Abstract
Caenorhabditis elegans, a cholesterol auxotroph, showed defects in larval development upon cholesterol starvation (CS) in a previous study. To identify cholesterol-responsive proteins likely responsible for the larval arrest upon CS, a comparative proteomic analysis was performed between C. elegans grown in normal medium supplemented with cholesterol (CN) and those grown in medium not supplemented with cholesterol (cholesterol starvation, CS). Our analysis revealed significant change (more than 2.2-fold, p < 0.05) in nine proteins upon CS. Six proteins were down-regulated [CE01270 (EEF-1A.1), CE08852 (SAMS-1), CE11068 (PMT-2), CE09015 (ACDH-1), CE12564 (R07H5.8), and CE09655 (RLA-0)], and three proteins were up-regulated [CE29645 (LEC-1), CE16576 (LEC-5), and CE01431 (NEX-1)]. RNAi phenotypes of two of the down-regulated genes, R07H5.8 (adenosine kinase) and rla-0 (ribosomal protein), in CN were similar to that of larval arrest in CS, and RNAi of a down-regulated gene, R07H5.8, in CS further enhanced the effects of CS, suggesting that down-regulation of these genes is likely responsible for the larval arrest in CS. All three up-regulated genes contain putative DAF-16 binding sites and mRNA levels of these three genes were all decreased in daf-16 mutants in CN, suggesting that DAF-16 activates expression of these genes.
Keywords: C. elegans development, cholesterol, proteomic analysis
INTRODUCTION
Cholesterol is required for many different physiological processses in mammals. Examples are as a component of cell membrane, as a precursor of hormones, and as a signaling molecule (Reviewed by Farese and Herz, 1998). Failure of cholesterol biosynthesis can cause severe developmental defects with teratogenic effects (Clayton, 1998). Caenorhabditis elegans is a cholesterol auxotroph, which requires exogenously supplied cholesterol for normal growth (Lozano et al., 1984). Cholesterol deprivation or starvation (CS) inhibits growth (Gerisch et al., 2001; Jeong et al., 2010). The effects accumulate with succeeding generations, with CS-mediated developmental defects worsening in the progeny until finally they are arrested at the larval stage (Jeong et al., 2010).
Larval arrest can be induced under various physiological conditions in C. elegans. To understand the molecular mechanism by which larval arrest is induced in the CS condition, cholesterol-responsive proteins, the levels of which are significantly changed in CS condition compared to normal medium condition (CN), were screened through a comparative proteomic analysis. Proteomic analysis with C. elegans proteome was well established to identify specific target proteins responding to different environmental cues (Jeong et al., 2009; Paik et al., 2006; Shim and Paik, 2010). Proteomes of worms grown in CN and in CS medium were compared in this study. Six metabolic proteins involved in methylation, phosphorylation, and protein synthesis were significantly down-regulated upon CS. Down-regulation of at least two of these proteins appeared to be involved in the larval arrest upon CS because RNA interference (RNAi) treatment of corresponding genes, R07H5.8 and rla-0, caused similar defects in CN.
MATERIALS AND METHODS
Nematode strains and general methods
Methods for maintenance and handling of C. elegans were as previously described (Brenner, 1974). Strain N2 was used as wild type for all analyses and the CF1038: daf-16(mu86) I mutant strain was also used. Strains were maintained at 20°C on Nematode Growth Medium (NGM) agar plates containing Escherichia coli strain OP50 supplemented with 5 μg/ml of cholesterol (CN, cholesterol normal feeding) or without cholesterol (CS, cholesterol starvation). Nematode strains were provided by the Caenorhabditis Genetics Center (University of Minnesota).
Sample preparation for two-dimensional (2D) gel electrophoresis analysis
Worms grown on seeded plates were harvested and treated with sodium hypochlorite to obtain large quantities of embryos. L1 stage worms were prepared by incubating the embryos in liquid medium without food, followed by growth at 20°C on 100 mm-diameter CN or CS plates seeded with a lawn of Escherichia coli OP50 until they reached young adult stage. Worm preparation was repeated to get the second generation (F2 generation). After growth in CN and CS for 48 h and 72 h, respectively, worms were harvested by washing the plates with M9 buffer and bacterial contamination was eliminated through centrifugation in 70% sucrose solution. Protein extraction and quantification were performed as previously described (Ahn et al., 2006).
