Abstract
Nuclear matrix attachment regions (MARs) regulate the higher-order organization of chromatin and affect the expression of their flanking genes. In this study, a tobacco MAR, TM6, was isolated and demonstrated to remarkably increase the expression of four different promoters that drive gusA gene and adjacent nptII gene. In turn, this expression enhanced the transformation frequency of transgenic tobacco. Deletion analysis of topoisomerase II-binding site, AT-rich element, and MAR recognition signature (MRS) showed that MRS has the highest contribution (61.7%) to the TM6 sequence-mediated transcription activation. Micrococcal nuclease (MNase) accessibility assay showed that 35S and NOS promoter regions with TM6 are more sensitive than those without TM6. The analysis also revealed that TM6 reduces promoter DNA methylation which can affect the gusA expression. In addition, two tobacco chromatin-associated proteins, NtMBP1 and NtHMGB, isolated using a yeast one-hybrid system, specifically bound to the TM6II-1 region (761 bp to 870 bp) and to the MRS element in the TM6II-2 (934 bp to 1,021 bp) region, respectively. We thus suggested that TM6 mediated its chromatin opening and chromatin accessibility of its flanking promoters with consequent enhancement of transcription.
Keywords: chromatin accessibility, DNA methylation, matrix attachment regions, transcription activation, transformation
INTRODUCTION
Nuclear matrix participates in the formation of structural and functional chromatin loop domains and has been implicated in cellular processes, such as DNA replication and transcription and RNA processing. Analysis of scaffold/matrix-associated DNA regions (S/MARs) could enhance the current understanding of higher-order chromatin organization (Luderus et al., 1992; Paul and Ferl, 1998). Although no consensus sequences strictly exist, characterized MARs are AT-rich or GC-rich and generally contain some regions that tend to produce single-stranded or base-unpaired sequences, one- or multi-unwinding sites and topoisomerase II-binding sites (Bode et al., 1992). MARs are widely distributed DNA sequences that exhibit high binding affinity for the nuclear matrix, to enhance gene expression in eukaryotic genomes (Avramova et al., 1995; Girod et al., 2007). In animals, MARs are essential for the transcription of a rearranged μ gene in transgenic B lymphocytes (Forrester et al., 1994). Activation of a promoter that controls transcription in transgenic mice also requires MARs that flank the intronic enhancer (Heng et al., 2004). In nuclear halos, the behavior of endogenous S/MARs relies on their physical attributes, however, whether these serve as rigid or flexible chromatin loop anchors remains incompletely understood. In plants, transgenic tobacco that have the chicken lysozyme MAR element known as the A element show the position-independent expression of transgenes (Mlynarova et al., 1995). In a similar manner, MARs significantly affect the expression of their flanking genes in vivo (Tikhonov et al., 2000). Increasing numbers of studies report that MARs can improve transgene expression levels, and demonstrat transgene expression and transgene silencing in transgenic plants (Brouwer et al., 2002; Li et al., 2008; Mlynarova et al., 2003).
In several cases, MARs are mapped on the boundaries of actively transcribed chromatin domains or on bracket transcription units to stimulate the transcription of flanked genes, protect the transcribed regions from the position effects of neighboring sequences (Loc and Stratling, 1988; Shewchuk et al., 2001), and augment the activity of enhancer elements by extending the distance over which they can function (Namciu and Fournier, 2004; Purbowasito et al., 2004). The latter activity has been linked to a specific DNA demethylation extension and chromatin core histone acetylation from MAR-associated enhancers to the promoters (Bode et al., 2003; Pikaart et al., 1998). Evidence indicates that the effects of MAR elements which depend on the promoter that is used to drive the transgene, can vary significantly in transgenic maize plants (Sidorenko et al., 2003). In mammals, the methylation state of the fibroblast genes that coincide with the MARs shows unique differences when compared with adjacent sites and with the corresponding sites of white cell genes (Ellis et al., 1988). In addition, MARs antagonize the methylation-dependent repression of long-range enhancer-promoter interactions (Forrester et al., 1999). In plants, the onset or release of transcriptional gene silencing (TGS) correlates with changes in repressive epigenetic markers (Gong et al., 2002; Mette et al., 2000). DNA demethylation prevents the TGS of transgenes, which is accompanied by RNA silencing (He et al., 2011; Penterman et al., 2007). A study revealed that the presence of MAR can prevent the RNA silencing of a tobacco transgene (Mlynarova et al., 2003). The relationship between MARs and DNA methylation of the flanking regions in plant systems has yet to be reported.
The interaction of chromatin with the nuclear matrix via MARs is considered to be of primary importance to the chromatin organization in all eukaryotic cells. Increasing amounts of evidence has show that nuclear matrix proteins specifically recognize the chromatin secondary structure, which may be a primary determinant of matrix binding. In animals, a special AT-rich binding protein, SATB, is an intensively investigated MAR-binding protein that acts as a global regulator of gene expression by recruiting various corepressor or coactivator complexes. This recruitment establishes a unique chromatin structure at the SATB genomic targets in a context-dependent manner (Ellies and Krumlauf, 2006; Han et al., 2008; Yang et al., 2009). Several MAR-binding proteins have been identified in higher plants. MFP1, a highly conserved MAR-binding filament-like protein located at the internal nuclear matrix, can bind double-stranded DNA (Harder et al., 2000; Jeong et al., 2003; Meier et al., 1996). MAF1 is novel plant protein located at the nuclear envelope. This protein interacts with the MAR-binding protein MFP1 and might connect the nuclear envelope and the internal nuclear matrix (Gindullis et al., 1999). AHM1 and AHL1 are two other nuclear matrix-localized and MAR-binding proteins that contain a single AT hook and a J domain-homologous region or plant and prokaryote conserved (PPC) domain. These proteins function between the intranuclear framework and MARs by recruiting specific partners as co-chaperones (Fujimoto et al., 2004; Morisawa et al., 2000). Other MAR-binding proteins, such as MARBP-1 and MARBP-2, may have multifunctional roles in chromatin organization and ribosome biogenesis (Hatton and Gray, 1999). Such MAR-binding proteins have yet to be identified to obtain a clearer understanding of the function of MAR in chromatin remodeling and gene expression.
This study focuses on the isolation and identification of plant MAR and determination of the mechanism that underlies MAR regulation of transcription of its flanking genes. TM6, a novel tobacco MAR, is involved in transcription enhancement by distorting the chromatin accessibility and reducing the methylation frequency of the MAR flanking promoter regions. Two TM6-binding proteins are isolated from tobacco that indicate the possible functions of TM6 in the chromatin accessibility and DNA methylation of the adjacent promoter to enhance transcription are discussed.
MATERIALS AND METHODS
Plant materials
Tobacco (Nicotiana tabacum L. cv. NC89) seeds were sterilized 0.1% mercuric chloride for 10 min and grown on full-strength MS medium (Murashige and Skoog, 1962) stratified at 4°C for 2 days in the dark prior to germination. Seedlings were grown in a growth chamber at 25°C with a 16 h light/8 h dark cycle (450 μmol photons · m−2 · s−1).
The tobacco suspension cell line BY-2 was grown in the dark at 25°C and shaken at 125 × g. Cell suspension cultures of BY-2 were subcultured every 7 days by transferring 5 ml aliquots to 100 ml fresh liquid MS media.
Nuclei were isolated from a tobacco cell line, and nuclear matrixes were isolated and characterized from the isolated nuclei as described previously (Xue et al., 2005).
Vector construction and plant and cell transformation
The structures of the transgene expression cassettes for plant transformation used in this study are shown (Fig. 2). The details for vector construction and plant and cell transformation are provided in the Supplemental Experimental Procedures.
Fig. 2.
