek1 kinase activity have biochemically distinct roles in partner choice, which in turn enhances the understanding of the mechanism associated with the precondition for homolog bias.
Keywords: homolog bias, meiotic recombination, Mek1 kinase, Shu1, Srs2
INTRODUCTION
Meiosis involves a complex progression of chromosomal events that result in the physical connection of homologous chromosomes. Meiotic DSBs occur immediately after the S-phase, when sisters are present. Nonetheless, these DSBs interact preferentially with homolog partner chromatids rather than with their sisters (Hunter and Kleckner, 2001; Kim et al., 2010; Schwacha and Kleckner, 1997). Interhomolog bias is a well-conserved pathway for the repair of homologous chromatids during meiosis. Although meiotic recombination has been shown to preferentially favor interhomolog bias through highly regulated establishment and maintenance machineries, elucidation of the molecular mechanism that drives its components is required to fully comprehend the mechanical processes.
During meiosis, recombination is initiated by programmed DSBs, and is catalyzed by the topoisomerase-like protein Spo11 (Keeney, 2001; Neale et al., 2005). Most of these DSBs establish interactions with a homolog partner (Bishop and Zickler, 2004; Hong et al., 2013; Hunter and Kleckner, 2001; Kim et al., 2010). The first DSB end is released and engaged in the partner duplex, and then is stabilized in single-end invasions (SEIs). This stable SEI is primed for DNA extension by polymerase, and forms double-Holliday Junctions (dHJs). In recent study, it was found that intersister SEIs form via hyper-resected DSB, and then associate with the second DSB end to yield intersister dHJs (Kim et al., 2010). Homology searching and crossing over must be directed to occur specifically between non-sister chromatids rather than between sister chromatids. The majority of these interhomolog interactions via SEIs and dHJs is designated for eventual maturation into a interhomolog crossover (IH-CO) via a tightly regulated process, while the remaining interhomolog interactions mature into interhomolog non-crossover (IH-NCO) products without any exchange of the flanking regions (Börner et al., 2004; Kim et al., 2010).
Recombinational repair of spontaneous DSBs is carried out by the RecA homolog Rad51, which loads onto the single-stranded DNA tail(s) of a DSB and then searches for a homologous partner DNA duplex (Krejci et al., 2012; Shinohara et al., 1992). Yeast Rad51 is assisted in this process by accessory factors that include two Rad51 paralogs, namely, Rad55 and Rad57, and the recently described ensemble of Psy3, Csm2, Shu1, and Shu2 (the PCSS complex) (Ball et al., 2009; Krejci et al., 2012; Sasanuma et al., 2013a; Shor et al., 2005; Tao et al., 2012; Qing et al., 2011). Srs2, a major anti-recombinase, is a 3′ to 5′ DNA helicase that interacts with and dissociates Rad51 from single-stranded DNA (ssDNA) in vitro (Antony et al., 2009; Liu et al., 2011). The Rad55-Rad57 heterodimer stabilizes Rad51 filaments to resist the disruption induced by Srs2 (Liu et al., 2011). Shu1 functions in Rad51-dependent homologous recombination, and the PCSS complex inhibits Srs2 to stabilize Rad51-ssDNA filaments (Mankouri et al., 2007). In the most recent study, core Psy3-Csm2 dimer was observed to demonstrate structural similarity to Rad51/RecA (Sasanuma et al., 2013a). Furthermore, because Shu2 and Srs2 physically interact with each other (Ito et al., 2001; Martín et al., 2006), it is possible that the PCSS complex promotes strand invasion by coordination with Rad51 to prevent Srs2 from disrupting Rad51 nucleofilaments (Mankouri et al., 2007). Alternatively, Psy3-Csm2, which is a component of the PCSS complex, could directly interact with ssDNA and stabilize Rad51 nucleoprotein filaments (Sasanuma et al., 2013a).
The template choice of meiotic DSB repair is highly regulated to favor interhomolog interaction after the occurrence of a DSB, but the underlying mechanism is still unclear. In the present study, the DNA physical analysis of recombination intermediates has been used to examine the function of Shu1 (the PCSS complex) in meiotic homologous recombination and its relationship with Mek1 and Srs2 to define the “partner choice” pathway.
MATERIALS AND METHODS
Strains
Saccharomyces cerevisiae strains utilized for this study are isogenic heterothallic derivatives of SK1, and are homozygous for ho::hisG, leu2::hisG, ura3Δ (ΔPst1-Sma1) (Hunter and Kleckner, 2001). The HIS4LEU2 locus has been described (Kim et al., 2010). The Shu1 promoter was replaced with the pCUP1-KanMX promoter using the pCUP1-3HA cassette (Janke et al., 2004). KKY281, 355, and 796 contain pCUP1-SHU1, srs2Δ, and shu1Δsrs2Δ mutations, respectively.