2D gel electrophoresis and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) analysis
After quantification, 1 mg of whole worm protein extract was applied to isoelectric focusing that was carried out at a total of 114100 Vh. Each IPG strip was equilibrated as previously described (Ahn et al., 2006). In the second dimension of electrophoresis, vertical sodium dodecyl sulfate gradient (7–13%) slab gel was used. Electrophoresis was conducted at a constant 67 mA/gel. The gel image was obtained using an ImageScanner (Amersham, USA), and analyzed with Melanie-4 (GeneBio, Switzerland). Three pairs of gels were analyzed and variance analysis of spot volume and comparisons of mean values of samples were performed using SAS statistical software, with a significance level of p < 0.05. More than 1.5-fold differentially expressed proteins were analyzed by MALDI-TOF MS as previously described (Kawasaki et al., 2011). The results from MS were further analyzed using MASCOT (http://www.matrixscience.com). Proteins with a score exceeding more than 67 and expression levels more than 2.2-fold difference were selected and further analyzed. Protein functions were cited from the wormbase (http://wormbase.org), the ExPASy database (http://www.expasy.org), and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
RNAi
Feeding RNAi was performed as previously described (Jeong et al., 2010). NGM agar plates with cholesterol (CN) and without cholesterol (CS) for RNAi were prepared by adding 0.2% lactose and 100 μg/ml ampicillin. Each bacterial clone for RNAi was precultured in LB medium containing 100 μg/ml ampicillin for 12–15 h at 37°C prior to seeding to the RNAi plates. The plates were incubated for 2–3 days at room temperature to induce double-stranded RNA (dsRNA) of target genes. Synchronized L1 worms of the test strain were grown on the CN or CS RNAi plates at 20°C until adulthood (first generation), and this process was repeated with their progeny (second generation), and the phenotypes in the first and the second generations in CN and CS plates were observed.
Analysis of embryonic lethality and larval development
N2 worms synchronized at the L1 stage were grown on NGM plates in the CN or CS condition. When they reached the L4 stage, hermaphrodites were individually cloned onto NGM plates with or without cholesterol at 20°C. The hermaphrodites were transferred to fresh plates at 16–24 h intervals. Laid embryos were judged as being dead if they did not hatch after 24 h at 20°C. Embryonic lethality was defined as the number of non-hatched embryos divided by the sum of non-hatched and hatched embryos. Percent larval development was defined as the fraction of hatched embryos that reached adulthood.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Wild-type N2 worms were grown at 20°C in NGM agar plates containing E. coli (OP50) with 5 μg/ml cholesterol (CN) or without cholesterol (CS). Synchronized young adult worms were collected into 300 μl of Trizol (Invitrogen, USA) and freeze-thawed twice in liquid nitrogen. Total RNA was isolated using PLG (Phase Lock Gel, Qiagen) with 1 μl LPA (Linear Polyacrylamide, Ambion Inc.). Reverse transcription and qPCR reaction were performed as previously described (Jeong et al., 2010). Primers for act-1, which was used as an internal control, were: forward primer, 5′-CCA GGA ATT GCT GAT CGT ATG CAG AA-3′; reverse primer, 5′-TGG AGA GGG AAG CGA GGA TAG A-3′ (GenBank accession no. NM_073418). Primers for lec-1 were: forward, 5′- CCA AAG TCC TAC CCA GTA CC-3′; reverse, 5′- CCA TTC GAG AAC GAG TTG AG-3′ (GenBank accession no. Z82081.1). Primers for lec-5 were: forward, 5′- GAA CAA GAA TAC AAG CTC CCA-3′; reverse, 5′- TCA TCT TCA GAC CAG TTT CC-3′ (GenBank accession no. U29244.3). Primers for nex-1 were: forward, 5′- CAT CGC TGG AGA TAC TTC TG-3′; reverse, 5′- GGT GAG GTA GGA TTT CTG GAG-3′ (GenBank accession no. U00064.1). The normalized RNA level of each gene was defined as the mRNA level of each gene averaged from triplicate measurements of two independent experiments and normalized with that of act-1, the internal control. PCR was performed using denaturation at 95°C for 10 min followed by 40 cycles consisting of 15 s at 95°C and 1 min at 60°C.
