Abstract
Tribless family proteins are pseudokinases that lack DFG (Asp-Phe-Gly) motif in the functional kinase domain, regulating Akt and BMP pathways, insulin metabolism, hypoxia, and ubiquitination. This report concerns expression patterns and functional roles of trb3 in zebrafish embryonic development. trb3 is evolutionarily well-conserved and located on zebrafish chromosome 11. Spatiotemporal expression studies show that trb3 transcripts are abundant throughout embryogenesis, but confined to mesendodermal cells during the late blastula phase. Over-expression of trb3 ventralizes the embryos while a knockdown of trb3 using morpholino alters positioning of the heart, liver, and pancreatic buds as well as gut looping. Furthermore, constitutive activation of TGF-β signaling with TARAM-A* (TGF-β-related type I receptor) significantly increases the level of trb3 transcripts during the late blastula phase. Over-expression of trb3 reduces the level of smurf1 transcripts, a member of TGF-β signaling. We thus propose that Trb3 governs left-right (LR) axis patterning as a component of TGF-β signaling in vertebrate embryonic development.
Keywords: left-right (LR) axis, mesendodermal cells, TGF-β, Tribless, zebrafish
INTRODUCTION
Members of the Tribbles family, Tribbles 1, 2 and 3 encode pseudokinases lacking catalytically active kinase domains. They have been highly conserved through the evolutionary process and TRIBBLES proteins are characterized with DLKLRK instead of DLKPEN in kinase motifs (Mayumi-Matsuda et al., 1999). TRIBBLES3 (TRB3) was identified in neuronal cells and is also called NIPK, SKIP3, and SINK (Bowers et al., 2003; Wu et al., 2003). TRB3 modulates NF-κB signaling, lipid metabolism and insulin metabolism (Du et al., 2003; Qi et al., 2006). It also regulates cell cycle via negative modulation of Slbo and String in Drosophila (Mata et al., 2002). TRB3 acts as an adopter/mediator molecule in the photomorphogenic protein 1 (COP1) that mediates degradation of acetyl-coenzyme A carboxylase (ACC) (Qi et al., 2006) and downregulates SMAD specific E3 ubiquitin regulatory factors 1/2 (Smurf1/2) (Chan at al., 2007; Hua et al., 2011). TRB3 is a key molecule in muscle differentiation via PKB/Akt signaling pathways (Kato and Du, 2007), a potent target of various physiological responses like hypoxia (Rzymski et al., 2008), ER stress (Avery et al., 2010), oxidative stress and nutrient deficiency in various cell types (Jousse et al., 2007). Tribbles family members have been reported to interact with BMPRII tail domain (BMPRII-TD) in the pulmonary artery smooth muscle cells (PASMCs) (Chan et al., 2007).
Despite the current manifests claiming that TRB3 is involved in BMP signaling and in various physiological roles, its roles in the vertebrate embryogenesis have not been elucidated. In this report we made an attempt to examine functional roles of Trb3 in zebrafish embryogenesis. This report explores the spatiotemporal expression patterns and developmental functions of trb3 in zebrafish embryos.
MATERIALS AND METHODS
Zebrafish care and embryos
Wild type zebrafish are obtained from the Korea Zebrafish Organogenesis Mutant Bank (ZOMB) and grown at 28.5°C. Embryos are obtained through natural spawning, raised, and staged as described in the Kimmel et al. (1995). Embryonic pigmentation was blocked by treating them with 0.002% PTU from the onset of somitogenesis.
Sequence analysis
Trb3 sequences are subjected for homology as described in the Kim et al. (2008).
Molecular cloning of trb3
Total RNA was isolated from the embryos at various stages using an easy BLUE total RNA extraction Kit (iNtRON Bio, Korea) according to manufacturer guidelines. cDNA was synthesized with Superscript™ III reverse transcriptase (Invitrogen Corp) as described in the Maddirevula et al. (2011). RT-PCR is performed using forward primer 5′-ACCATGAGTATGAACCT GTCAACT-3′; reverse primer 5′-GAATTCCTGACAGTCCTCA GTCTGG-3′ with HiPi Pus DNA polymerase (Elpis Bio, Korea). For over-expression studies ORF of trb3 was sub-cloned into the pCS2+ vector in between BamH1 and EcoR1.
Morpholinos, in vitro transcription, and microinjections
Translation blocking oligonucleotides, morpholinos were obtained from Gene-Tools, USA, and dissolved in water. Ten ng of trb3-specific morpholinos [5′-taggagttgacaggttcatactcat-3′], control morpholinos [5′-tagCaCttgaGagCttcataGtcat-3′] were injected into zebrafish embryos at 1 cell stage. trb3, TARAM-A*, lefty1/antivin and smurf1-gfp mRNAs were synthesized with Ambion mMESSAGE mMACHINE kit as per the company directions. The synthesized mRNAs were dissolved in nuclease-free water, diluted in 0.5% phenol red solution for microinjection, which was conducted with Picopump microinjection device (World Precision Instruments).
Whole mount in situ Hybridization (WISH)
Embryos were fixed in 4% paraformaldehyde (PFA) overnight, and dehydrated in 100% methanol. Embryos after 24 hpf were digested with 10 μg/ml protease K. WISH is performed with minor modifications in washing as described in (Thisse et al., 1993). smurf1 antisense probes were synthesized from the partial region (472 bps) within ORF. The partial region is amplified with forward primer 5′-ATGTCGAATCCTGGGACTCG-3′; reverse primer 5′-TCTCTACCAGTCCTCGACA-3′ and cloned into pGEM®-T Vector (Promega). trb3, cas, ntl, gata5, mixer, cmlc2, foxa3 (Field et al., 2003), chd and evel (Joly et al., 1993) antisense probes were synthesized with DIG RNA Labeling Kit (SP6/T7) (Roche).
