Abstract
During skeletal development, both osteogenic and chondrogenic programs are initiated from multipotent mesenchymal cells, requiring a number of signaling molecules, transcription factors, and downstream effectors to orchestrate the sophisticated process. Col10a1, an important downstream effector gene, has been identified as a marker for maturing chondrocytes in higher vertebrates, such as mammals and birds. In zebrafish, this gene has been shown to be expressed in both osteoblasts and chondrocytes, but no study has reported its role in osteoblast development. To initially delineate the osteogenic program from chondrogenic lineage development, we used the zebrafish col10a1 promoter to establish a transgenic zebrafish expressing a GFP reporter specifically in osteoblast-specific bone structures that do not involve cartilaginous programs. A construct harboring a ∼2.2-kb promoter region was found to be sufficient to drive the reporter gene in osteoblast-specific bone structures within the endogenous col10a1 expression domain, confirming that separable cis-acting elements exist for distinct cell type-specific expression of col10a1 during zebrafish skeletal development. The ∼2.2-kb col10a1:GFP transgenic zebrafish marking only bone structures derived from osteoblasts will undoubtedly be an invaluable tool for identifying and characterizing molecular events driving osteoblast development in zebrafish, which may further provide a differential mechanism where col10a1 is involved in the development of chondrocytes undergoing maturation in other vertebrate systems.
Keywords: bone, col10a1, osteoblast, model system, transgenic zebrafish
INTRODUCTION
Collagen type 10A1 (COL10A1) is a short alpha-chain collagen and is considered as a reliable marker for chondrocyte maturation in higher vertebrates [reviewed in (Komori, 2010; van der Kraan and van den Berg, 2012)]. Due to its expression in hypertrophic chondrocytes, which are closely related to osteoarthritis, much attention has been drawn to the regulation of COL10A1. Analyses of the Col10a1 promoter sequences in higher vertebrates have shown that the promoter contains multiple binding sites for various transcription factors that may collectively act to induce cell type-specific col10a1 expression (Beier et al., 1996; 1997; Dourado and LuValle, 1998; Jacenko et al., 1993). One of the best-studied upstream regulators of COL10A1 is RUNX2, a master regulator that directs pluripotent mesenchymal cells toward development into osteoblast and chondrocyte lineages. Numerous studies have shown the dynamic nature of RUNX2 expression during skeletal development in higher vertebrates, including birds and mammals (Drissi et al., 2003; Enomoto et al., 2000; Higashi-kawa et al., 2009; Li et al., 2011; Zheng et al., 2003; 2009). During chondrocyte maturation, RUNX2 upregulates COL10A1 expression by directly binding to core sequences located in the promoter, showing that the RUNX2-COL10A1 pathway is an indispensable step that leads to chondrocyte hypertrophy (van der Kraan and van den Berg, 2012). During osteoblast differentiation, RUNX2 is initially required to specify the osteoblast lineage program but needs to be downregulated during osteoblast maturation (Komori, 2010). Consistent with its role in skeletal development, Runx2-knockout mice develop a cartilage-only skeleton with no bone components (Komori et al., 1997; Otto et al., 1997). In addition to RUNX2 as a positive regulator, other transcription factors such as c-FOS and DLX also regulate Col10a1 expression in a tissue-specific manner (Hassan et al., 2004; Riemer et al., 2002). Therefore, detailed analysis on the regulation of Col10a1 expression in both osteogenic and chondrogenic lineages may provide valuable information regarding the upstream regulators of Col10a1 acting specifically to induce either bone structures or cartilages.
In contrast to the pattern seen in tetrapods, col10a1 expression is detected in both chondrocytes and osteoblasts in zebrafish (Eames et al., 2012). Along with the advantages of using zebrafish as a model organism, the generation of a transgenic zebrafish that expresses a reporter gene in a lineage should facilitate the identification of the molecular pathways leading to osteoblast- and chondrocyte-specific differentiation.
In this study, we generated a stable transgenic zebrafish by using the col10a1 promoter that drives GFP reporter expression in bone structures derived directly from osteoblasts. Since the endogenous zebrafish col10a1 is uniquely expressed in both osteoblasts and chondrocytes, this bone-specific transgenic zebrafish will provide a useful tool for identifying molecular pathways that regulate osteoblast-specific programs not involving transient chondrocyte development.