RESULTS
Proteomic analysis of proteins extracted from worms grown in CS
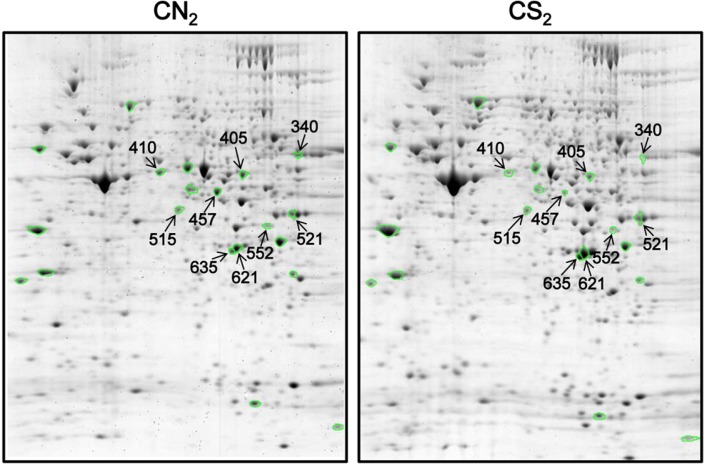
Worms grown in CS showed defects in developmental process-ses (Jeong et al., 2010). Especially, their larval development was affected and these defects became more profound when worms were grown in CS for consecutive generations. We performed a comparative proteomic analysis using total protein extracts from worms in the second generation grown in either CN (CN2) or CS (CS2). A total of 606 protein spots were matched in three pairs of gels and proteins altered in their expression levels more than 1.5-fold up or down in CS compared to CN with a significance level of p < 0.05 were further identified by MS. These 27 proteins are summarized in supplementary Table 1 and each protein spot on the gel is marked with a circle in Fig. 1. Among the 27 proteins, nine protein spots, which were more than 2.2-fold up- or down-regulated in CS compared to CN with a MASCOT score exceeding more than 67, were selected for further analyses (Fig. 1). These nine proteins were annotated to be involved in either metabolism or protein synthesis (Table 1). Identified metabolic proteins down-regulated in CS included CE08852 (SAMS-1, S-adenosylmethionine synthetase), CE11068 (PMT-2, SAM-dependent methyltransferase), CE09015 (ACDH-1, Short-chain acyl-CoA dehydrogenase), and CE12564 (R07H5.8, adenosine kinase). Proteins functioning in protein synthesis that were also down-regulated in CS were CE01270 (EEF-1A.1, eukaryotic translation elongation factor EF-1-alpha) and CE09655 (RLA-0, ribosomal protein, large subunit). CE29645 (LEC-5, galectin), CE16576 (LEC-1, galectin), and CE01431 (NEX-1, annexin) were up-regulated in CS.
Fig. 1.
2D gel images of proteins extracted from worms grown for two generations with (CN2) or without (CS2) cholesterol. Nine protein spots that were more than 2.2-fold up- or down-regulated in CS2 compared to CN2 are marked with arrows and ID numbers in the gel images.
Table 1.
Identified proteins that showed differential expression levels in worms grown in CS
| Spot ID | Gene name | Protein identified | Accession no. | Theoretical mass (kDa)/pI | Match % | Scorea |
|---|---|---|---|---|---|---|
| Metabolism | ||||||
| 405↓ | sams-1 | S-adenosylmethionine synthetase | CE08852 | 43.8/6.02 | 34 | 101 |
| 410↓ | pmt-2 | SAM-dependent methyltransferase | CE11068 | 49.9/5.61 | 32 | 152 |
| 457↓ | acdh-1 | Short-chain acyl-CoA dehydrogenase | CE09015 | 47.6/6.42 | 45 | 166 |
| 515↓ | R07H5.8 | Adenosine kinase | CE12564 | 37.9/5.67 | 69 | 229 |
| Protein synthesis | ||||||
| 340↓ | eef-1A.1 | Eukaryotic translation elongation factor EF-1-alpha | CE01270 | 51.1/9.07 | 26 | 71 |
| 552↓ | rla-0 | Ribosomal protein, large subunit | CE09655 | 33.9/6.27 | 40 | 96 |
| Other | ||||||
| 521↑ | lec-5 | Galectin | CE29645 | 35.5/7.74 | 34 | 103 |
| 621↑ | lec-1 | Galectin | CE16576 | 31.8/6.12 | 77 | 253 |
| 635↑ | nex-1 | Annexin | CE01431 | 35.7/6.13 | 62 | 203 |
Proteins that showed more than 2.2-fold different expression levels with p-value < 0.05 were selected. Up-regulated proteins in worms grown in CS medium are marked with ↑, and down-regulated ones are with ↓.
MASCOT was used as a decision database.