Meiotic time course and DNA cross-linking
Meiotic time courses were performed as described previously (Kim et al., 2010). Strains were patched on YPG plates (1% yeast extract, 2% peptone, 3% glycerol, and 2% bacto agar) for 24 h. Cells were then streaked on YPD plates (1% yeast extract, 2% peptone, 2% glucose, and 2% bactoagar) and grown for two days. A single colony from the YPD plate was then inoculated into 3 mL YPD liquid medium (1% yeast extract, 2% peptone, and 2% glucose) and cultured for 24 h at 30 °C in a shaking incubator. To synchronize the cells at the G1 stage, a 1:500 dilution of the YPD cultures was added into SPS liquid medium [1% potassium acetate, 1% bactopeptone, 0.5% yeast extract, 0.17% yeast nitrogen base without amino acids, 0.5% ammonium sulfate, 0.05 M potassium biphtalate, and 2 drops/L anti-foam (Sigma), pH to 5.5] and cultured for 18 h. The SPS cultures were washed with SPM medium (0.2% potassium acetate, 0.02% raffinose, and 2 drops/L antifoam) and then resuspended in 200 ml of SPM medium. Meiotic cells were harvested at different time points and resuspended in 50 mM Tris-HCl and 50 mM EDTA. Cells were cross-linked with psoralen (Sigma) under UV light at 365 nm (6 mW/cm2) for 10 min. Non-cross-linked cells were suspended in 50 mM Tris-HCl and 50 mM EDTA buffer without psoralen. To express Shu1 protein, in the pCUP1-SHU1 strain, 30 μM CuSO4 was directly added into the SPM culture after 2 h. Then, yeast cells were harvested at the time points indicated. Meiotic cells from synchronized meiotic cultures were fixed in a buffer (40% ethanol and 0.1M sorbitol) and stained with DAPI. Spore formation and DAPI foci were counted for each time point.
Genomic DNA extraction
Cells were then spheroplasted in Zymolase (100T, USBiological) before being lysed via guanidine-phenol extraction. Isolated genomic DNA was precipitated in ethanol and dried at 4°C. DNA pellets were dissolved in 50 mM Tris-HCl and 1 mM EDTA (Schwacha and Kleckner, 1994). DNA concentration was measured using the Picogreen assay kit (Invitrogen).
DNA physical analysis
For 1D gel analysis, 2 μg of genomic DNA was digested with XhoI enzyme (Enzynomics) for 3 h and precipitated in 100% ethanol. DNA samples were loaded onto 0.6% Seakem LE agarose gel in TBE buffer at ∼2 V/cM for 24 h. Two-dimensional (2D) gel electrophoresis was performed as described elsewhere (Kim et al., 2010; Schwacha and Kleckner, 1994). Genomic DNA was digested as described above, and loaded onto 0.4% Seakem Gold agarose gel without ethidium bromide in TBE buffer at ∼1 V/cM for 21 h. Gels were stained with 0.5 μg/ml ethidium bromide for 30 min. The lanes containing the DNA of interest were cut out, and the gel slices were placed onto 2D gel trays. SeaKem LE agarose (0.8%) with 0.5 μg/ml ethidium bromide was poured around the gel trays at 4°C. Gel electrophoresis was carried out at ∼6 V/cM for 6 h at 4°C. For crossover and non-crossover gel analysis, genomic DNA (2 μg) was digested with XhoI and NgoMIV, and then analyzed on 1D gel electrophoresis as shown in above. After transferring the gel onto membrane using 0.6% NaOH solution, Southern blot analysis was performed using 32P-dCTP-labeled radioactive nucleotides reacted with Random primer labeling mixture (Agilent technologies). Hybridizing signals were acquired using a Bio-Rad phosphoimager and quantified with Quantify One software (Bio-Rad).
MMS sensitivity test
Cells were cultured in YPD liquid medium for overnight at 30°C. Saturated YPD-cultures were serial diluted with YPD liquid and spotted on YPD plates containing 0.02% MMS (Sigma).