MATERIALS AND METHODS
Constructs
To isolate the zebrafish col10a1 promoter, we searched for the col10a1 gene sequence in a genome database (http://www.ensembl.org/index.html). Based on the sequence, a forward primer (5′-GGTCGACGCAATGACAAGAACCTGTTCTTGGCC-3′) and a reverse primer (5′-GGATCCACCTACCGGCATACTTGGGTTCCA-3′) were designed, and the ∼2.2-kb promoter region was amplified using a standard PCR protocol. The PCR product was inserted into the pGEM T-easy vector (Promega), and the insert was recovered and subcloned into the mini Tol2 mGFP vector (detailed information and protocol regarding construct construction are available upon request) (Jung et al., 2010; Kim et al., 2008; Park et al., 2000).
Zebrafish care and transgenesis
The zebrafish and their embryos were handled (Jung et al., 2012) and staged according to a standard protocol (Kimmel et al., 1995). Mini Tol2 col10a1:mGFP constructs were injected together with transposase mRNA at the 1-cell stage of zebrafish embryos, as previously described. The injected embryos were selected based on transient GFP expression, and the sorted embryos were raised to sexual maturity. Each zebrafish was outcrossed to wild-type zebrafish, and the resulting embryos were selected again based on the GFP expression in the bone structures (F1). The F3- and F4-generation embryos were analyzed in this study.
Alizarin Red staining
Embryos at various developmental stages were fixed with 4% paraformaldehyde at 4°C overnight and washed 3 times with PBT. The pigments were removed with a bleaching solution (1% KOH, 3% H2O2 in PBT), and the embryos were rinsed 3 times with PBT. The samples were processed for Alizarin Red staining (0.4 ml of Alizarin Red S solution in 10 ml of 0.5% KOH) overnight at room temperature. Alizarin Red S solution was obtained by dissolving 0.1 g of Alizarin Red (Sigma, A5533) in 100 ml of 96% ethanol. After developing Alizarin Red bone staining, the background staining was removed by treating samples with 1% KOH overnight at room temperature.
In situ hybridization
In situ hybridization was performed as described previously (Choe et al., 2009). The col10a1 antisense probe was synthesized using a standard protocol.
RESULTS
Zebrafish col10a1 is expressed in all bone structures during development
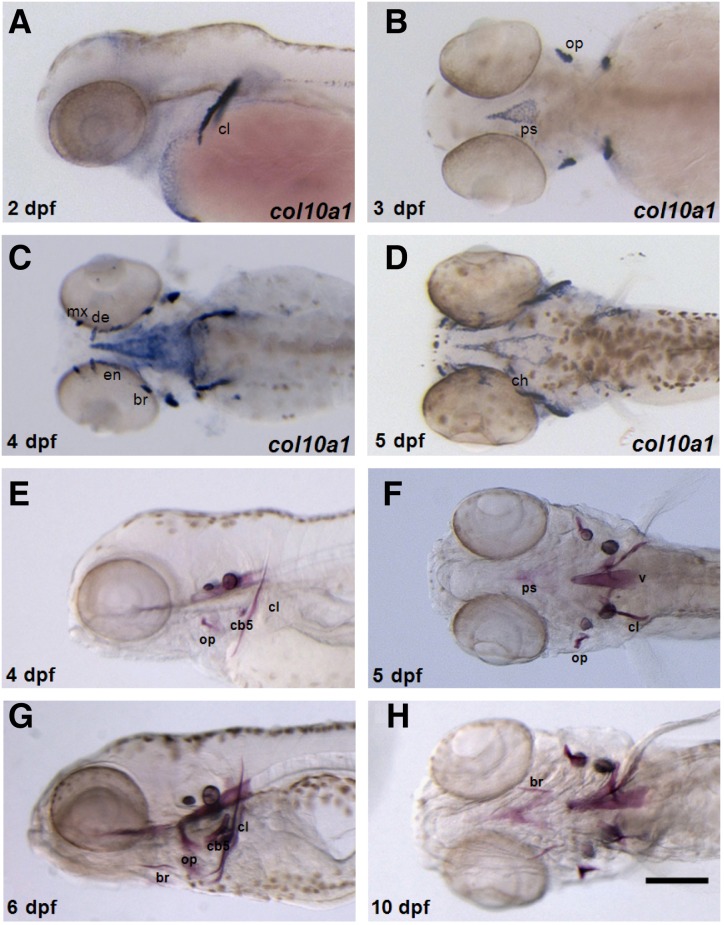
A previous study suggested that col10a1 is expressed in both chondrocyte and osteoblast lineages during zebrafish development (Eames et al., 2012). In order to confirm col10a1 expression in zebrafish, we performed in situ hybridization by using various stages of developing zebrafish embryos. At 2 days post fertilization (dpf), col10a1 expression was first detected in the cleithrum (cl), which is formed by intramembranous ossification (Fig. 1A). This expression was maintained during the later stages of development. At 3 dpf, the intramembranous opercle (op) and parasphenoid (ps) showed col10a1 expression, which was also maintained during the later stages of development (Fig. 1B). At 4 dpf, col10a1 expression was also seen in the maxilla (mx), dentary (de), branchiostegal ray (br), and entopterygoid (en) (Fig. 1C). At 5 dpf, col10a1 expression was additionally detected in the cartilaginous ceratohyal (ch) and hyomandibula (hm) (Fig. 1D). Thus, col10a1 expression was found in both osteoblasts and chondroblasts in zebrafish. Next, we performed Alizarin Red staining to determine the extent to which col10a1 expression is related to osteoblast differentiation. Alizarin Red-positive signals seemed to be somewhat delayed by 1–2 days after initial col10a1 expression, but the signals became visible in bones belonging to a subset of col10a1-expressing structures (Figs. 1E–1H). These results suggest a strong correlation between col10a1 expression and bone induction during zebrafish development.