Down-regulation of two of the genes may be responsible for the larval developmental defect in CS
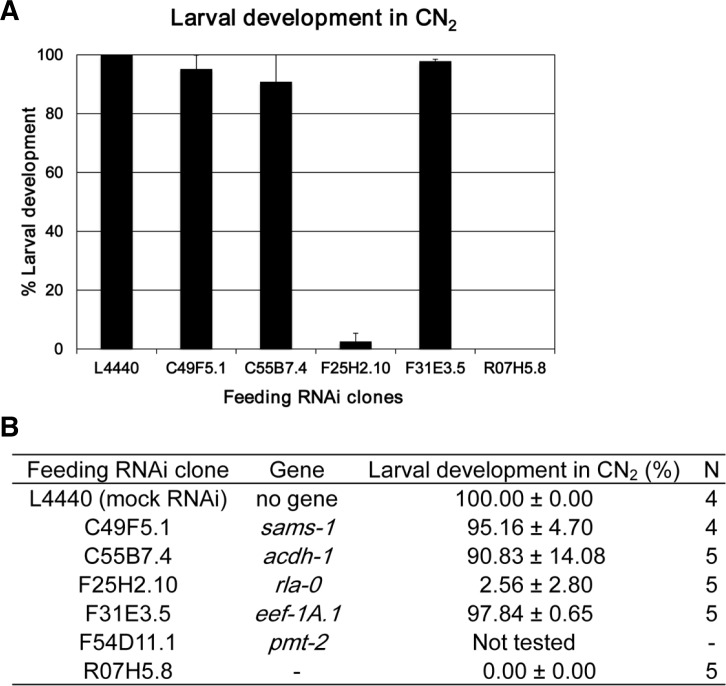
To examine whether any of the down-regulated proteins were responsible for the defects observed in worms grown in CS, RNAi phenotypes of five of the six genes, which encode CS down-regulated proteins, were analyzed in CN condition for two generations. In the first generation (CN1), none of these RNAi-treated worms showed significant levels of larval arrest (data not shown). However, in the second generation (CN2), RNAi of two of the genes, F25H2.10 (rla-0) and R07H5.8, caused significant levels of larval developmental arrest (Fig. 2). Because the RNAi phenotypes of these two genes exactly mimicked the CS-induced larval arrest phenotype, that is, less significant defect in the first generation but more severe defect in the second generation, we assume that down-regulation of one of the two genes could be responsible for the larval developmental defect observed in worms grown in CS. As we could not obtain an RNAi clone of pmt-2, we could not evaluate whether pmt-2 is also involved in the larval arrest upon CS or not.
Fig. 2.
Percent larval development of worms treated with RNAi against CS down-regulated genes for two generations in CN condition. Synchronized L1 wild-type N2 worms were grown on CN plates seeded with bacterial clones for feeding RNAi against five of the six genes that were down-regulated in CS until the L4 stage (first generation), and these L4 worms were individually transferred to respective fresh feeding RNAi plates for three to four consecutive days until they finish laying self-fertilized eggs. Then, percent larval development of hatched progeny (second generation) after each feeding RNAi was collectively scored and displayed as a bar graph (A) and summarized as a table (B). Error bars in (A) show S.D. N in (B) is the number of L4 mother worms used to measure percent larval development of their progeny after each RNAi. F54D11.1 (pmt-2) was not tested. L4440 was used as a mock feeding RNAi control.
RNAi of a down-regulated gene and CS treatment additively induced larval arrest
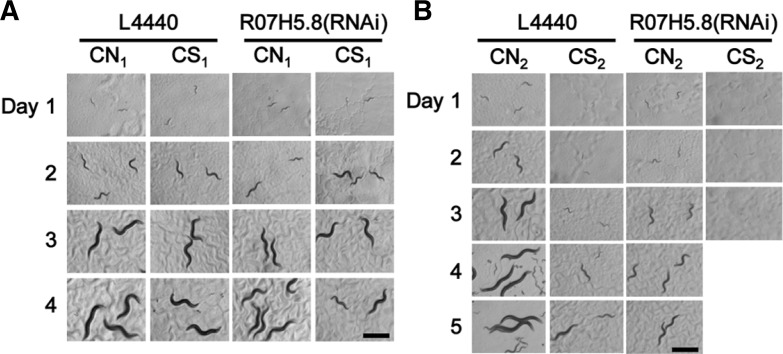
RNAi of one of the two genes, R07H5.8, which caused significant larval arrest in CN2, was also tested in CS condition to see if it might increase sensitivity to CS. Worms were grown in CS with or without RNAi treatment of R07H5.8, an adenosine kinase gene, and resulting first (CN1 and CS1) and second (CN2 and CS2) generations of worms were observed. Worms treated with R07H5.8 RNAi showed growth retardation and larval arrest from the first generation in CS (Fig. 3A), while in CN condition, R07H5.8(RNAi) treated worms showed these defects only in the second generation (Fig. 3B). These results suggest that CS treatment and reduction of R07H5.8 activity additively affect the induction of larval arrest. Although in the CS2 condition, percent larval development was significantly different with (0%, n = 5; n, numbers of mothers) or without (12.3% ± 0.02, n = 5) R07H5.8 (RNAi) treatment, embryonic lethality was not so much different: 2.7% ± 0.01 (n = 5) with R07H5.8 RNAi and 1.9% ± 0.01 (n = 5) without R07H5.8 RNAi, suggesting that R07H5.8 can be a specific target for controlling larval development in CS.