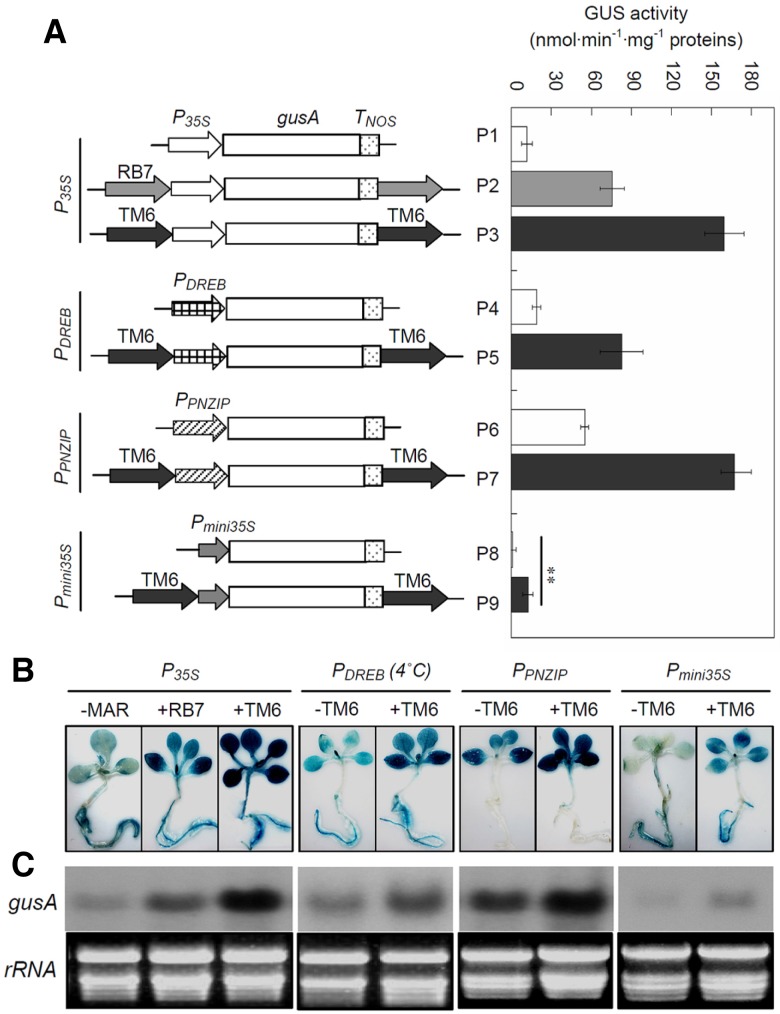
Schematic representation of the constructs used in this study and their effects on the GUS activity of independent transgenic tobacco. (A) The various constructs, fused to different promoters, are indicated on the left. CaMV 35S promoter (P35S), PNZIP promoter of the Pharbits nil PNZIP gene (PPNZIP), DREB promoter of the dehydration-responsive element binding gene (PDREB), the −49 to +5 region of 35S promoter (Pmini35S), gusA cDNA for β-glucuronidase. The average GUS activity from 20 transgenic tobacco lines is given as nanomoles of methylumbelliferone generated per minute per milligram of protein, with error bars representing ± SE from three experiments. (B) The transgenic tobacco with nine constructs corresponding to (A) were stained with GUS staining solution overnight and decolorized with 70% ethanol. “−”, transgenic lines without TM6. “+”, transgenic lines with TM6 or RB7. (C) The messenger RNA accumulation of the gusA gene in transcription levels in transgenic tobacco corresponding to (B) The ethidium bromide-stained total RNA is shown as a loading control.
GUS activity assays and histochemical staining
GUS activity assays were performed as described previously (Yan et al., 2012; Zhang et al., 2007). The details for GUS activity assays and histochemical staining are provided in the Supplemental Experimental Procedures.
Seed germination
Surface sterilized seeds of transgenic lines were grown in a medium containing 100-, 400-, 800- and 1,000 mg · l−1 kanamycin for 14 days in a growth chamber maintained at 25°C under a 16 h light/8 h dark cycle, respectively. At least 100 seeds for each genotype were sterilized and sown on MS medium and the germination results were calculated based on at least three independent experiments.
Nuclei extraction and treatment with micrococcal nuclease
Nuclei were collected from 100 g of the four-leaf tobacco leaves and endonuclease digestion were performed as described previously (Fukuda and Nishikawa, 2003). The details for nuclei extraction and treatment with micrococcal nuclease are provided in the Supplemental Experimental Procedures.
Yeast one-hybrid screening
Two regions of the TM6, TM6II-1 and TM6II-2 were synthesized by PCR and inserted into the EcoRI-MluI site of the pHIS2 vector (Supplementary Table S1). Tobacco cDNA libraries were prepared from tobacco leaves using BD SMART™ cDNA synthesis. We cotransformed yeast strain Y187 with double-strand cDNA, pGADT7-Rec2 and TM6II-1-pHIS2/TM6II-2-pHIS2, and restreaked the yeast cells on SD/-His-Leu-Trp + 60 mM 3-AT plates. The yeast screening procedure was performed according to the manufacturer’s protocol (CLONTECH Match-maker one-hybrid system). At last, 20 colonies were restreaked on the selection medium to eliminate false-positive colonies, and finally obtained six yeast colonies.
6× His-tagged protein expression and purification
pET32a-NtMBP1 and pET32a-NtHMGB proteins were transformed into E. coli BL21 (DE3), and recombinant proteins were expressed and purified by nickel-nitrilotriacetic acid agarose (Ni-NTA) affinity chromatography according to the protocol (Qiagen, Germany). Then recombinant proteins were dialyzed in 20 mM HEPES-KOH, pH 7.9, 1 mM MgCl2, 50 mM KCl, 1 mM DTT, 20% glycerol and 0.02% NP-40 overnight.
Electrophoretic mobility shift assay (EMSA)
The digoxigenin (DIG) gel shift was used for protein-DNA binding assays (Roche, http://www.roche-applied-science.com). The details for electrophoretic mobility shift assay (EMSA) are provided in the Supplemental Experimental Procedures.
Bisulfite genomic sequencing and PCR amplification
Genomes of the transgenic tobacco lines were treated with EZ DNA Methylation-Gold™ Kit (ZYMO, http://www.zymoresearch.com) for methylated DNA detection following the manufacturer’s protocol. The PCR primers were designed by MethPrimer (Li and Dahiya, 2002) and shown in Supplementary Table S1. PCR products were cloned into pGEM-T Easy vector (Promega, http://www.promega.com) following the manufacturer’s protocol. The chloroplast genome of every sample was conducted by BSP to calculate the conversation efficient. Conversion efficiency was greater than 98% for each bisulphate-treated sample. Sequenced results were calculated by Cy-MATE (http://www.cymate.org/).
RESULTS
Isolation and characterization of TM6
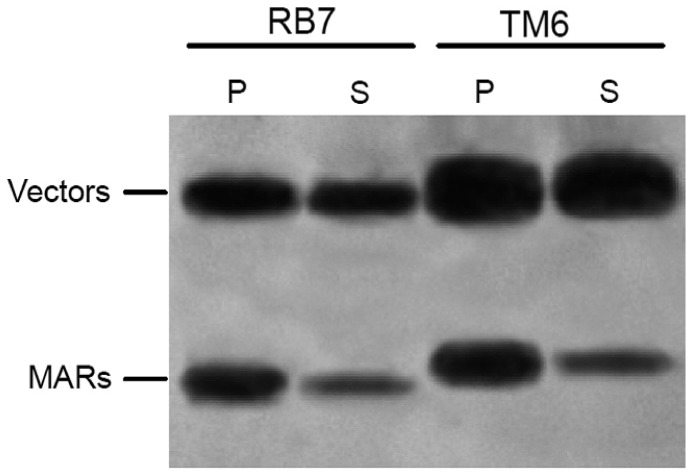
A library of random tobacco DNA fragments copurified with a tobacco nuclear matrix was constructed to isolate new MARs from tobacco. TM6 (GenBank accession No. KC555564), a 1,193 bp MAR fragment which contains one MRS, one topoisomerase II-binding site, and five AT-rich elements (Supplementary Fig. S1) was attached to the nuclear matrix and thus enriched by the purification procedure, and then isolated from the library as described previously (Xue et al., 2005; Zhang et al., 2004; 2007). To identify the binding ability of TM6 to the nuclear matrix, the in vitro binding assay was performed. RB7, an identified tobacco MAR (Ascenzi et al., 2001), was used as a control. The binding strength of MARs to the nuclear matrix was demonstrated by the binding efficiency ratios of the binding MARs and the corresponding vector. The signal strength of TM6 and RB7 to the corresponding vectors were 5.76 and 2.92, respectively, indicating that TM6 could strongly bind to nuclear matrix and even better than RB7 (Fig. 1).