Fig. 1.
Spatiotemporal col10a1 expression and bone formation in developing zebrafish embryos. (A–D) Endogenous col10a1 expression was visualized by in situ hybridization. Embryos are shown in lateral (A) or ventral views (B–D) with the anterior region to the left. (A) col10a1 is expressed in the cleithrum at 2 dpf. (B) At 3 dpf, col10a1 expression is also found in the parasphenoid and opercle. (C) col10a1 expression is present in the maxilla, branchiostegal ray, and entopterygoid at 4 dpf. (D) At 5 dpf, col10a1 expression is also found in the cartilaginous ceratohyal. (E–H) Alizarin Red staining showed the head skeleton. Embryos are shown in the lateral (E, G) or ventral views (F, H) with the anterior region to the left. (E) At 4 dpf, the cleithrum, opercle, and ceratobranchial 5 bone structures are visible. At 5 dpf, the parasphenoid bone is formed (F), and at 6 dpf, the branchiostegal rays are visualized (G). These bone structures are detected at 10 dpf (H). cl, cleithrum; op, opercle; ps, parasphenoid; v, vertebrate; br, branchiostegal ray; mx, maxilla; en, entopterygoid; ch, ceratohyal; hm, hyomandibula; cb5, ceratobranchial 5. Scale bar, 200 μm.
The zebrafish col10a1 promoter was used to generate constructs for producing a transgenic zebrafish line to study osteoblast lineage development
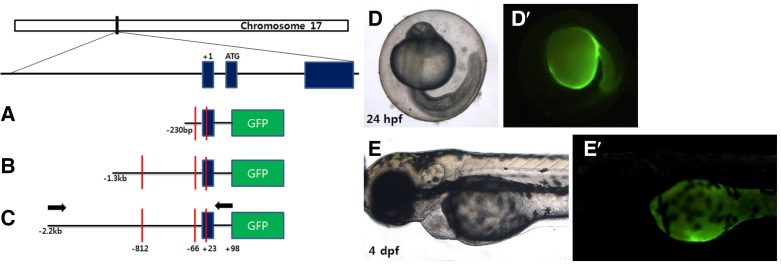
To identify the col10a1 promoter region responsible for osteoblast-specific gene expression in zebrafish, we first analyzed the nucleotide sequence of the col10a1 promoter. As reported previously (Simoes et al., 2006), we found that the col10a1 promoter contains 2 Runx2 consensus binding sequences (PuACCPu-CA/TGTGGT) upstream of the predicted transcriptional start site (TSS, −812 and −66 positions) and an additional binding site at the +23 position from the TSS (Fig. 2). Moreover, we also identified other consensus sequences for transcription factors implicated in skeletal development (e.g., RAR/RXR and ERE). Based on this information obtained from the amino acid sequence and on the fact that Runx2 is essentially required for normal osteoblast development, we generated several transgenic constructs by using different lengths of the col10a1 promoter regions, including the putative Runx2-binding sites (Fig. 2): the first construct contained the immediate proximal promoter containing 2 Runx2-binding sites at −66 and +23; the second construct included an ∼1.3-kb promoter sequence containing 3 binding sites at −812, −66, and +23 bp; and the third construct harbored an ∼2.2-kb region containing the same Runx2-binding sites as those present in the second construct and an additional upstream ∼900-bp fragment. It has been suggested that the col10a1 promoter contains positive and negative elements for its gene expression in vitro (Simoes et al., 2006); therefore, these constructs were used to test whether different parts of the col10a1 promoter are necessary to render promoter activity in the bone primordia.