RESULTS
Experimental system
Meiotic recombination is evaluated by one-dimensional (1D) and two-dimensional (2D) gel electrophoresis of recombination intermediates at the well-characterized hot spot of HIS4LEU2 locus (Fig. 1; Hunter and Kleckner, 2001; Kim et al., 2010; Oh et al., 2007), where DSBs happen in a single hot spot. DSBs occur at two parental “Mom” and “Dad” sites in the HIS4LEU2 locus during meiosis (Fig. 1A). Synchronized meiotic cells were harvested at specific time points from the SPM culture. Genomic DNA was carefully extracted using the guanidine-phenol extraction method (Kim et al., 2010; Koszul et al., 2008). XhoI restriction site polymorphisms can distinguish “Mom” and “Dad” diagnostic fragments from parental and recombinant chromosomes. The DNA from individual time points was separated by 1D gel electrophoresis to analyze DSBs and crossovers. In wild-type (WT) meiosis, a DSB interacts with a partner and, after a regulated differentiation, yields either interhomolog crossovers (IH-COs) or a non-exchange interhomolog noncrossovers (IH-NCOs) (Fig. 1A; Börner et al., 2004; Kim et al., 2010). IH-COs and IH-NCOs are detected at 4.6 and 4.3 kb, respectively, after double-digestion with XhoI and NgoMIV restriction enzymes (Figs. 1A and 1C). At the HIS4LEU2 locus (Fig. 1A), IH and IS joint molecules have distinguishable molecular weights and shapes (Fig. 1B) and thus can be differentiated at both the SEI and dHJ stages based on their mobility in 2D gels (Fig. 1B; Hong et al., 2013; Hunter and Kleckner, 2001; Kim et al., 2010). These species are normally detected by proper psoralen-UV cross-linking procedures (Fig. 1B). Cell samples from each time point were treated with psoralen and then exposed to UV light to produce cross-linked DNA interstrands, which are joint molecules (i.e., SEIs and dHJs) (Fig. 1B; Supplementary Fig. 1).
Fig. 1.
Experimental assay system. (A) Physical map of HIS4LEU2 hot spot locus. Parental homologs are distinguished by XhoI restriction site polymorphism. DSBs occur at site HIS4LEU2 during meiosis. For diagnostic analysis of IH-COs and IH-NCOs, DNA digested with both XhoI and NgoMIV restriction enzymes was analyzed on the gel electrophoresis. Probe A was used to detect recombination intermediates by Southern hybridization analysis. DSB, double-strand break; IH-CO, interhomolog crossover; IH-NCO, interhomolog non-crossover. (B) Joint molecules detected at HIS4LEU2 hot spot by two-dimensional gel electrophoresis analysis. (C) One-dimensional gel analysis displaying IH-COs and IH-NCOs.
Absence of Shu1 delays meiotic recombination and reduces crossover recombinants
Psy3, Csm2, Shu1, and Shu2 (hereafter referred to as the “PCSS complex”) were originally identified from a genetic screen as mutational suppressors of the top3Δ or sgs1Δ growth defect in Saccharomyces cerevisiae (Mankouri et al., 2007; Shor et al., 2005). The proteins of the PCSS complex physically interact and fall in the same epistasis group (Ito et al., 2001; Shor et al., 2005). Furthermore, these single mutants show similar levels of sensitivity to methylmethane sulfonate (MMS) (Mankouri et al., 2007), implying that the PCSS complex components play together in vivo in the same pathway. To observe the conditional effect of Shu1, we constructed a copper-specific conditional expression allele by replacing the Shu1 promoter with the CUP1 promoter, which is repressed in the absence of copper (Fig. 2; Supplementary Fig. 2). Furthermore, to inactivate Mek1 meiosis-specific serine/threonine kinase, we used mek1as allele that could be inactivated by adding 1-NA-PP1 inhibitor in synchronized SPM cultures (Kim et al., 2010). DSB and crossover products can also be assayed and quantified at each time point using 1D gel analysis (Figs. 2A–2C). Meiosis-specific DSBs were observed in pCUP1-SHU1 mek1as strains, and then compared during all the recombination processes (Figs. 2A and 2B). In the absence of Shu1, DSBs appeared at WT timing, which was delayed by ∼2 h compared to normal turnover timing, and eventually disappeared. In contrast, when copper is added to pCUP1-SHU1 mek1as(-IN) after 2 h, when DNA replication is completed and DSBs have formed but no later species are present, the timing and levels of DSBs, COs, and divisions correspond to those of the WT/mek1as(-IN) strain (Kim et al., 2010). The absence of Shu1 protein reduced the absolute CO levels to ∼13% and delayed the CO formation (Fig. 2C). Next, we investigated the meiotic division using DAPI staining. We observed that about 50% of the cells reached the MI stage at 7 h after transfer to the sporulation medium at 30°C in a culture incubator, and they rapidly finished the nuclear division after 8 h in mek1as(-IN) (Fig. 2D). In contrast, the division of pCUP1-SHU1(-Cu) was delayed by ∼2 h compared to that of pCUP1-SHU1(+Cu), and reached ∼90% of the pCUP1-SHU1(+Cu) division level (Fig. 2D). In general, cells that have defective complete meiotic division also have a defective DNA recombination; thus, Shu1 is required specifically for efficient homologous recombination during meiosis. As reported previously, Mek1 kinase plays a critical role in regulating the timing and kinetics of meiotic recombination. Inactivation of Mek1 kinase activity exhibits dramatically shorter SEI and dHJ life span (Kim et al., 2010). The addition of 1-NA-PP1 (inactivation of Mek1 kinase; +IN) in the absence or presence of copper alters the progression of recombination to WT level in terms of timing and kinetics of recombination, but reduces the CO levels (Figs. 2A and 2C). In the absence of Mek1 kinase activity, shortened DSB turnover and reduced absolute CO levels (3%) are observed. Thus, our finding suggests that Mek1 kinase activity mediates all cellular processes of meiotic recombination in the absence or presence of Shu1.