Fig. 1.
In vitro binding assays of tobacco matrix attachment regions (MARs) to the rice nuclear matrix. The nuclear matrix was treated with the restriction enzymes EcoRI and HindIII. The released vector and MAR fragments were end-labeled with α-[32P] dCTP. P pellet, S supernatant, RB7 a known strong MAR isolated from the 3′ flanking region of the RB7 root-specific gene of tobacco (used as control here).
TM6 enhances transcription activation in transgenic tobacco independent of promoters
Eight pBI121-derived plant expression vectors were constructed and employed in this study (Fig. 2A). The gusA gene, which was controlled by four different promoters, namely, P35S (CaMV 35S), promoter of a gene that is predicted to encode a Pharbitis nil leucine zipper (PPNZIP) (Zheng et al., 1998), promoter of a dehydration-responsive element-binding gene (PDREB) (Shan et al., 2007), and Pmini35S (-46 to -1 region of the 35S promoter) (Odell et al., 1985), was used to evaluate the effect of TM6 on downstream gene expression. For each constructs, twenty independent transgenic tobacco lines were selected for the β-glucuronidase (GUS) activity assays. When the expression cassettes were flanked by TM6, the average GUS activities that were driven by the four promoters increased 12.7-, 4.3-, 9.8-, and 2.9-fold, respectively (Table 1). Northern blot results indicated that, compared with the controls, TM6 can significantly enhance mRNA accumulation under these four promoters (Fig. 2C).
Table 1.
Effects of TM6 in different tissues of transformants
|
P35S
|
PGhDREB
|
PPNZIP
|
Pmini35S
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plant tissue | GUS activity | N-fold increasec | Average fold | GUS activity | N-fold increasec | Average fold | GUS activity | N-fold increasec | Average fold | GUS activity | N-fold increasec | Average fold | ||||
| (−)a | (+)b | (−)a | (+)b | (−)a | (+)b | (−)a | (+)b | |||||||||
| Leaves | 15.5 | 211 | 14.2** | 12.7** | 5.1 | 16.7 | 3.3* | 4.3* | 58.3 | 164.9 | 2.9* | 2.9* | 1.3 | 15.4 | 11.8** | 9.8** |
| Roots | 10.2 | 114 | 11.2** | 6.9 | 37.9 | 5.5* | -d | -d | -d | 0.9 | 7.0 | 7.8** | ||||
Mean GUS activity (nmol · min−1·mg−1 protein) of all transformants without TM6.
Mean GUS activity of transformants with TM6.
Mean GUS activity for TM6 transformants divided by mean GUS activity for control transformants.
Mean GUS activity of ≤ 0.1 nmol·min−1·mg−1 protein. Asterisks denote significant differences at the P < 0.05(*) or P < 0.01(**) level.
Interestingly, the gusA expression driven by PPNZIP, a tissue-specific promoter, increased approximately 3-fold by TM6 in transgenic tobacco leaves. However, no GUS activity was detected in nonphotosynthetic tissues at either the mRNA or the protein level (Fig. 2B). Moreover, the gusA expression driven by PDREB, a cold-inducible promoter, increased 4.3-fold by TM6 in transgenic tobacco under cold stress conditions, whereas a 1.7-fold increase was observed under normal growth conditions. In transgenic tobacco that contain PMINI35S, TM6 can dramatically enhance GUS activity to the level of the full-length 35S promoter (P35S) (P < 0.05).
TM6 increases the transformation frequency by enhancing the nptII expression in transgenic tobacco
To investigate the effect of TM6 on the transformation frequency in tobacco, kanamycin-resistant shoots that appeared at the edge of transformed leaf disks were observed during the first two weeks of the experiment (Table 2). Compared with the control (without MARs), kanamycin-resistant shoots appeared 5 and 2 days earlier in transformed leaf disks that contained TM6 and RB7, respectively. The shoot numbers per leaf disk in the TM6, RB7 and control disks were 7.6, 5.3 and 2.0, respectively. The transformation frequency of the disk that contained TM6 increased by up to 3.8-fold.
Table 2.
Influence of TM6 on the frequency of Agrobacterium-mediated tobacco transformation
| Construct | No. of leaf disks infected | No. of resistant shoots | Frequency (shoot no. per leaf disk) | Mean time of the first shoot formation (days) |
|---|---|---|---|---|
| 35S::gusA | 45 | 91 ± 15 | 2.0 ± 0.3 | 15 ± 3 |
| RB7::35S::gusA::RB7 | 53 | 280 ± 27* | 5.3 ± 0.5* | 12 ± 2* |
| TM6::35S::gusA::TM6 | 51 | 389 ± 43* | 7.6 ± 0.4** | 10 ± 2* |
Data for kanamycin resistant shoot formation were calculated after cultivation on selection medium for 25 days and presented as means ± SE at triplicates. Ten leaf disks were infected per plate. Asterisks denote significant differences at the P < 0.05(*) or P < 0.01(**) level
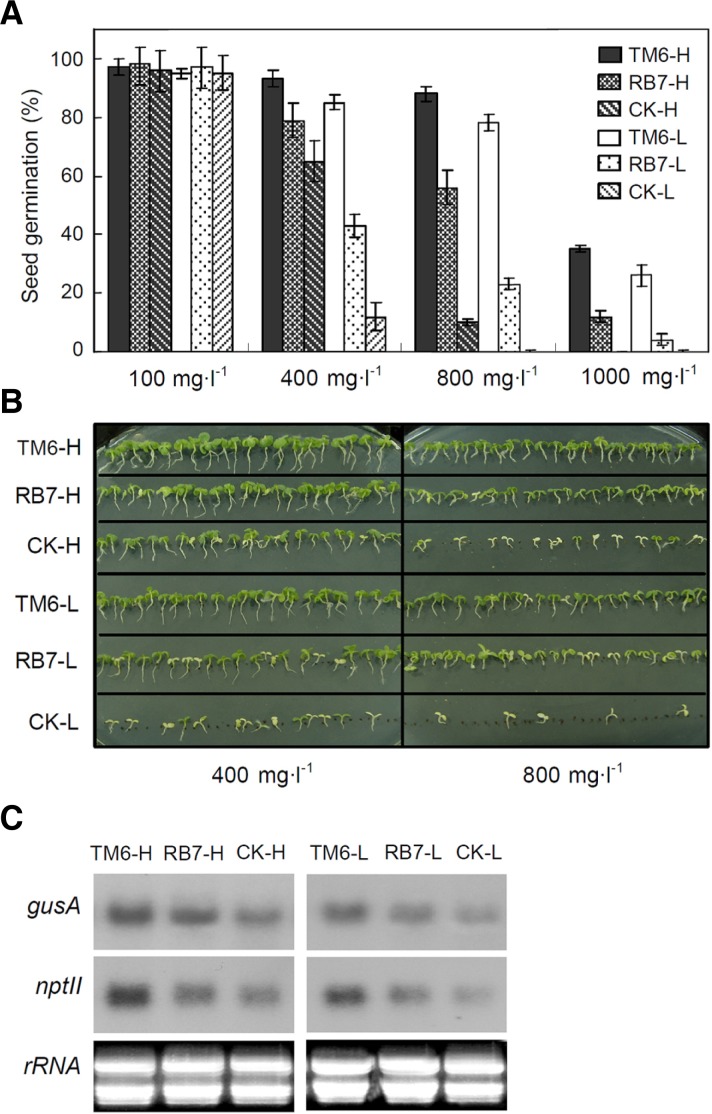
To investigate the mechanism behind the TM6-induced increase in transformation frequency, seed germination and Northern blot analysis of transgenic tobacco were performed. To determine the seed survival percentage, tobacco seeds of T2 generation transgenic plants with TM6, RB7, or the control were grown on medium containing 100, 400, 800 or 1,000 mg · l−1 kanamycin (Fig. 3A). Twenty independent lines of each construct were classified into high (H) and low (L) groups based on their GUS activities: TM6-H, RB7-H, and CK-H; and TM6-L, RB7-L, and CK-L. As the kanamycin concentration increased, lines containing TM6, TM6-H, and TM6-L exhibited higher survival percentages than the controls without MARs. Interestingly, when the kanamycin concentration in the medium was increased to 800 mg · l−1 (Fig. 3B), the average survival percentage of TM6-H and TM6-L seedlings still reached 94%. The mRNA accumulation of nptII in TM6-H and TM6-L was significantly higher than that in the other transgenic groups (Fig. 3C). These results indicate that TM6 acts as a strong MAR to increase the transformation efficiency of transgenic tobacco by improving nptII expression.