Fig. 2.
Schematic view of the transgenic constructs. Col10a1 is located on chromosome 17 and is composed of 3 exons with the translational start codon present in exon 2 as depicted. The first construct contains 2 predicted Runx2-binding sites and includes the col10a1 promoter region from the −230 position to the +98 position, relative to the transcriptional start site (TSS) fused to the GFP open reading frame (A). The second construct contains 3 putative Runx2-binding sites and includes a ∼1.3-kb promoter fragment (B). The third construct contains 3 Runx2-binding sites and an additional ∼900-bp region upstream of the second construct, yielding an ∼2.2-kb col10a1 promoter region (C). The arrows in (C) indicate a primer set to initially isolate the promoter region by PCR. Runx2-binding sites are marked by red bars. (D–E’) Microinjection of transposase mRNA together with the construct (A) or (B) did not induce transient GFP expression at 1 dpf or 4 dpf as indicated.
Transient GFP expression with various promoter lengths helped identify the col10a1 promoter domain responsible for col10a1 expression in zebrafish bone
Since Tol2-based transgenesis has been reported to effectively introduce a transgene in the zebrafish genome, resulting in germ-line transmission, we used the Tol2 backbone in our constructs to determine their ability to induce the expression of the GFP reporter (Figs. 2A–2C). Each construct along with transposase mRNA was microinjected into the 1-cell stage of zebrafish embryos, and transient GFP expression was analyzed to test whether the expression of the GFP reporter recovered within the endogenous col10a1 expression domain during zebrafish development.
Embryos injected with the first construct harboring an immediate proximal promoter region (from approximately −230 bp to +98) did not induce any GFP expression when analyzed at ∼24 h post fertilization (hpf) and 4 dpf during development (Figs. 2A and 2D). Similarly, the second ∼1.3-kb construct containing the promoter region with 3 predicted Runx2-binding sites also seemed to be inactive because no GFP expression was detected in the injected embryos (Figs. 2B and 2E). Since both constructs contain a promoter region with at least 2 Runx2 consensus binding sequences that are thought to be important for col10a1 expression, it was somewhat surprising to detect no obvious GFP-expressing structures during the entire course of embryonic development.
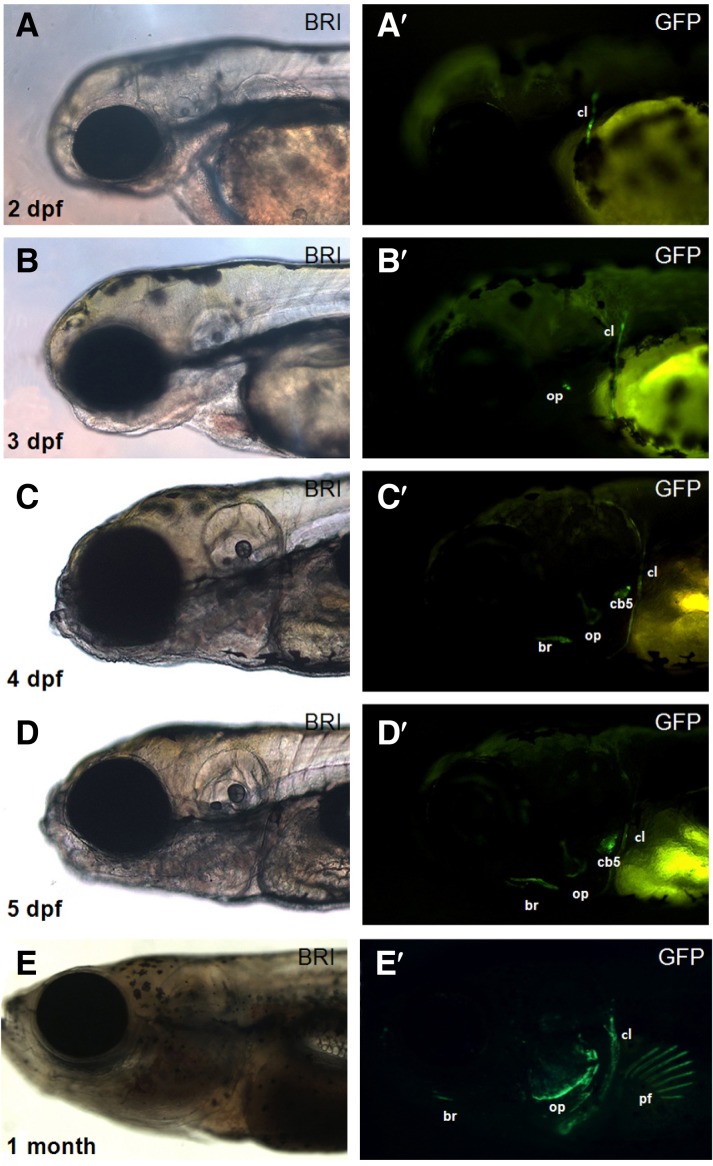