Fig. 2.
Meiotic recombination in pCUP1-SHU1 mek1as strain. (A) One-dimensional gel analysis of pCUP1-SHU1 mek1as cells in various conditions. Copper and/or inhibitor were added to aliquots of the same pre-meiotic culture, and recombination intermediates were quantified at the HIS4LEU2 construct and analyzed as described elsewhere (Kim et al., 2010). -IN, absence of 1-NA-PP1; +IN, presence of 1-NA-PP1, Mom, mom species; Dad, dad species; COs, crossover species; DSBs, double-strand breaks; JMs, joint molecules. (B, C) Quantitative analysis of DSBs and crossover products during the same time points shown in (A). Percentage of total DNA in DSBs and COs is plotted as a function of time. (D) MI + MII represent meiotic division progression as estimated by the number of DAPI foci. (E) Representative two-dimensional gels of SEIs and dHJs. Corresponding images of the entire gels are shown in Supplementary Fig. 3. (F, G) Quantitative analysis of joint molecules in pCUP1-SHU1 cells with or without 1-NA-PP1 and copper. DNA event represents DNA species as percent of the total hybridizing signal at specific time points after the transfer to sporulation medium. SEI, single end invasion; IH-dHJ, interhomolog double Holliday Junction; IS-dHJ, intersister double Holliday Junction. (H) Levels of IH-COs and IH-NCOs in the pCUP1-SHU1 mek1as strain. COs and NCOs were assayed with a HIS4LEU2 tester construct containing BamHI (“Mom”) and NgoMIV (“Dad”) sites. IH-COs and IH-NCOs are shown for each time point sample as percentages of the total DNA.
Shu1 is required for the establishment of homolog bias
Joint molecules are strongly visible in 2D gels (Hong et al., 2013; Hunter and Kleckner, 2001; Kim et al., 2010). The expression of Shu1 protein exhibits prominent IH-SEIs and IH-dHJs, indicating normal kinetics of recombination. Meiotic division is about 95% after 24 h (Fig. 2). However, pCUP1-SHU1(-Cu) mek1as(-IN) mutants form 2 prominent arc signals that are similar to downward DSB hyper-resection, while many of the IH-SEI signals overlap the SEI region (Fig. 2E; Supplementary Fig. 3). These unique arc shapes correspond to “Mom” and “Dad” IS-SEIs, as shown in Red1-/Mek1kinase- (Kim et al., 2010). The maximum levels of SEIs are similar to those of pCUP1-SHU1(+Cu) mek1as(-IN), but the observed turnover delays of ∼2 h is similar to that of COs and meiotic division. IS-dHJs are more numerous than IH-dHJs, and two dHJ signals appear and disappear coordinatively with the same kinetics as SEIs (Figs. 2F and 2G). The most striking feature is that the joint molecules of pCUP1-SHU1(-Cu) mek1as(+IN) are highly reduced compared to those of pCUP1-SHU1(-Cu) mek1as(+IN), although the levels of DSB are the same in both pCUP1-SHU1(+Cu) mek1as(+IN) and pCUP1-SHU1(+Cu) mek1as(+IN) (Figs. 2B, 2F, and 2G); SEIs and dHJs levels are ∼50% lower than those in pCUP1-SHU1 (-Cu) mek1as(+IN) (Figs. 2F and 2G). The pCUP1-SHU1(-Cu) mek1as(-IN) strain has increased IS-dHJ levels. Further, Shu1 is also required for efficient DSB repair through sister chromatids in mek1as(+IN). In the absence of Mek1 kinase activity, elimination of Rad51 abolishes SEIs and dHJs (Hong et al., 2013), and in this study, elimination of Shu1 reduces SEIs and dHJs (Hong et al., 2013; Figs. 2E–2G) while DSBs are efficiently repaired to intersister-COs or interhomo-log-NCOs. These results imply that Shu1 affects the efficiency of homologous recombination in both homolog and sister interaction under different circumstances. In the absence of Shu1, the homolog bias is not properly established in the “partner choice” stage, and the meiotic recombination process is delayed. From these results, we conclude that pCUP1-SHU1(-Cu) strains convert many of the DSBs to IS-SEIs and generate intersister crossover, while IH-dHJs can be still changed to interhomolog crossover.