Fig. 3.
Seed germination and nptII expression of T2 generation transgenic tobacco. A, Seed germination of the transgenic tobacco grown on Murashige and Skoog medium with different concentrations of kanamycin (100-, 400-, 800- and 1000 mg · l−1) was recorded at 10 days. Values are expressed as the mean of four replications ± SE. At least 100 seeds per transgenic line with TM6/RB7 or without MAR (CK) were measured in each replicate, respectively. H, transgenic lines with higher GUS activity than average. L, transgenic lines with lower GUS activity than average. B, Photographs of 14-day-old transgenic tobacco seedlings grown on MS with 400 mg · l−1 or 800 mg · l−1 kanamycin. C, mRNA accumulation of the nptII and gusA genes in T2 generation transgenic tobacco lines. The picture represents one of three independent experiments that gave similar results.
TM6 increases MNase sensitivity of the 35S and NOS promoter regions in transgenic plants
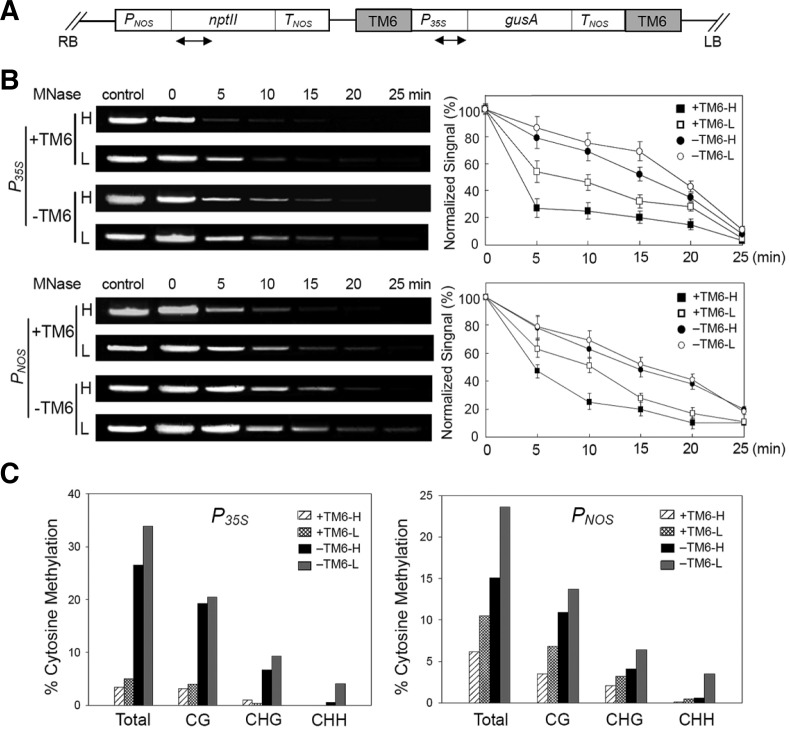
To investigate the effect of TM6 on the expression of its flanking nptII and gusA genes, the study focused on the possible remodeling of nucleosomes near the TM6 chromatin (Fig. 4A). Given that MNase preferentially digests nucleosome-free DNA regions (Wolffe, 1995), nuclease accessibility was used to indicate nucleosome remodeling (Rao et al., 2001). Nuclei were isolated from transgenic lines with and without TM6 and then incubated with MNase for time-course digestion. Genomic DNA was extracted for PCR assay using primer pairs that encompassed the 35S and NOS promoter regions. Nuclease digestions of fifteen independent transformants were performed in four groups for each line, with each group containing the same weight of five independent transgenic cell lines. Aliquots of the amplified products from the three groups of transgenic events were combined for electrophoresis. NOS and 35S promoters adjacent to TM6 degraded more rapidly than the controls (Fig. 4B). Only a slight difference was observed between the digestion rates of the 35S and NOS promoter regions: the TM6-H values for these two regions at 5 min exhibited a 2-fold difference. These results suggest that TM6 might be necessary to the nucleosome remodeling of its adjacent promoter region, which is responsible for promoting of transcription.
Fig. 4.
Influence of TM6 elements on the accessibility of 35S and NOS promoter regions to Mnase and DNA methylation pattern. (A) Schematic representation of the transgene construct. The location of TM6 MAR, the chimeric neomycin phosphotransferase gene (nptII) and the gusA gene are indicated. The primer pairs used in the assays are indicated by arrow-heads. PNOS, the promoter of the nptII gene. TNOS, the terminators of the gusA and nptII genes. RB and LB, the right and left T-DNA borders respectively. (B) Isolated nuclei from transgenic lines were incubated with MNase for the times indicated. Control refers to a reaction without MNase. Total DNA was prepared from nuclei at various times and amplified by PCR to determine the sensitivity to MNase of 35S and NOS promoter regions under 0-, 5-, 10-, 15-, 20- and 25-min treatments. Lane H, transgenic lines with high GUS activity; Lane L, transgenic lines with low GUS activity. “−”, transgenic lines without TM6. “+”, transgenic lines with TM6. Amplified DNA products were separated on a 1% agarose gel, visualized by staining with ethidium bromide and quantified with a fluorimager. Quantification of the DNA products is shown on the right. The amount of DNA present at each time was normalized to that at time 0. Values are means of at least six replications, with error bars representing ± SE. (C) Bisulfite sequencing analysis of promoter methylation status of 35S and NOS promoter for the corresponding locus in transgenic lines with TM6, +TM6-H and +TM6-L, as well as the transgenic lines without TM6, -TM6-H and -TM6-L. For each region, forty independent top-strand clones were sequenced from each sample. Each assay was obtained from at least five independent lines and repeated three times.
TM6 decreases the DNA methylation of the promoter regions
The promoter regions of silenced transgenes are sometimes subjected to hypermethylation of the cytosine residues in their DNA strands (Finnegan et al., 1998). Bisulfite sequencing was used to investigate the DNA methylation of promoter regions adjacent to TM6. The 860 bp nucleotide sequence of the 35S and full-length NOS promoter connected to the 5′ coding region of the gusA gene was amplified by bisulfite sequencing. The representative status of DNA methylation in the target sequences of 35S and NOS promoters from transgenic lines with (+TM6) and without TM6 (-TM6) are demonstrated (Supplementary Figs. S2 and S3). Hypomethylation of both the 35S and the NOS promoters was detected in transgenic lines with TM6. The average percentages of total DNA methylation in the 35S and NOS promoters with TM6 were 26% and 11% lower than those without TM6, respectively (Fig. 4C). Bisulfite sequencing results show that the hypomethylation occurs in the CG, CHG, and CHH (H is A, T, or C) contexts. Moreover, the high-GUS activity group -TM6-H exhibited 26.5% methylation frequency, whereas 33.9% methylation frequency was detected in the low-GUS activity group -TM6-L in the 35S promoters (Supplementary Table S2). Overall, the DNA methylation percentage in transgenic lines without TM6 partially determines the gusA accumulation level. This behavior suggests that the decrease in the DNA methylation percentage at the 35S promoter region is one reason for the TM6-induced enhancement of gusA gene expression.