In contrast, the third construct (∼2.2-kb col10a1:GFP), which contained 3 Runx2-binding sequences and an additional 900-bp region upstream of the second construct, induced robust GFP expression in the cl at 2 dpf and in the op at 3 dpf (Figs. 3A and 3B). GFP expression in the ceratobranchial (cb) and branchiostegal ray (br) was also evident at 4 dpf (Figs. 3C and 3D). Importantly, this GFP expression pattern was maintained at least until the embryos were 1 month old (Fig. 3E). Although some embryos showed ectopic GFP expression in non-col10a1 domains such as the pectoral fin and dorsal fin (Fig. 3E), the GFP expression pattern in most embryos injected with the ∼2.2-kb col10a1:GFP construct was strikingly similar to the endogenous col10a1 expression in the bone structures of developing zebrafish, suggesting that an early genome integration event of the introduced construct had occurred to activate the bone-specific ∼2.2-kb col10a1 promoter.
Fig. 3.
Transient GFP expression generated using the ∼2.2-kb col10a1:GFP construct in developing zebrafish embryos. (A–D) A bright field image and an image showing GFP expression are shown as a pair for embryos at each stage. The ∼2.2-kb col10a1 promoter induces GFP expression in the cl at 2 dpf (A, A′); in the cl and op at 3 dpf (B, B′); and in the cl, op, br, and cb5 at 4 dpf (C-D′). This expression pattern was maintained until the zebrafish was at least 1 month old (E). Ectopic GFP expression, for example, in the pectoral fins, was detected in some embryos (E′). cl, cleithrum; op, opercle; br, branchiostegal ray; cb5, ceratobranchial 5; pf, pectoral fin.
We noted that the ∼2.2-kb construct contained 3 putative Runx2-binding sequences that were also present in the ∼1.3-kb construct and that only the ∼2.2-kb construct induced GFP expression in transient transgenic zebrafish. Since Runx2 has been found to be essentially required for both bone and cartilage development, this result suggests that Runx2 may not be the sole regulator but requires other transcription factors (or positive cis-regulatory elements) to induce col10a1 expression in osteoblast-associated structures in zebrafish, further emphasizing the collective roles of various regulators in directing cell-type specific col10a1 expression. Nonetheless, all the constructs that we used failed to generate col10a1-dependent GFP expression in chondrogenic tissues in developing zebrafish, suggesting that cartilage-specific col10a1 expression may be suppressed or not sufficiently activated in our constructs.