The absence of Shu1 reduces COs and NCOs
The formation of IH-COs versus IH-NCOs was also monitored over time by 1D gel analysis that allows the detection of products diagnostic of the two species (Fig. 1C; Storlazzi et al., 1995). Here, we analyzed IH-COs and IH-NCOs at HIS4LEU2 in 1D gel. When Shu1 proteins are expressed after 2.5 h, IH-CO and IH-NCO levels are about 4% and 2.7%, respectively, which are similar to the corresponding values in WT (Fig. 2H; Kim et al., 2010). However, in the absence of Shu1, both IH-COs and IH-NCOs occur at ∼50% lower levels than in WT because most of IS-dHJs are resolved to IS-COs. Thus, we conclude that Shu1 is required for establishing homolog bias, and it also acts in both IH-COs and IH-NCOs pathways. After the addition of the inhibitor in the absence or presence of copper, IH-CO and IH-NCO formation are severely reduced, and thus could reflect a direct effect on the early step of recombination prior to CO decision (Fig. 2H).
Shu1 promotes interhomolog recombination independent of Srs2
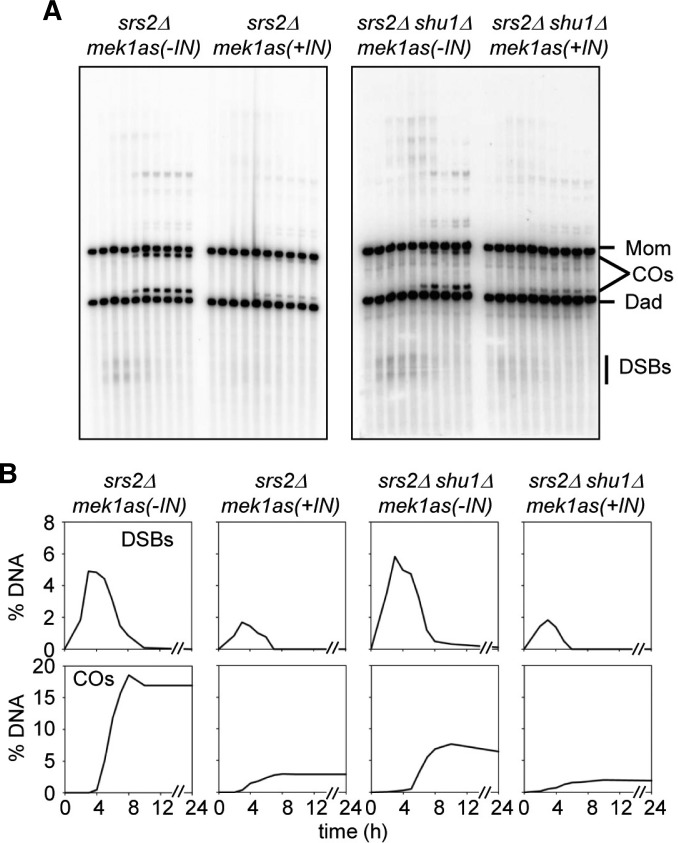
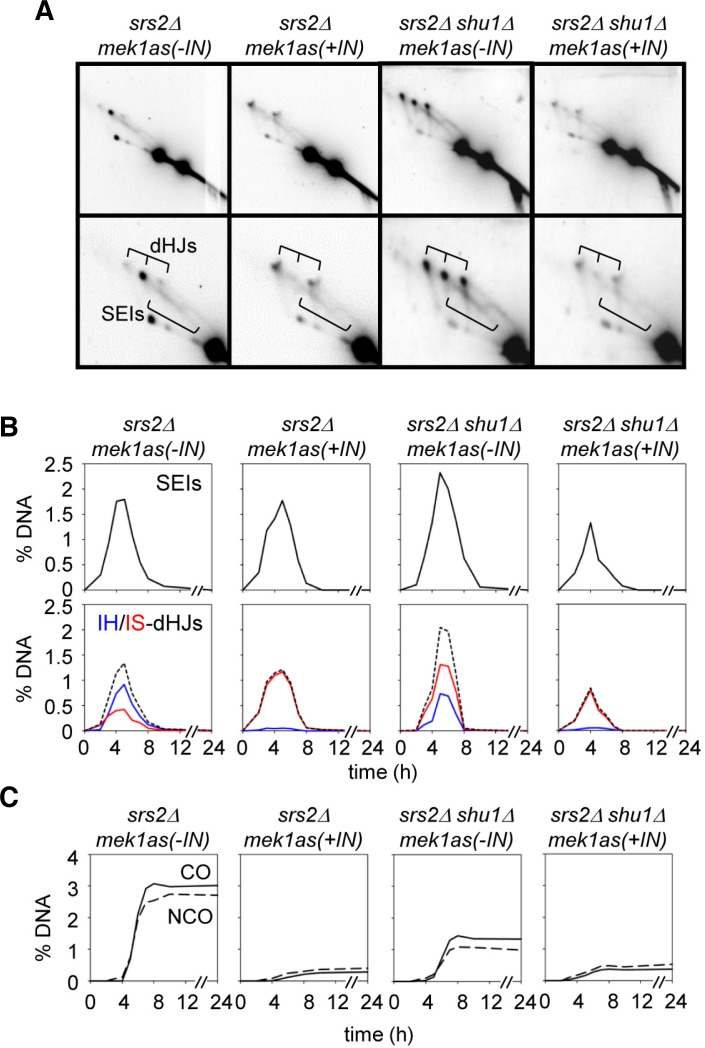
Srs2, a nucleic acid helicase belonging to the SF1 family, is a 3′-5′ DNA helicase and prevents homologous recombination by disrupting Rad51 nucleofilaments (Aboussekhra et al., 1992; Rong and Klein, 1993). The PCSS complex has distinct roles in meiotic recombination, promoting homolog bias and strand exchange in meiosis and mitosis, respectively (above; Hong et al., 2013; Ito et al., 2001; Sasanuma et al., 2013a; Shor et al., 2005). In mitosis, Shu1 inhibits Srs2 by removing Rad51-ssDNA filaments, and functions as an accessory factor of Rad51 (Bernstein et al., 2011; Tao et al., 2012). The deletion of the Srs2 gene slightly reduces Rad51 foci and exhibits reduced spore viability (Sasanuma et al., 2013b). The absence of Srs2 caused ∼5% DSBs at 4 h and normally processed ∼18% of crossover products (Fig. 3B). However, a srs2Δshu1Δ double mutant exhibits ∼2-h delay in DSB turnover and 