Fig. 3.
Photographs of R07H5.8 RNAi treated worms grown for two generations in CN and CS. Photographs were taken serially for four days in the first generation (A) and for five days in the second generation (B) as indicated on the top and at the left of each panel. The worms in the second generation treated with R07H5.8 RNAi in CS were all dead on day 3, thus their day 4 and day 5 pictures are not shown. L4440 was used as a mock RNAi control. Scale bars, 500 μm.
Galectin and annexin are possible targets of DAF-16
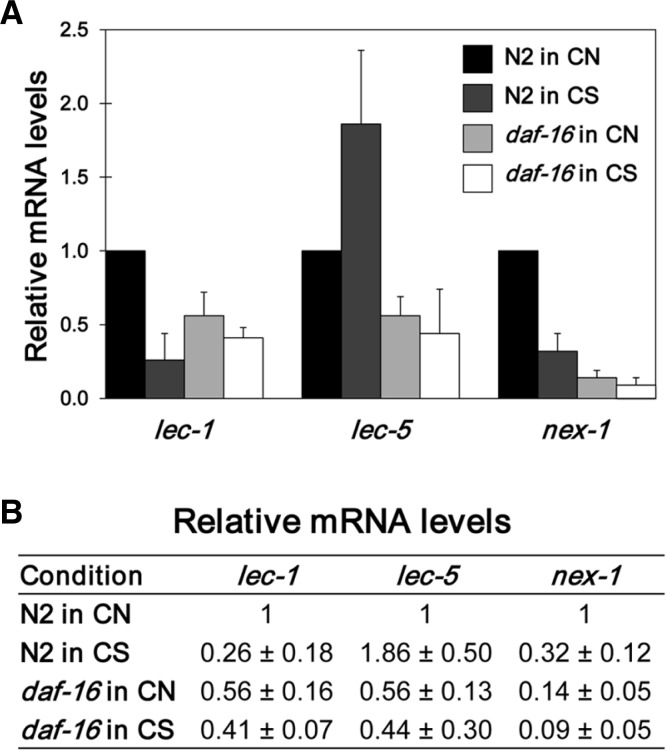
Levels of LEC-1, LEC-5, and NEX-1 were significantly increased upon CS treatment (Table 1). In our previous study, we reported that DAF-16, a FOXO transcription factor, is translocated to the nucleus and activated upon CS treatment (Jeong et al., 2010). To examine the possibility that these three genes are regulated by the DAF-16 transcription factor, we examined their promoter sequences to ascertain whether they contain putative DAF-16 binding sites, as previously reported (Oh et al., 2006). As summarized in Table 2, two sites in lec-1, two sites in lec-5, and eight sites in nex-1 were identified within their −4 kb upstream regions. This finding suggests that DAF-16 can affect the expression of these genes upon CS. To verify this idea, we measured the mRNA level of each gene in both wild-type N2 and daf-16(mu86) deletion mutant worms (Fig. 4). In CN condition, all three genes showed decreased levels of mRNA in the daf-16 mutant compared to N2 as expected, but the levels were not reduced to zero. On the other hand, unlike their protein levels, N2 worms grown in CS showed decreased levels of mRNA compared to CN for lec-1 and nex-1 genes, while lec-5 mRNA level was increased 1.86-fold upon CS compared to CN (Fig. 4). These results suggest that although DAF-16 is required for the activation of these three genes in CN, DAF-16 is probably not the sole transcription factor that regulates them. Further, in CS, although lec-5 is likely transcriptionally activated by DAF-16, lec-1 and nex-1 are up-regulated post-transcriptionally rather than transcriptionally by as yet unidentified factors.
Table 2.
Position of putative DAF-16 binding sites
| Gene | DAF-16 binding sequences
|
||||
|---|---|---|---|---|---|
| GTAAACAA | GTAAATAA | TTGTTTAC | TTATTTAC | CTTATCA | |
| lec-1 (W09H1.6) | −226 | −197 | |||
| lec-5 (ZK1248.16) | −3545 | −1438 | |||
| −3612 | −3645 | ||||
| nex-1 (ZC155.1) | −3403 | −3020 | −2726 | −414 | |
| −163, −150 | |||||
Sequences of putative DAF-16 binding sites were adapted from a previous report (Oh et al., 2006). The 5′ upstream sequence of each gene was searched by using softwares in UCSC Genome Bioinformatics (http://genome.ucsc.edu/), European Bioinformatics Institute (http://www.ebi.ac.uk), and wormbase (http://www.wormbase.org/).