Transgenic zebrafish with the ∼2.2-kb col10a1:GFP construct expressed GFP in the region where endogenous col10a1 was also expressed
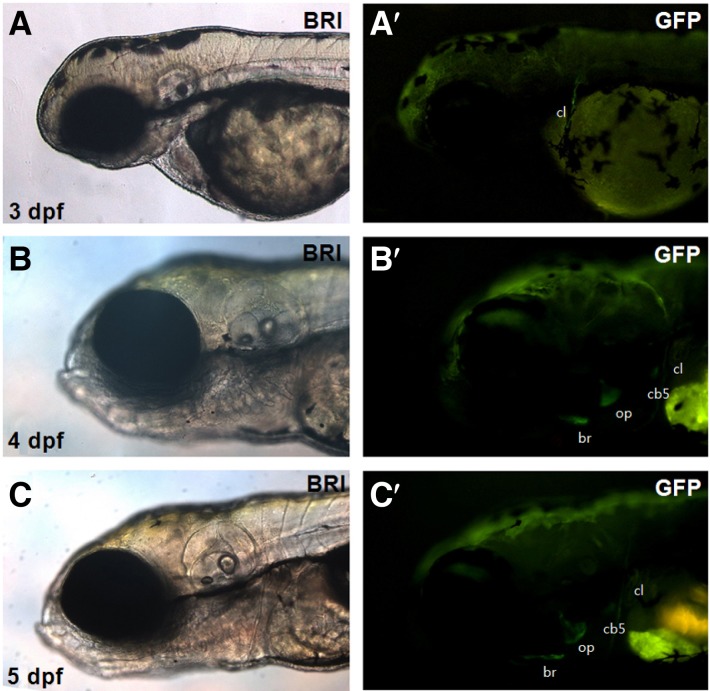
Next, we examined GFP expression in the stable transgenic zebrafish (F3 and F4) in which the third construct (i.e., ∼2.2-kb col10a1:GFP) was integrated in the genome. In particular, we compared the expression pattern of the GFP transgene with that of the endogenous col10a1 in bone structures. We observed strong GFP expression starting at 3 dpf in the cl, op, br, and cb where endogenous Col10a1 presumably acts to induce bone formation directly from osteoblasts (Fig. 4) (Cubbage, 1996; Li et al., 2009). This GFP expression pattern was faithfully maintained in the same structures during the entire course of development (Fig. 4 and not shown), and, importantly, the ectopic GFP expression in the pectoral and dorsal fin regions observed in the transient transgenic zebrafish completely disappeared in the stable transgenic line. We noted that GFP expression in the transgenic zebrafish is restricted to a subset of endogenous col10a1 expression domains and that all the GFP-expressing structures did not originate from the chondrocyte-associated skeleton but were derived directly from osteoblasts.
Fig. 4.
GFP expression in the stable transgenic zebrafish with the ∼2.2-kb col10a1:GFP. Expression of the GFP reporter in the stable transgenic zebrafish with col10a1:GFP was detected in the cl at 3 dpf (A-A′); in the cl, op, br, and cb5 at 4 dpf (B-B′); and in the cl, op, br, and cb5 at 5 dpf (C-C′). Images taken from both the bright and fluorescent field channels are shown as a pair at each stage. cl, cleithrum; op, opercle; br, branchiostegal ray; cb5, ceratobranchial 5.
Taken together, we have generated a bone-specific transgenic zebrafish by using the ∼2.2-kb col10a1 promoter that drives GFP expression in a subset of endogenous col10a1-expressing structures. Unlike other vertebrates such as mammals and birds, col10a1 expression is atypically observed in osteoblasts during zebrafish development. Furthermore, no study describing the exact role of col10a1 during osteogenic lineage development has been published yet. Therefore, our col10a1:GFP transgenic zebrafish will provide a useful tool for identifying upstream regulators and/or downstream effectors of col10a1 acting specifically on osteogenic pathways during zebrafish development.
DISCUSSION
Previous studies have suggested that the col10a1 promoter of vertebrates may contain several components that direct its tissue-specific gene expression, such as the proximal basal regulatory region, and negative and positive regulatory elements (Higashikawa et al., 2009; Li et al., 2011; Simoes et al., 2006; Zheng et al., 2003). Our study also supports the existence of different regulatory elements within the zebrafish col10a1 promoter at least for the osteoblast-specific program. In particular, the use of a transgenic construct containing an ∼2.2-kb fragment of the col10a1 promoter successfully induced the expression of the GFP reporter in osteoblast-specific bone structures, while other constructs (both the ∼1.3-kb fragment and the immediate proximal promoter region alone) did not induce the expression of the GFP reporter (Figs. 2 and 3). Therefore, the zebrafish col10a1 promoter may contain a positive regulatory element between −2.1 and −1.2 kb from the transcriptional start site for activating its unique expression in the osteoblast lineage. Additionally, a negative element present in the −1.2-kb region may act when no positive signal is delivered, which could be a general mechanism for suppressing col10a1 expression in most cell types. Consistent with this possibility, the ∼1.1-kb col10a1 promoter (similar to our ∼1.3-kb construct) showed only a small amount of transactivation of the luciferase reporter in the Xenopus A6 kidney epithelial cell line (Higashikawa et al., 2009; Li et al., 2011; Simoes et al., 2006; Zheng et al., 2003). Therefore, tissue-specific col10a1 expression in vertebrates may depend on the activity of different elements within the promoter. Further investigation on the molecular interplay among the transcription factors implicated in osteoblast differentiation, including Osterix, β-catenin, HDACs, Twist1, Twist2, ATF4, and Dlx, should shed light on the mechanism underlying the regulation of col10a1 expression during development.