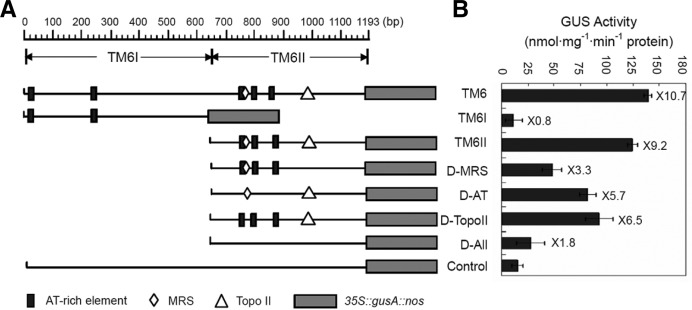
Site-specific deletion of TM6 affects the enhancement of transgene expression
Sequence analysis indicated that the 1,193 bp TM6 contains three essential elements: one MRS, one topoisomerase II-binding site, and five AT-rich elements. To determine whether or not these sites are functional elements of TM6, the sites were deleted and then introduced into the flanking regions of gusA expression cassettes (Fig. 5A). The GUS activities in the transgenic BY-2 cells with TM6II (651 bp to 1,193 bp) increased by 9.2-fold compared with the control (Fig. 5B). These results are similar to the effects of the full-length TM6, indicating that TM6II contains elements that are essential to the TM6 function of regulating the expression of flanking genes.
Fig. 5.
Site-specific deletion analysis of TM6. (A) The schematic representation shows the distribution of important elements in full-length TM6. Rectangles represent the AT-box (TTTTATTATT). Rhombuses represent the MRS element (TAWAWWWNNAWWRTA ANNWWG). Triangles represent the topoisomerase II-binding site (GTCAAAATATAATAG). D-MRS, D-AT, D-TopoII and D-All, the different element-deletion constructs of the TM6 II. (B) GUS activities from transgenic BY-2 cells for different constructs are shown. GUS activities were measured by enzymatic conversion of 4-methylumbelliferone, which was quantified with a spectrofluorimeter. The activity is expressed as nmol 4-methylumbelliferone min−1 · mg−1 protein. Values are means of triplicates ± SE (n ≥ 15). Fold increase by TM6 and various deletions as related to the control is indicated on the right of the bars.
In contrast to the effects of TM6II on GUS expression, the average GUS activity levels exhibited by constructs with any of the deleted TM6II sequences significantly declined (t-test, P < 0.05). Deletion of the MRS element resulted in a 5.9-fold reduction of GUS activity, which suggests that MRS is necessary to transgene expression. In addition, deletion of the AT-rich elements and of the topoisomerase II-binding site resulted in 3.5- and 2.7-fold reductions in expression, respectively. When all four elements were deleted, the gusA gene exhibited a 7.4-fold decrease in expression, nearly similar to that of the control. These results demonstrate that the four elements all contribute to the function of TM6II.
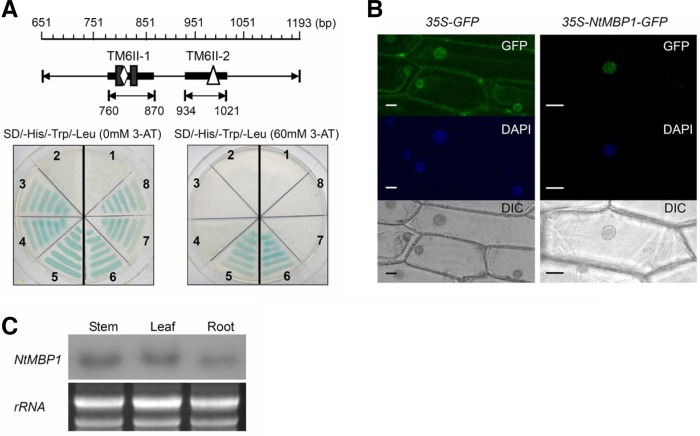
Isolation of cDNA encoding DNA-binding proteins that interact with the TM6II regions
To determine the effects of TM6 on the chromatin structure, a yeast one-hybrid screening of a tobacco leaf cDNA library was conducted using TM6II-1 (760 bp to 870 bp) and TM6II-2 (934 bp to 1,021 bp) fragments as the target binding sequences to individually isolate new MAR-binding proteins for their functional elements (Fig. 6A). A total of 220 positive colonies from selective-medium plates (SD/-His-Leu-Trp + 60 mM 3-AT) were obtained. When the 3-AT concentration was increased from 30 mM to 60 mM, two clones that normally grow on the SD/-His-Leu-Trp + 60 mM 3-AT plates were isolated (Fig. 6A). Sequence analysis indicates that these two cDNAs encode proteins with DNA-binding domains, and these two cDNA fragments, designated as NtMBP1 (NTU06712) and NtHMGB (EF051129), encode proteins that contain DNA-binding domains.
Fig. 6.
Characterization of NtMBP1 and NtHMGB. (A) Isolation of NtMBP1 and NtHMGB by yeast one-hybrid assay using TM6II-1 or TM6II-2 as a bait, respectively. 1 and 2, negative controls transformed with nonrecombinant plasmid for NtHMGB and NtMBP1, respectively; 5, NtMBP1; 6, NtHMGB; 3 and 4, putative positive clones baited by TM6II-1; 7 and 8, putative positive clones baited by TM6II-2. (B) Subcellular localization of the NtMBP1 protein. The fusion constructs for NtMBP1-GFP and the GFP control plasmid were introduced into onion epidermis cells by biolistic bombardment transformation. The transformed cells were cultured on the MS medium at 28°C for 24 h. For DAPI staining, cells were stained in 0.2 mg·l−1 DAPI for 15 min, washed three times in PBS solution and observed under a fluorescence microscope. Bars = 20 μm. (C) The NtMBP1 mRNA accumulation in different organs. The ethidium bromide-stained total RNA is shown as a loading control.
The NtMBP1 cDNA contains a single ORF of 546 amino acid residues and encodes putative proteins with a predicted molecular mass of 57.2 kDa. Comparison of the amino acid sequences of NtMBP1 and the homologues from other plant species (Supplementary Fig. S4) shows that NtMBP1 consists of a C-terminal domain containing five AT-hook motifs and an additional plant-specific linker histone H1 domain at the N-terminal. To verify the possible role of NtMBP1 protein as a nuclear factor, its subcellular localization was examined. The NtMBP1 ORF was fused to the upstream region of the green fluorescent protein gene to act as a fluorescent marker. 35S-GFP or 35S-NtMBP1-GFP plasmids were introduced into the onion epidermal cells by particle bombardment, and the GFP fluorescence was visualized using fluorescence microscopy. The GFP fluorescence of the control was distributed throughout the cells, whereas that of the NtMBP1-GFP fusion protein was localized in the nuclei (Fig. 6B). These results indicate that NtMBP1 is a nuclear protein. Northern blot analysis was then performed to determine the NtMBP1 expression pattern in tobacco seedlings. NtMBP1 transcripts were found in the leaves, stems, and in the roots (Fig. 6C).