Fig. 4.
Relative mRNA levels of lec-1, lec-5, and nex-1 genes in N2 and daf-16 mutant worms in CN and CS. Wild-type N2 and daf-16(mu86) mutant worms were synchronized at L1 stage and grown in CN and CS conditions, and then young adult worms were collected to perform qRT-PCR as described in materials and methods. Relative mRNA level of each gene in each condition was defined as the mRNA level of each gene in each condition averaged from triplicate measurements of two independent experiments and normalized with that of act-1, the internal control. Then relative values are shown using CN N2 value as 1.0.
DISCUSSION
Previous studies by us and other groups showed that CS affects larval development of C. elegans (Gerisch et al., 2001; Jeong et al., 2010). In the present study, we have identified genes likely responsible for the defects of larval development in CS condition using proteomic and RNAi analyses. Six genes involved in metabolism and protein synthesis were identified as down-regulated genes in CS. However, we were unable to distinguish whether reduction of levels of these proteins was caused directly by CS or indirectly as a consequence of reduction of overall metabolic rates of worms grown in CS. We compared our results with that of a previous report, which analyzed gross protein expression patterns during wild-type C. elegans development (Tabuse et al., 2005). In that report, the authors categorized protein expression patterns into six groups. Among the nine proteins we identified, six proteins were also analyzed by them. In that report, five of the six down-regulated proteins in CS were classified into either group I, V, or VI. Group I proteins were expressed stably throughout the development. Expression of group V proteins markedly increased after hatching and remained high throughout postembryonic stages. Expression of group VI proteins was gradually increased after hatching and remained high through the rest of the postembryonic stages (Tabuse et al., 2005). According to this classification, the five down-regulated proteins in CS are stably or highly expressed during postembryonic stages in wild-type C. elegans. If so, down-regulation of these proteins upon CS may possibly interfere with postembryonic development of C. elegans. To know whether down-regulation of these proteins has a critical effect on larval development of C. elegans, we observed the RNAi phenotypes of these genes for two generations in CN condition. RNAi phenotypes of sams-1 (Gottschalk et al., 2005), pmt-2 (Sönnichsen et al., 2005), R07H5.8 (Sönnichsen et al., 2005), eef-1A.1 (Simmer et al., 2003), and rla-0 (Rual et al., 2004) were previously reported that they showed larval developmental defects. Unlike those previous reports, we found that RNAi of two out of six CS down-regulated genes, F25H2.10 (rla-0) and R07H5.8, caused severe larval arrest phenotype in the second generation in CN. In addition, when we treated RNAi of R07H5.8 in CS condition, worms displayed more severe defects than CS alone even in the first generation. These results indicate that R07H5.8 and F25H2.10 (rla-0) are certainly critical targets of CS response in C. elegans.
The sams-1 and pmt-2 genes are among the significantly down-regulated genes in CS condition. SAMS-1, S-adenosyl-methionine synthetase, converts methionine to S-adenosyl-methionine (SAM) by adding an adenosyl group of adenosine triphosphate to methionine (Chiang et al., 1996). SAM is a key methyl donor for various methyltransferases including PMT-2, a SAM-dependent methyltransferase. Reduction of SAMS-1 and PMT-2 should reduce methylation levels of their substrates in worms grown in CS. It was reported that cholesterol depletion caused accumulation of methylated sterols (Matyash et al., 2004). This finding suggests that SAMS-1 and PMT-2 are not involved in sterol methylation because their levels were decreased in CS. It was also reported that SAMS-1 and PMT-1, another SAM-dependent methyltransferase, are down-regulated under stressful environmental condition and by pod-2 (acetyl- CoA carboxylase) RNAi (Li et al., 2011). Under these conditions, reduction of SAMS-1 and PMT-1 induced the accumulation of triacylglycerol lipid droplets in the intestine of worms (Li et al., 2011). Therefore, in CS condition, reduction of SAMS-1 and PMT-2 might also alter lipid metabolism and this change of lipid metabolism might affect larval development.
The mRNA level of acdh-1, which encodes a short-chain acyl-CoA dehydrogenase, was reported to be down-regulated in starved worms (www.wormbase.org). In our previous study, CS treatment caused shortening of worm life span (Jeong et al., 2010). In our current study, we found a significant reduction of ACDH-1 protein. As RNAi of acdh-1 was reported to shorten worm life span (www.wormbase.org), reduction of ACDH-1 may be responsible for the short life span of worms grown in CS.