7% of CO level, which is similar to that of shu1Δ single mutation. This phenotype is similar to that of Shu1- cells, implying that Srs2 does not influence homologous recombination during meiosis in the absence of Shu1. In mek1as(+IN), both srs2Δ and srs2Δshu1Δ exhibit fast DSB turnover, and the relative CO levels are reduced to 3% (Fig. 3B). Next, we analyzed joint molecule formation in srs2Δ and srs2Δshu1Δ mutant cells (Figs. 4A and 4B). In srs2Δ single deletion cells, interhomolog dHJs are the prominent species (0.91%), while IS-dHJs are about 0.42%. The IH:IS dHJ ratio is about 2:1, which is slightly reduced in homolog bias (Fig. 4B). Furthermore, SEIs/dHJs appear with normal timing in srs2Δ, peak at 5 h, and persist for ∼1 h longer than in WT. Interestingly, a srs2Δshu1Δ double mutant exhibits the same defect of shu1Δ in homolog bias that the IH:IS dHJ ratio is about 1:2.5 (Fig. 4B and Supplementary Fig. 4). As shown for Red1-/Mek1-, homolog bias is completely altered to sister bias even in srs2Δmek1as(+IN) cells (Figs. 4A and 4B; Hong et al., 2013; Kim et al., 2010). In both srs2Δ mek1as(+IN) and srs2Δshu1Δ mek1as(+IN), intersister dHJs are prominent and the ratio of IH-dHJ versus IS-dHJ is about 1:9, with JMs developed in normal timing (Fig. 4B). This finding implicates that the role of Shu1 is essential in the establishment of homolog bias and the Srs2 helicase is not required for partner choice in the absence of Shu1.
Fig. 3.
DSB and CO formation in srs2Δ and srs2Δ shu1Δ mek1as strains. (A) One-dimensional analysis of srs2Δmek1as and srs2Δ shu1Δ mek1as strains. (B) Quantitative analysis of DSBs and COs using one-dimensional gels.
Fig. 4.
Joint molecule formation of srs2Δ and srs2Δ shu1Δ mek1as strains. (A) Representative two-dimensional gel analysis of srs2Δ mek1as and srs2Δ shu1Δ mek1as strains. (B) Quantitative analysis of SEIs and dHJs using two-dimensional gels. (C) Levels of IH-COs and IH-NCOs in srs2Δ and srs2Δ shu1Δ in the absence or presence of Mek1 kinase activity.