The zebrafish has emerged as a powerful developmental model organism due to the ease of microscopic observations at the embryonic level as well as other advantages in cellular and molecular studies. In recent years, numerous chemical screens have been performed using developing embryos of zebrafish, exemplifying its value as a live model organism [reviewed in (Lieschke and Currie, 2007)]. The transgenic zebrafish generated in this study will provide an invaluable tool in addition to the previously-reported bone-specific transgenic zebrafish lines that are used to dissect the molecular pathways involved in osteogenic activities (Hammond and Moro, 2012). Since endogenous col10a1 is expressed in both osteoblasts and chondrocytes in zebrafish but only in chondrocytes in mammals, our bone-specific transgenic zebrafish can be used, for example, to delineate the molecular regulators acting in osteoblast function/differentiation from those in chondrocyte development. Recently, a col10a1 transgenic line whose GFP expression is almost similar to endogenous col10a1 was reported (Mitchell et al., 2013). Since reporter expression was also found in chondrocytes, this col10a1 transgenic line could be used to develop an osteoarthritis model, which completely distinguishes the use of the transgenic zebrafish from the one generated in this study.
In summary, we have established a transgenic zebrafish by using a defined length of the col10a1 promoter that drives the GFP reporter in zebrafish head bones induced specifically from osteoblasts.
Acknowledgments
We thank Dr. K. Kawakami for providing the Tol2 transposon vector. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. 2011-0030719). The authors declare no conflict of interests.
REFERENCES
- Beier F, Lammi MJ, Bertling W, von der Mark K. (1996). Transcriptional regulation of the human type X collagen gene expression. Ann N Y Acad Sci. 785, 209–211 [DOI] [PubMed] [Google Scholar]
- Beier F, Vornehm S, Poschl E, von der Mark K, Lammi MJ. (1997). Localization of silencer and enhancer elements in the human type X collagen gene. J Cell Biochem. 66, 210–218 [DOI] [PubMed] [Google Scholar]
- Choe SK, Lu P, Nakamura M, Lee J, Sagerstrom CG. (2009). Meis cofactors control HDAC and CBP accessibility at Hox-regulated promoters during zebrafish embryogenesis. Dev Cell 17, 561–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubbage CC, Mabee PM. (1996). Development of the cranium and paired fins in the zebrafish Danio rerio (Ostariophysi, Cyprinidae). J Morphol. 229, 121–160 [DOI] [PubMed] [Google Scholar]
- Dourado G, LuValle P. (1998). Proximal DNA elements mediate repressor activity conferred by the distal portion of the chicken collagen X promoter. J Cell Biochem. 70, 507–516 [DOI] [PubMed] [Google Scholar]
- Drissi MH, Li X, Sheu TJ, Zuscik MJ, Schwarz EM, Puzas JE, Rosier RN, O’Keefe RJ. (2003). Runx2/Cbfa1 stimulation by retinoic acid is potentiated by BMP2 signaling through interaction with Smad1 on the collagen X promoter in chondrocytes. J Cell Biochem. 90, 1287–1298 [DOI] [PubMed] [Google Scholar]
- Eames BF, Amores A, Yan YL, Postlethwait JH. (2012). Evolution of the osteoblast: skeletogenesis in gar and zebrafish. BMC Evol Biol. 12, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto H, Enomoto-Iwamoto M, Iwamoto M, Nomura S, Himeno M, Kitamura Y, Kishimoto T, Komori T. (2000). Cbfa1 is a positive regulatory factor in chondrocyte maturation. J Biol Chem. 275, 8695–8702 [DOI] [PubMed] [Google Scholar]
- Hammond CL, Moro E. (2012). Using transgenic reporters to visualize bone and cartilage signaling during development in vivo. Front Endocrinol. 3, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. (2004). Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol. 24, 9248–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashikawa A, Saito T, Ikeda T, Kamekura S, Kawamura N, Kan A, Oshima Y, Ohba S, Ogata N, Takeshita K, et al. (2009). Identification of the core element responsive to runt-related transcription factor 2 in the promoter of human type X collagen gene. Arthritis Rheum. 60, 166–178 [DOI] [PubMed] [Google Scholar]