Levels of LEC-1 and LEC-5 were increased in CS. There are numerous potential galectin genes in C. elegans (Nemoto-Sasaki et al., 2008), however their functions during the development are yet to be determined. Lectins in other organisms such as in Drosophila have been reported to function during development (Seppo and Tiemeyer, 2000). Expression of lectins was increased by triggering innate immune system in Drosophila (Pace et al., 2002), and lectin level was also increased upon stress in human serum (Iwamoto et al., 2010). The levels of galectins may also increase in response to some stresses including CS in C. elegans. It is also interesting to find that the NEX-1 level was increased upon CS. NEX-1 binds to phospholipids that are important components of cell membrane (Satoh et al., 2000). NEX-1 may function for maintenance of membrane integrity. Because cholesterol is also an important component of cell membrane, CS condition may be detrimental to membrane integrity, and up-regulation of NEX-1 may be required to maintain the membrane integrity. Further analysis of membrane components in CN and CS conditions may provide more clues about the molecular mechanism of larval arrest upon CS. Furthermore, NEX-1 functions as a ligand that mediates efficient clearance of apoptotic cells by colocalizing with phosphatidylserine (Arur et al., 2003). Therefore, up-regulation of NEX-1 is likely to promote engulfment of cell corpses generated by developmental defects. However, unlike CS condition, when sterol biosynthesis inhibitor, azacoprostane was treated to cause cholesterol deprivation in C. elegans, NEX-1 was significantly decreased (Choi et al., 2003), suggesting that although both CS and drug treatment aimed for cholesterol deprivation, their physiological impacts were different.
In CS condition, DAF-16 was translocated to the nucleus where it was likely activated (Jeong et al., 2010). We analyzed three up-regulated genes, lec-1, lec-5, and nex-1, and found that their mRNA expression levels were dependent on daf-16 activity in CN, indicating that these genes are at least partially regulated by DAF-16 at the transcriptional level. Nevertheless, although LEC-1, LEC-5, and NEX-1 proteins were all up-regulated in CS, only mRNA level of lec-5 was up-regulated in CS (Fig. 4). These results suggest that lec-1 and nex-1 are up-regulated at post-transcriptional levels rather than at transcriptional levels in CS, resulting in difference of expression levels between mRNA and protein. Similar discrepancy between mRNA levels and protein levels has been reported also by others (Gygi et al., 1999). Since DAF-16 may possibly repress gene expression (Jeong et al., 2010), we also examined promoter sequences of six down-regulated genes. Among them, only sams-1 contained one DAF-16 binding site at -1,333 bp region (data not shown). Whether this site is actually contributing to the expression of sams-1 remains to be determined.
Here we identified cholesterol-responsive metabolic proteins, some of which are essential for C. elegans larval development, by using both proteomic analysis and reverse genetic analysis. How they are regulated by cholesterol signaling at either transcriptional or post-transcriptional level remains to be elucidated.
Supplementary Material
Acknowledgments
Nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). We thank Yonsei Proteome Research Center for analyzing mass spectrometry and protein identification, and Dr. Junho Lee at Seoul National University for materials. This study was supported by Konkuk University in 2010 to Y.-H. Shim and by the 2013 KU Brain Pool Program of Konkuk University to I. Kawasaki.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Ahn DH, Singaravelu G, Lee S, Ahnn J, Shim YH. Functional and phenotypic relevance of differentially expressed proteins in calcineurin mutants of Caenorhabditis elegans. Proteomics. 2006;6:1340–1350. doi: 10.1002/pmic.200500315. [DOI] [PubMed] [Google Scholar]
- Arur S, Uche UE, Rezaul K, Fong M, Scranton V, Cowan AE, Mohler W, Han DK. Annexin I is an endogenous ligand that mediates apoptotic cell engulfment. Dev. Cell. 2003;4:587–598. doi: 10.1016/s1534-5807(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, McCann PP. S-adenosylmethionine and methylation. FASEB J. 1996;10:471–480. [PubMed] [Google Scholar]