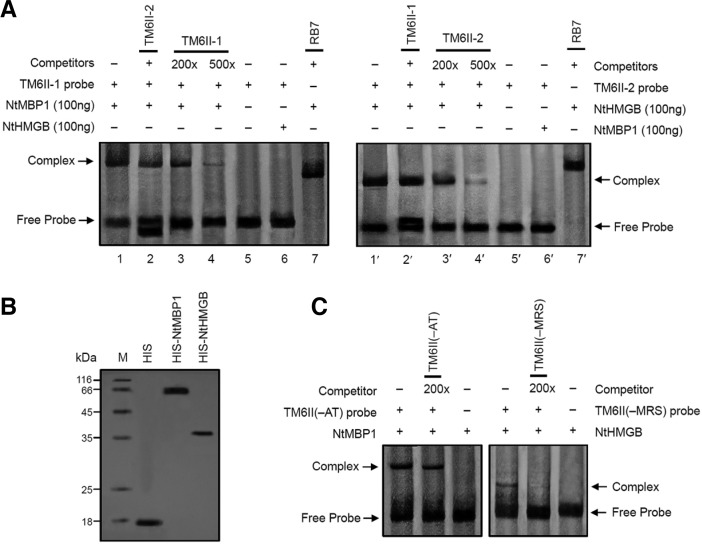
The NtMBP1 and NtHMGB proteins can specifically bind to the TM6 regions
An electrophoretic mobility shift assay (EMSA) to confirm the specific binding of TM6II to NtMBP1 or NtHMGB was performed. Recombinant NtMBP1 and NtHMGB proteins were expressed in Escherichia coli as His-tag fusion proteins and then purified by affinity chromatography (Fig. 7B). The TM6II-1 fragment specifically bound to NtMBP1 but not to NtHMGB, whereas the TM6II-2 fragment specifically bound only to NtHMGB (Fig. 7A). As a negative control, RB7 did not show any binding activity with NtMBP1/NtHMGB alone. Moreover, the removal of MRS from TM6II significantly decreased TM6II binding to NtHMGB. A nearly immobile D-MRS:NtHMGB complex was efficiently out-competed by 200 molar excess of unlabeled D-MRS, indicating that MRS significantly affects the specific binding of NtHMGB to TM6II (Fig. 7C). Deletion of the AT-rich element in TM6II appears to have no significant effect on the binding of NtMBP1 to D-AT, thus implying that other elements in TM6II serve as the NtMBP1 binding site.
Fig. 7.
NtMBP1 and NtHMGB specifically bind to TM6 regions. (A) Interaction of NtMBP1 and NtHMGB proteins with DIG-labeled TM6II-1 and TM6II-2 fragments, respectively. Lane 1/1′, the reaction of TM6II-1/TM6II-2 with NtMBP1/NtHMGB protein; lane 2, the TM6II-1 and NtMBP1 reaction competed with TM6II-2; lane 2′, the TM6II-2 and NtHMGB reaction completed with TM6II-1; lanes 3/3′ and 4/4′, reactions with NtMBP1/NtHMGB protein, plus 200- and 500-fold excess of the unlabelled TM6II-1/TM6II-2, respectively; lane 5/5′, control reaction without NtMBP1/NtHMGB protein; lane 6/6′, control reaction with in vitro expressed NtHMGB/NtMBP1 protein; lanes 7/7′, control reaction with RB7. (B) Identification of NtMBP1 and NtHMGB. The purified proteins (0.1 mg each), NtMBP1 and NtHMGB, were separated on an SDS-polyacryl-amide gel and stained with Coomassie Brilliant Blue (left). The molecular weight marker (M) is indicated on the left-hand side. His-tag was used as the marker. (C) Gel retardation assay of DIG-labeled D-MRS/D-AT probe with NtMBP1/NtHMGB protein. Binding reactions contained no or a molar excess (200-fold) of unlabeled D-MRS and D-AT fragments as competitors, respectively. Details of the electrophoresis are described in Experimental procedures.
DISCUSSION
TM6 acts as a bidirectional enhancer that increases the transcription of its flanking genes
A number of animal studies have shown that MARs can serve as boundaries of chromosomal domains (Gombert et al., 2003; Heng et al., 2001), protecting genes from the cooperative spreading of condensed chromatin or reducing DNA:DNA pairing interactions within complex loci (Stief et al., 1989). MARs dynamically associate with the nuclear matrix by induction of the active transcription within this particular DNA loop to regulate the expression of their flanking genes (Iarovaia et al., 2005). The TM2 MAR, which flanks the gusA-gene cassette and is located downstream of the selective marker (nptII) cassette, enhances β-glucuronidase expression in a bidirectionally efficient manner and improves the transformation frequency by increasing nptII gene expression in transgenic rice and tobacco plants (Xue et al., 2005; Zhang et al., 2004; 2007). In addition, no directional effect of the MARs is observed on the transcription enhancement of the target gene expression (Zhang et al., 2009). RB7 MAR reportedly increases transgene GFP expression by increasing the percentage of cells expressing the trans-gene (likelihood) and increasing the level of cell expression (magnitude) when flanking the GFP cassette and upstream of the selective marker (nptII) cassette (Halweg et al., 2005). The current results clearly show that the presence of a novel potential tobacco MAR, TM6, between the nptII- and gusA-gene cassettes significantly increases the expression of the gusA gene, the number of kanamycin-resistant tobacco shoots, and the survival percentage of transgenic seeds on medium containing high kanamycin concentrations by enhancing the nptII gene expression in transgenic tobacco (Fig. 3). Therefore, TM6 acts as a bidirectional enhancer to increase the average expression levels of its flanking genes, which in turn contributes to the high transformation frequency by enhancing the expression of selective marker genes. Thus, expression vectors that contain MAR are often a critical requirement for the broad use of any gene transfer technique.
MARs increase the flanking gene transcription by affecting the promoter regions
In animals, MARs function as cis-acting elements that promote chromatin accessibility or chromatin remodeling (Fernandez et al., 2001; Petrov et al., 2006) and antagonize the methylation-dependent repression of long-range enhancer-promoter interactions (Chong et al., 2002; Lawton et al., 2008). In plants, MARs in the upstream region of a tobacco basic class I chitinase gene (CHN50) affect the accessibility of the promoter region to MNase (Fukuda and Nishikawa, 2003). In this study, TM6 caused a preferential nuclease accessibility to the adjacent 35S and NOS promoters in transgenic tobacco, which could affect the transcription of gusA and nptII genes by chromatin remodeling (Fig. 4).
TGS is often associated with an increased cytosine methylation level in the affected promoters (Gong et al., 2002). DNA methylation at specific CpG sites in the promoter leads to changes in the DNA-protein interactions at those sites and, subsequently, to the transcriptional repression of the gene (Griswold and Kim, 2001). Non-CpG methylation is also required for TGS in both siRNA-dependent and -independent pathways (Chinnusamy and Zhu, 2009; He et al., 2011). Reduced DNA methylation is clearly associated with the transcriptional activation of specific genes both in animals and plants (Liu et al., 2010; Villagra et al., 2002). Another study showed that the 35S promoter region is a methylation hot spot that can spread methylation toward its upstream and downstream regions (Mishiba et al., 2005). To investigate the mechanism by which TM6 regulates its flanking gene transcription, the methylation frequencies of the 35S and NOS promoter regions were determined. TM6 dramatically decreased the methylation frequencies in both CpG and non-CpG contexts of the 35S and NOS promoters in transgenic tobacco, respectively (Fig. 4C). To the best of the authors’ knowledge, this finding is the first evidence to confirm that MAR affects the DNA methylation frequency in the flanking regions in plants. However, further study is required to clarify the mechanism by which TM6 affects the methylation of adjacent DNA.
MAR and MAR-binding proteins affect the chromatin structure
Chromatin remodeling can be mediated by the interaction between MAR and MAR-binding proteins (Yamasaki et al., 2007). Several MAR-binding proteins have been isolated (Fujimoto et al., 2004; Han et al., 2008; Hatton and Gray, 1999; Meier et al., 1996; Morisawa et al., 2000). In this study, TM6 fragments were used as bait to isolated two MAR-binding proteins by the yeast one- hybrid approach. NtMBP1, an unknown nuclear-located protein, contains a linker histone H1 domain at the N-terminal and five conserved AT-hook motifs at the C-terminal. Interestingly, a unique glutamine-rich region was observed in the middle of NtMBP1. However, further investigation is required (Supplementary Fig. S4). NtHMGB, as a homolog of the AtHMGB protein family, can function as a dynamic chromatin modulator to increase the structural flexibility of DNA, thereby promoting the assembly of nucleoprotein complexes that control DNA-dependent processes, including transcription (Grasser et al., 2006). HMGB proteins can interact with histone H1 on the active chromatin domain to create an open chromatin structure for the transcription of vicinal genes (Cato et al., 2008; El Gazzar et al., 2009).