- Jacenko O, LuValle PA, Olsen BR. (1993). Spondylometaphyseal dysplasia in mice carrying a dominant negative mutation in a matrix protein specific for cartilage-to-bone transition. Nature 365, 56–61 [DOI] [PubMed] [Google Scholar]
- Jung SH, Kim S, Chung AY, Kim HT, So JH, Ryu J, Park HC, Kim CH. (2010). Visualization of myelination in GFP-transgenic zebrafish. Dev Dyn. 239, 592–527 [DOI] [PubMed] [Google Scholar]
- Jung SH, Kim HS, Ryu JH, Gwak JW, Bae YK, Kim CH, Yeo SY. (2012). Her4-positive population in the tectum opticum is proliferating neural precursors in the adult zebrafish brain. Mol Cells 33, 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Kang KH, Kim CH, Choi SY. (2008). Real-time imaging of mitochondria in transgenic zebrafish expressing mitochondrially targeted GFP. Biotechniques 45, 331–334 [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. (1995). Stages of embryonic development of the zebrafish. Dev Dyn. 203, 253–310 [DOI] [PubMed] [Google Scholar]
- Komori T. (2010). Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 339, 189–195 [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al. (1997). Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- Li N, Felber K, Elks P, Croucher P, Roehl HH. (2009). Tracking gene expression during zebrafish osteoblast differentiation. Dev Dyn. 238, 459–466 [DOI] [PubMed] [Google Scholar]
- Li F, Lu Y, Ding M, Napierala D, Abbassi S, Chen Y, Duan X, Wang S, Lee B, Zheng Q. (2011). Runx2 contributes to murine Col10a1 gene regulation through direct interaction with its cis-enhancer. J Bone Miner Res. 26, 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. (2007). Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 8, 353–367 [DOI] [PubMed] [Google Scholar]
- Mitchell RE, Huitema LF, Skinner RE, Brunt LH, Severn C, Schulte-Merker S, Hammond CL. (2013). New tools for studying osteoarthritis genetics in zebrafish. Osteoarthritis Cartilage 21, 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, et al. (1997). Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89, 765–771 [DOI] [PubMed] [Google Scholar]
- Park HC, Kim CH, Bae YK, Yeo SY, Kim SH, Hong SK, Shin J, Yoo KW, Hibi M, Hirano T, et al. (2000). Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev Biol. 227, 279–293 [DOI] [PubMed] [Google Scholar]
- Riemer S, Gebhard S, Beier F, Poschl E, von der Mark K. (2002). Role of c-fos in the regulation of type X collagen gene expression by PTH and PTHrP: localization of a PTH/PTHrP-responsive region in the human COL10A1 enhancer. J Cell Biochem. 86, 688–699 [DOI] [PubMed] [Google Scholar]
- Simoes B, Conceicao N, Viegas CS, Pinto JP, Gavaia PJ, Hurst LD, Kelsh RN, Cancela ML. (2006). Identification of a promoter element within the zebrafish colXalpha1 gene responsive to runx2 isoforms Osf2/Cbfa1 and til-1 but not to pebp2-alphaA2. Calcif Tissue Int. 79, 230–244 [DOI] [PubMed] [Google Scholar]
- van der Kraan PM, van den Berg WB. (2012). Chondrocyte hypertrophy and osteoarthritis: role in initiation and progression of cartilage degeneration?. Osteoarthritis Cartilage 20, 223–232 [DOI] [PubMed] [Google Scholar]
- Zheng Q, Zhou G, Morello R, Chen Y, Garcia-Rojas X, Lee B. (2003). Type X collagen gene regulation by Runx2 contributes directly to its hypertrophic chondrocyte-specific expression in vivo. J Cell Biol. 162, 833–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Keller B, Zhou G, Napierala D, Chen Y, Zabel B, Parker AE, Lee B. (2009). Localization of the cis-enhancer element for mouse type X collagen expression in hypertrophic chondrocytes in vivo. J Bone Miner Res. 24, 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]