- Choi BK, Chitwood DJ, Paik YK. Proteomic changes during disturbance of cholesterol metabolism by azacoprostane treatment in Caenorhabditis elegans. Mol. Cell. Proteomics. 2003;2:1086–1095. doi: 10.1074/mcp.M300036-MCP200. [DOI] [PubMed] [Google Scholar]
- Clayton PT. Disorders of cholesterol biosynthesis. Arch Dis Child. 1998;78:185–189. doi: 10.1136/adc.78.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese RV, Herz J., Jr Cholesterol metabolism and embryogenesis. Trends Genet. 1998;14:115–120. doi: 10.1016/s0168-9525(97)01377-2. [DOI] [PubMed] [Google Scholar]
- Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev. Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- Gottschalk A, Almedom RB, Schedletzky T, Anderson SD, Yates JR, 3rd, Schafer WR. Identification and characterization of novel nicotinic receptor-associated proteins in Caenorhabditis elegans. EMBO J. 2005;24:2566–2578. doi: 10.1038/sj.emboj.7600741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Taguchi C, Sasaguri K, Kubo K, Horie H, Yamamoto T, Onozuka M, Sato S, Kadoua T. The galectin-1 level in serum as a novel marker for stress. Glycoconj J. 2010;27:419–425. doi: 10.1007/s10719-010-9288-z. [DOI] [PubMed] [Google Scholar]
- Jeong PY, Na K, Jeong MJ, Chitwood D, Shim YH, Paik YK. Proteomic analysis of Caenorhabditis elegans. Methods Mol Biol. 2009;519:145–169. doi: 10.1007/978-1-59745-281-6_10. [DOI] [PubMed] [Google Scholar]
- Jeong MW, Kawasaki I, Shim YH. A circulatory transcriptional regulation among daf-9, daf-12, and daf-16 mediates larval development upon cholesterol starvation in Caenorhabditis elegans. Dev Dynam. 2010;239:1931–1940. doi: 10.1002/dvdy.22322. [DOI] [PubMed] [Google Scholar]
- Kawasaki I, Jeong MW, Shim YH. Regulation of sperm-specific proteins by IFE-1, a germline-specific homolog of eIF4E, in C. elegans. Mol. Cells. 2011;31:191–197. doi: 10.1007/s10059-011-0021-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Na K, Lee HJ, Lee EY, Paik YK. Contribution of sams-1 and pmt-1 to lipid homoeostasis in adult Caenorhabditis elegans. J Biochem. 2011;149:529–538. doi: 10.1093/jb/mvr025. [DOI] [PubMed] [Google Scholar]
- Lozano R, Chitwood DJ, Lusby WR, Thompson MJ, Svoboda JA, Patterson GW. Comparative effects of growth inhibitors on sterol metabolism in the nematode Caenorhabditis elegans. Comp Biochem Physiol. 1984;79:21–26. doi: 10.1016/0742-8413(84)90156-7. [DOI] [PubMed] [Google Scholar]
- Matyash V, Entchev EV, Mende F, Wilsch-Brauninger M, Thiele C, Schmidt AW, Knölker HJ, Ward S, Kurzchalia TV. Sterol-derived hormone(s) controls entry into diapause in Caenorhabditis elegans by consecutive activation of DAF-12 and DAF-16. PLoS Biol. 2004;2:e280. doi: 10.1371/journal.pbio.0020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemato-Sasaki Y, Hayama K, Ohya H, Arata Y, Kaneko MK, Sitou N, Hirabayashi J, Kasai K. Caenorhabditis elegans galectins LEC-1-LEC-11: structural features and sugar-binding properties. Biochim. Biophys. Acta. 2008;1780:1131–1142. doi: 10.1016/j.bbagen.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- Pace KE, Lebestky T, Hummel T, Arnoux P, Kwan K, Baum LG. Characterization of a novel Drosophila melanogaster galectin. J Biol Chem. 2002;277:13091–13098. doi: 10.1074/jbc.M112105200. [DOI] [PubMed] [Google Scholar]
- Paik YK, Jeong SK, Lee EY, Jeong PY, Shim YH. C. elegans: an invaluable model organism for the proteomics studies of the cholesterol-mediated signaling pathway. Expert Rev. Proteomics. 2006;3:439–453. doi: 10.1586/14789450.3.4.439. [DOI] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A, Hazuki M, Kojima K, Hirabayashi J, Matsumoto I. Ligand-binding properties of annexin from Caenorhabditis elegans (Annexin XVI, Nex-1) J Biochem. 2000;128:377–381. doi: 10.1093/oxfordjournals.jbchem.a022764. [DOI] [PubMed] [Google Scholar]
- Seppo A, Tiemeyer M. Function and structure of Drosophila glycans. Glycobiology. 2000;10:751–760. doi: 10.1093/glycob/10.8.751. [DOI] [PubMed] [Google Scholar]
- Shim YH, Paik YK. Caenorhabditis elegans proteomics comes of age. Proteomics. 2010;10:846–857. doi: 10.1002/pmic.200900542. [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Tabuse Y, Nabetani T, Tsugita A. Proteomic analysis of protein expression profiles during Caenorhabditis elegans development using two-dimensional difference gel electrophoresis. Proteomics. 2005;5:2876–2891. doi: 10.1002/pmic.200401154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.