Site-specific deletion analysis shows that most of the TM6 sequence-mediated transcriptional activation (85.1%) can be attributed to the functional elements in TM6. Moreover, the MRS element, which specifically binds to NtHMGB in vitro, may be the key to the expression of TM6 flanking genes. EMSA results suggest that the MRS element specifically binds to the NtHMGB protein (Fig. 7C). However, NtMBP1 only weakly binds to the AT-rich element in TM6. These results indicate that NtMBP1 can interact with other unidentified cis-elements to influence the chromatin structure. In addition, some chromatin architectural factors, such as helicases and topoisomerases, can bind to their specific sites in the MAR sequences to form an expanding single-strand region (Fiorini et al., 2006). In turn, this single-strand region facilitates the binding of transcription factors to the promoter regions, which leads to the efficient transcription in animals (Zlatanova et al., 2000). To obtain a clearer understanding of the mechanism and function of TM6, NtMBP1 and NtHMGB mutants should be analyzed and the specific binding element of NtMBP1 must be identified. In conclusion, this study shows that the functional elements in MAR sequences respond to specific chromatin architectural factors, such as HMG proteins and topoisomerases, to open nearby chromatin structures and subsequently recruit other structural and transcriptional factors.
Supplementary Material
Acknowledgments
We thank Dr J. Haseloff (MRC Laboratory of Molecular Biology, Cambridge, UK) for the GFP construction pBINmGFP5-ER. This work was supported by the National Basic Research Program (Grant No. 2012CB114200), the National Natural Science Foundation (Grant No. 30970225) and the Genetically Modified Organisms Breeding Major Projects (Grant No. 2011ZX08009-003-002) in China.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Ascenzi R, Ingram JL, Massel M, Thompson WF, Spiker S, Weissinger AK. (2001). The role of cell differentiation state and HMG-I/Y in the expression of transgenes flanked by matrix attachment regions. Transgenic Res. 10, 465–470 [DOI] [PubMed] [Google Scholar]
- Avramova Z, SanMiguel P, Georgieva E, Bennetzen JL. (1995). Matrix attachment regions and transcribed sequences within a long chromosomal continuum containing maize Adh1. Plant Cell 7, 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. (1992). Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science 255, 195–197 [DOI] [PubMed] [Google Scholar]
- Bode J, Goetze S, Heng H, Krawetz SA, Benham C. (2003). From DNA structure to gene expression: mediators of nuclear compartmentalization and dynamics. Chromosome Res. 11, 435–445 [DOI] [PubMed] [Google Scholar]
- Brouwer C, Bruce W, Maddock S, Avramova Z, Bowen B. (2002). Suppression of transgene silencing by matrix attachment regions in maize: a dual role for the maize 5′ ADH1 matrix attachment region. Plant Cell 14, 2251–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cato L, Stott K, Watson M, Thomas JO. (2008). The interaction of HMGB1 and linker histones occurs through their acidic and basic tails. J Mol Biol. 384, 1262–1272 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu JK. (2009). RNA-directed DNA methylation and demethylation in plants. Sci China C Life Sci. 52, 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S, Kontaraki J, Bonifer C, Riggs AD. (2002). A Functional chromatin domain does not resist X chromosome inactivation: silencing of cLys correlates with methylation of a dual promoter-replication origin. Mol Cell Biol. 22, 4667–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Gazzar M, Yoza BK, Chen X, Garcia BA, Young NL, McCall CE. (2009). Chromatin-specific remodeling by HMGB1 and linker histone H1 silences proinflammatory genes during endotoxin tolerance. Mol Cell Biol. 29, 1959–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellies DL, Krumlauf R. (2006). Bone formation: the nuclear matrix reloaded. Cell 125, 840–842 [DOI] [PubMed] [Google Scholar]
- Ellis GC, Grobler-Rabie AF, Hough FS, Bester AJ. (1988). Location and methylation pattern of a nuclear matrix associated region in the human pro alpha 2(I) collagen gene. Biochem Biophys Res Commun. 157, 500–506 [DOI] [PubMed] [Google Scholar]
- Fernandez LA, Winkler M, Grosschedl R. (2001). Matrix attachment region-dependent function of the immunoglobulin mu enhancer involves histone acetylation at a distance without changes in enhancer occupancy. Mol Cell Biol. 21, 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Genger RK, Peacock WJ, Dennis ES. (1998). DNA methylation in plants. Annu Rev Plant Physiol Plant Mol Biol. 49, 223–247 [DOI] [PubMed] [Google Scholar]
- Fiorini A, Gouveia Fde S, Fernandez MA. (2006). Scaffold/Matrix Attachment Regions and intrinsic DNA curvature. Biochemistry (Mosc.) 71, 481–488 [DOI] [PubMed] [Google Scholar]
- Forrester WC, van Genderen C, Jenuwein T, Grosschedl R. (1994). Dependence of enhancer-mediated transcription of the immunoglobulin mu gene on nuclear matrix attachment regions. Science 265, 1221–1225 [DOI] [PubMed] [Google Scholar]
- Forrester WC, Fernandez LA, Grosschedl R. (1999). Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer-promoter interactions. Genes Dev. 13, 3003–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S, Matsunaga S, Yonemura M, Uchiyama S, Azuma T, Fukui K. (2004). Identification of a novel plant MAR DNA binding protein localized on chromosomal surfaces. Plant Mol Biol. 56, 225–239 [DOI] [PubMed] [Google Scholar]
- Fukuda Y, Nishikawa S. (2003). Matrix attachment regions enhance transcription of a downstream transgene and the accessibility of its promoter region to micrococcal nuclease. Plant Mol Biol. 51, 665–675 [DOI] [PubMed] [Google Scholar]
- Gindullis F, Peffer NJ, Meier I. (1999). MAF1, a novel plant protein interacting with matrix attachment region binding protein MFP1, is located at the nuclear envelope. Plant Cell 11, 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod PA, Nguyen DQ, Calabrese D, Puttini S, Grandjean M, Martinet D, Regamey A, Saugy D, Beckmann JS, Bucher P, et al. (2007). Genome-wide prediction of matrix attachment regions that increase gene expression in mammalian cells. Nat Methods 4, 747–753 [DOI] [PubMed] [Google Scholar]
- Gombert WM, Farris SD, Rubio ED, Morey-Rosler KM, Schubach WH, Krumm A. (2003). The c-myc insulator element and matrix attachment regions define the c-myc chromosomal domain. Mol Cell Biol. 23, 9338–9348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814 [DOI] [PubMed] [Google Scholar]
- Grasser M, Lentz A, Lichota J, Merkle T, Grasser KD. (2006). The Arabidopsis genome encodes structurally and functionally diverse HMGB-type proteins. J Mol Biol. 358, 654–664 [DOI] [PubMed] [Google Scholar]
- Griswold MD, Kim JS. (2001). Site-specific methylation of the promoter alters deoxyribonucleic acid-protein interactions and prevents follicle-stimulating hormone receptor gene transcription. Biol Reprod. 64, 602–610 [DOI] [PubMed] [Google Scholar]
- Halweg C, Thompson WF, Spiker S. (2005). The rb7 matrix attachment region increases the likelihood and magnitude of transgene expression in tobacco cells: a flow cytometric study. Plant Cell 17, 418–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. (2008). SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature 452, 187–193 [DOI] [PubMed] [Google Scholar]
- Harder PA, Silverstein RA, Meier I. (2000). Conservation of matrix attachment region-binding filament-like protein 1 among higher plants. Plant Physiol. 122, 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton D, Gray JC. (1999). Two MAR DNA-binding proteins of the pea nuclear matrix identify a new class of DNA-binding proteins. Plant J. 18, 417–429 [DOI] [PubMed] [Google Scholar]
- He XJ, Chen T, Zhu JK. (2011). Regulation and function of DNA methylation in plants and animals. Cell Res. 21, 442–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng HH, Krawetz SA, Lu W, Bremer S, Liu G, Ye CJ. (2001). Re-defining the chromatin loop domain. Cytogenet Cell Genet. 93, 155–161 [DOI] [PubMed] [Google Scholar]