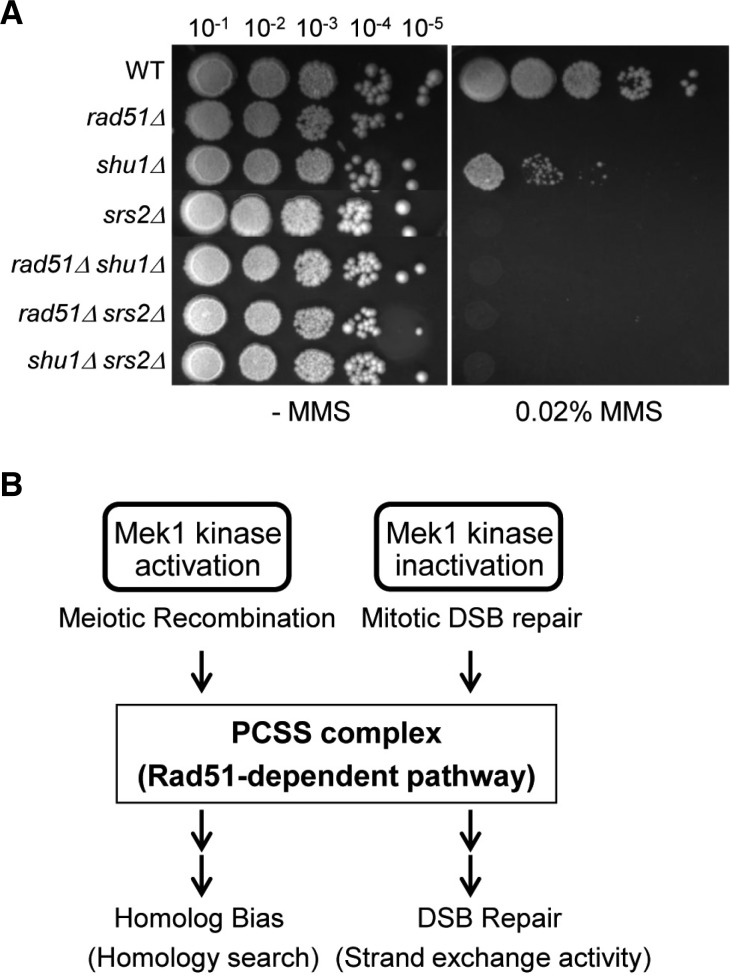
In Srs2 deletion mutants, both CO and NCO levels are slightly reduced by 3% and 2.6%, respectively (Fig. 4C). This result indicates that srs2Δshu1Δ double mutant exhibits the same defect of a Shu1- mutant in CO and NCO formation. Moreover, in mitosis, Shu1 involves in the same pathway as Srs2 (Bernstein et al., 2011). The srs2Δshu1Δ double mutant show similar MMS sensitivity to srs2Δ, indicating that both Srs2 and Shu1 exhibits an epistatic relationship in mitotic DSB repair (Fig. 5A). Taken together, these experiments also demonstrate that Shu1 influences the homologous recombination upstream of the Srs2 pathway.
Fig. 5.
Genetic relationship of Shu1, Srs2, and Rad51. (A) The specified were cultured overnight at equal concentration and spotted onto YPD or YPD-plus 0.02% MMS. The plates were then incubated at 30°C for 2 days. (B) Role of the PCSS complex for homolog bias and strand exchange in Rad51-dependent recombination.
DISCUSSION
The present work elucidates the interplay between recombination mediators and chromosome axis components during the stage at which a DSB decides to select a homolog. Additional information also emerges regarding the roles of Shu1 at post-DSB stage and the roles of Shu1 and Mek1 in promoting strand exchange, but at different regulation sites. Chromosome axis components, Red1/Mek1/Hop1, determine partner selection in a program-appropriate way (Kim et al., 2010). In the absence of Mek1 kinase activity, cohesin channels the reaction into the sister, as appropriate for mitotic DSB repair (Kim et al., 2010). Therefore, the sister is automatically used preferentially as a partner because of its spatial proximity; unique meiosis-specific features promote the homolog bias. The present findings suggest that the basic role of Shu1 (or the PCSS complex) for homolog bias is to support Rad51-dependent homologous recombination (Fig. 5B). The effects described above come into play during the establishment of homolog bias. Additional features ensure that homolog bias is maintained during the IH-SEI to IH-dHJ transition of IH-CO formation (Kim et al., 2010). Previously, it has been reported that Rad51 functions to establish homolog bias (Schwacha and Kleckner, 1997). We further reported that Rad51 is assisted by its known mediators, not only Rad55 and 57, but also by the PCSS complex, which comprises Psy3, Csm2, Shu1, and Shu2 (Hong et al., 2013; this study). Since the hierarchy of dependencies on these factors for homolog bias is the same as that seen for these same molecules in the execution of Rad51-mediated strand exchange for DSB repair, these findings imply that the entire Rad51 ensemble has been recruited to this new meiotic role. The homology search process and the ensuing strand exchange involve the loading of Rad51 onto a single-stranded DNA in one binding site followed by interactions with an intact duplex partner DNA in the second site (Hong et al., 2013).
At the time of initiation, Spo11-mediated DSBs occur in intimate association with this complex (Panizza et al., 2011). At stages of CO/NCO differentiation, CO recombination interactions exhibit preferential association with domains in which Mek1 is hyperabundant (Joshi et al., 2009). The present work further implicates that Mek1 has a comprehensive role during the partner selection stage. In the absence of Mek1 kinase activity, all the features of meiotic recombination at this stage are absent, initiating a process that corresponds closely to mitotic DSB repair using sister chromatids (Hong et al., 2013).