- Heng HH, Goetze S, Ye CJ, Liu G, Stevens JB, Bremer SW, Wykes SM, Bode J, Krawetz SA. (2004). Chromatin loops are selectively anchored using scaffold/matrix-attachment regions. J Cell Sci. 117, 999–1008 [DOI] [PubMed] [Google Scholar]
- Iarovaia OV, Akopov SB, Nikolaev LG, Sverdlov ED, Razin SV. (2005). Induction of transcription within chromosomal DNA loops flanked by MAR elements causes an association of loop DNA with the nuclear matrix. Nucleic Acids Res. 33, 4157–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SY, Rose A, Meier I. (2003). MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res. 31, 5175–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton BR, Carone BR, Obergfell CJ, Ferreri GC, Gondolphi CM, Vandeberg JL, Imumorin I, O’Neill RJ, O’Neill MJ. (2008). Genomic imprinting of IGF2 in marsupials is methylation dependent. BMC Genomics 9, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Dahiya R. (2002). MethPrimer: designing primers for methylation PCRs. Bioinformatics 18, 1427–1431 [DOI] [PubMed] [Google Scholar]
- Li J, Brunner AM, Meilan R, Strauss SH. (2008). Matrix attachment region elements have small and variable effects on transgene expression and stability in field-grown Populus. Plant Biotechnol J. 6, 887–896 [DOI] [PubMed] [Google Scholar]
- Liu Q, Wang J, Miki D, Xia R, Yu W, He J, Zheng Z, Zhu JK, Gong Z. (2010). DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell 22, 2336–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loc PV, Stratling WH. (1988). The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 7, 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderus ME, de Graaf A, Mattia E, den Blaauwen JL, Grande MA, de Jong L, van Driel R. (1992). Binding of matrix attachment regions to lamin B1. Cell 70, 949–959 [DOI] [PubMed] [Google Scholar]
- Meier I, Phelan T, Gruissem W, Spiker S, Schneider D. (1996). MFP1, a novel plant filament-like protein with affinity for matrix attachment region DNA. Plant Cell 8, 2105–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. (2000). Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19, 5194–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiba K, Nishihara M, Nakatsuka T, Abe Y, Hirano H, Yokoi T, Kikuchi A, Yamamura S. (2005). Consistent transcriptional silencing of 35S-driven transgenes in gentian. Plant J. 44, 541–556 [DOI] [PubMed] [Google Scholar]
- Mlynarova L, Jansen RC, Conner AJ, Stiekema WJ, Nap JP. (1995). The MAR-mediated reduction in position effect can be uncoupled from copy number-dependent expression in transgenic plants. Plant Cell 7, 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarova L, Hricova A, Loonen A, Nap JP. (2003). The presence of a chromatin boundary appears to shield a trans-gene in tobacco from RNA silencing. Plant Cell 15, 2203–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisawa G, Han-Yama A, Moda I, Tamai A, Iwabuchi M, Meshi T. (2000). AHM1, a novel type of nuclear matrix-localized, MAR binding protein with a single AT hook and a J domain-homologous region. Plant Cell 12, 1903–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namciu SJ, Fournier RE. (2004). Human matrix attachment regions are necessary for the establishment but not the maintenance of transgene insulation in Drosophila melanogaster. Mol Cell Biol. 24, 10236–10245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua NH. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313, 810–812 [DOI] [PubMed] [Google Scholar]
- Paul AL, Ferl RJ. (1998). Higher order chromatin structures in maize and Arabidopsis. Plant Cell 10, 1349–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. (2007). DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA 104, 6752–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A, Pirozhkova I, Carnac G, Laoudj D, Lipinski M, Vassetzky YS. (2006). Chromatin loop domain organization within the 4q35 locus in facioscapulohumeral dystrophy patients versus normal human myoblasts. Proc Natl Acad Sci USA 103, 6982–6987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaart MJ, Recillas-Targa F, Felsenfeld G. (1998). Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 12, 2852–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purbowasito W, Suda C, Yokomine T, Zubair M, Sado T, Tsutsui K, Sasaki H. (2004). Large-scale identification and mapping of nuclear matrix-attachment regions in the distal imprinted domain of mouse chromosome 7. DNA Res. 11, 391–407 [DOI] [PubMed] [Google Scholar]
- Rao S, Procko E, Shannon MF. (2001). Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J Immunol. 167, 4494–4503 [DOI] [PubMed] [Google Scholar]
- Shan DP, Huang JG, Yang YT, Guo YH, Wu CA, Yang GD, Gao Z, Zheng CC. (2007). Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytol. 176, 70–81 [DOI] [PubMed] [Google Scholar]
- Shewchuk BM, Cooke NE, Liebhaber SA. (2001). The human growth hormone locus control region mediates long-distance transcriptional activation independent of nuclear matrix attachment regions. Nucleic Acids Res. 29, 3356–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko L, Bruce W, Maddock S, Tagliani L, Li X, Daniels M, Peterson T. (2003). Functional analysis of two matrix attachment region (MAR) elements in transgenic maize plants. Transgenic Res. 12, 137–154 [DOI] [PubMed] [Google Scholar]
- Stief A, Winter DM, Stratling WH, Sippel AE. (1989). A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature 341, 343–345 [DOI] [PubMed] [Google Scholar]
- Tikhonov AP, Bennetzen JL, Avramova ZV. (2000). Structural domains and matrix attachment regions along colinear chromosomal segments of maize and sorghum. Plant Cell 12, 249–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villagra A, Gutierrez J, Paredes R, Sierra J, Puchi M, Imschenetzky M, van Wijnen A, Lian J, Stein G, Stein J, et al. (2002). Reduced CpG methylation is associated with transcriptional activation of the bone-specific rat osteocalcin gene in osteoblasts. J Cell Biochem. 85, 112–122 [PubMed] [Google Scholar]
- Wolffe AP. (1995). Chromatin: structure and function. 2nd ed., (London: Academic Press; ). [Google Scholar]
- Xue H, Yang YT, Wu CA, Yang GD, Zhang MM, Zheng CC. (2005). TM2, a novel strong matrix attachment region isolated from tobacco, increases transgene expression in transgenic rice calli and plants. Theor Appl Genet. 110, 620–627 [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Akiba T, Yamasaki T, Harata K. (2007). Structural basis for recognition of the matrix attachment region of DNA by transcription factor SATB1. Nucleic Acids Res. 35, 5073–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Liu P, Wu CA, Yang GD, Xu R, Guo QH, Huang JG, Zheng CC. (2012). Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol Cell 48, 521–531 [DOI] [PubMed] [Google Scholar]
- Yang YT, Yu YL, Yang GD, Zhang JD, Zheng CC. (2009). Tissue-specific expression of the PNZIP promoter is mediated by combinatorial interaction of different cis-elements and a novel transcriptional factor. Nucleic Acids Res. 37, 2630–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KW, Wang JM, Zheng CC. (2004). The potential role of nuclear matrix attachment regions (MARs) in regulation of gene expression. Sheng Wu Gong Cheng Xue Bao 20, 6–9 [PubMed] [Google Scholar]
- Zhang MM, Ji LS, Xue H, Yang YT, Wu CA, Zheng CC. (2007). High transformation frequency of tobacco and rice via Agrobacterium-mediated gene transfer by flanking a tobacco matrix attachment region. Physiol Plant 129, 644–651 [Google Scholar]
- Zhang J, Lu L, Ji L, Yang G, Zheng C. (2009). Functional characterization of a tobacco matrix attachment region-mediated enhancement of transgene expression. Transgenic Res. 18, 377–385 [DOI] [PubMed] [Google Scholar]
- Zheng CC, Porat R, Lu P, O’Neill SD. (1998). PNZIP is a novel mesophyll-specific cDNA that is regulated by phytochrome and the circadian rhythm and encodes a protein with a leucine zipper motif. Plant Physiol. 116, 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, Caiafa P, Van Holde K. (2000). Linker histone binding and displacement: versatile mechanism for transcriptional regulation. FASEB J. 14, 1697–1704 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.