Yeast cells enter meiosis in response to nutrient deprivation. However, if nutrients reappear in the environment prior to the end of prophase, the meiotic program is aborted and cells return smoothly to the mitotic cell cycle, carrying out a regular mitotic division in which intact sister chromatids segregate regularly to opposite poles. Recently, in vivo studies have revealed a strong dependence of Dmc1 activity on Rad51 (Cloud et al., 2012; Hong et al., 2013). A DSB preferentially selects a homolog partner in both Rad51- and Dmc1-promoted recombination, in the absence or presence of the meiotic axis components Red1/Mek1/Hop1. This finding suggests that interhomolog bias is an intrinsic feature of the recombination mechanism per se. Logically, it is easier to think that homolog bias arises because the use of the sister is precluded, rather than having a DSB that specifically identifies unique features of the homolog.
Creation of a DSB-mediated link between the DSB donor chromosome and a recipient partner chromosome likely involves the release of only one DSB end from the initiation complex. This “leading” end searches for, and establishes, a stable interaction with a homolog partner duplex. The other “lagging” end is not brought into the reaction until at a much later stage (Kim et al., 2010). The present results provide a general logic for the interplay between chromosome structure and recombination biochemistry for recombination partner choice in meiosis. However, future studies are needed to properly elucidate the basic nature of partner selection and the ensuing DSB/homolog partner interactions not only in vivo, but also in vitro.
Supplementary Material
Acknowledgments
We would like to thank all the members of the Kim’s Laboratory for their comments. This research was supported by the National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning (2012R1A1A1010578; 2012 M3A9C6050367).
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semi-dominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball LG, Zhang K, Cobb JA, Boone C, Xiao W. The yeast Shu complex couples error-free post-replication repair to homologous recombination. Mol Microbiol. 2009;73:89–102. doi: 10.1111/j.1365-2958.2009.06748.x. [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Reid RJ, Sunjevaric I, Demuth K, Burgess RC, Rothstein R. The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase. Mol. Biol. Cell. 2011;22:1599–1607. doi: 10.1091/mbc.E10-08-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, Hunter N. Crossover/non-crossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- Cloud V, Chan YL, Grubb J, Budke B, Bishop DK. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science. 2012;337:1222–1225. doi: 10.1126/science.1219379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Sung Y, Yu M, Lee M, Kleckner N, Kim KP. The logic and mechanism of homologous recombination partner choice. Mol. Cell. 2013;51:440–453. doi: 10.1016/j.molcel.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Joshi N, Barot A, Jamison C, Börner GV. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 2009;5:e1000557. doi: 10.1371/journal.pgen.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J, Kleckner N. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell. 2010;143:924–937. doi: 10.1016/j.cell.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008;133:1188–1201. doi: 10.1016/j.cell.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer WD. Rad51 paralogues Rad55-Rad57 balance the anti-recombinase Srs2 in Rad51 filament formation. Nautre. 2011;479:245–248. doi: 10.1038/nature10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Ngo HP, Hickson ID. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol. Biol. Cell. 2007;18:4062–4073. doi: 10.1091/mbc.E07-05-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V, Chahwan C, Gao H, Blais V, Wohlschlegel J, Yates JR, 3rd, McGowan CH, Russell P. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 2006;25:2564–2574. doi: 10.1038/sj.emboj.7601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR, Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011;146:372–283. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Qing Y, Yamazoe M, Hirota K, Dejsuphong D, Sakai W, Yamamoto KN, Bishop DK, Wu X, Takeda S. The epistatic relationship between BRCA2 and the other RAD51 mediators in homologous recombination. PLoS Genet. 2011;7:e1002148. doi: 10.1371/journal.pgen.1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong L, Klein HL. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J Biol Chem. 1993;268:1252–1259. [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Sasanuma H, Tawaramoto MS, Lao JP, Hosaka H, Sanda E, Suzuki M, Yamashita E, Hunter N, Shinohara M, Nakagawa A, et al. A new protein complex promoting the assembly of Rad51 filaments. Nat Commun. 2013a;4:1676. doi: 10.1038/ncomms2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H, Furihata Y, Shinohara M, Shinohara A. Remodeling of the Rad51 DNA strand-exchange protein by the Srs2 helicase. Genetics. 2013b;194:859–872. doi: 10.1534/genetics.113.150615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Shor E, Weinstein J, Rothstein R. A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair. Genetics. 2005;169:1275–1289. doi: 10.1534/genetics.104.036764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Xu L, Cao L, Kleckner N. Crossover and noncrossover recombination during meiosis: timing and pathway relationships. Proc. Natl. Acad. Sci. USA. 1995;92:8512–8516. doi: 10.1073/pnas.92.18.8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Li X, Liu Y, Ruan J, Qi S, Niu L, Teng M. Structural analysis of Shu proteins reveals a DNA binding role essential for resisting damage. J Biol Chem. 2012;287:20231–20239. doi: 10.1074/jbc.M111.334698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.