RESULTS
Characterization of zebrafish tribbles3 (trb3)
Because TRB3 is a critical element in various signaling pathways (Du et al., 2003; Qi et al., 2006) we investigated its embryological roles in a zebrafish animal model. We searched for orthologues of human TRIBBLES3 (TRB3) in the zebrafish genome at Zv9 (www.ensembl.org) and identified that zebrafish tribbles3 (trb3) contains 3 exons within the range of 4.66 kb. Identified contig region was further searched at the NCBI tBLAST and confirmed as zebrafish Trb3. We isolated the orthologue of human TRB3 (GenBank accession no; NM_021158), trb3 (GenBank accession no.; NM_212869) from the cDNA library of zebrafish embryos at 30% epiboly. Comparative analysis of the deduced amino acid sequences of Trb3 versus human TRB3 found that Trb3 shares 52% homology (Fig. 1) with the human TRB3 while it shares 98% homology in the pseudokinase domain located between the 67th and 314th amino acid residues (Fig. 1). It has been reported that human TRB3 contains MEK1 binding domains for control of MEK1/ERK pathways as well as COP1 binding domains for physical interaction with COP1 in lipid metabolism (Lee et al., 2012; Qi et al., 2006; Yokoyama and Nakamura, 2011). Trb3 contains both MEK1 binding domains and COP1 binding domains at the C-Terminal region (Fig. 1; Red and Green box). Taken together, zebrafish trb3 is a homologue of human TRB3 and highly conserved in the functional domains pseudokinase, COP1 interacting and MEK1 binding domains.
Fig. 1.
Amino acid sequence homology study of Tribbles proteins 1, 2 and 3 (ENSDARP00000101 435, ENSDARP00000089195, ENS DARP00000036401) in comparison with humans (protein access number: NP_066981.2). Blue box indicates pseudokinas domain.
Spatiotemporal distribution of trb3 transcripts in the embryos
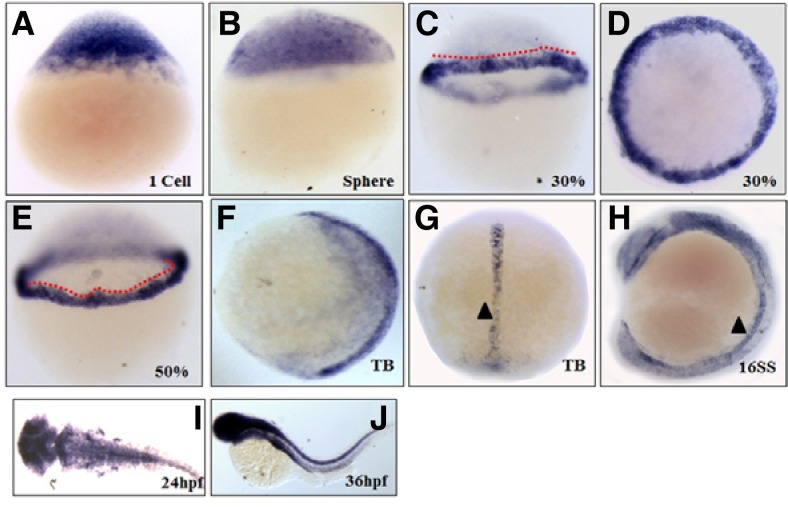
As an initial approach to circumvent biological functions of Trb3, the spatiotemporal expression profile of trb3 was analyzed with RT-PCR (Supplementary Fig. 1) and WISH using a DIG-labeled trb3-specific anti-sense probe in the first cell stage through 4 days of post-fertilization (dpf) embryos. trb3 transcripts are present in the first cell stage and in sphere (Figs. 2A and 2B), and become confined to the blastoderm marginal cells during the late blastula (30% epiboly) and early gastrula stage (50% epiboly) (Figs. 2C–2E). At the end of gastrula stage, the transcripts are more specifically restricted to the axial cells and developing notochord at the tail bud (Figs. 2F and 2G). trb3 transcripts appear more prominent in the notochord and developing nervous system at the 16 somites stage (Fig. 2H), and later in the forebrain, tectum, midbrain-hindbrain boundary, and hindbrain at 24 hpf (Fig. 2I). Interestingly, they become intensified in the nervous system and developing endoderm-derived organs, such as the intestine at 36 hpf (Fig. 2J). These cell- and tissue-specific expression patterns of trb3 in the embryos suggest that Trb3 may have a critical role in early embryogenesis.
Fig. 2.
Spatiotemporal distribution of trb3 transcripts in the embryos. WISH with anti-sense trb3 probe at one cell stage (A), Sphere (B), 30% epiboly (C, D), 50% epiboly (E), Tail bud (F, G), 10 somite stage (H), 24 hpf (I) and 36 hpf (J). (A–C, E, F, and H) Lateral views with dorsal side right. (D, I) dorsal views. (J) Lateral views with anterior part left. Dotted lines indicate transcripts restriction to marginal cells. Arrow head indicates notochord.
Because trb3 transcripts are restricted to the blastodermal marginal cells at 30% epiboly (Fig. 2C) we analyzed in-depth expression profiles of trb3 in cells using two color WISH germ layered specific markers, such as a cas (endodermal marker), ntl (mesoderm marker), gata5 and bon/mixer (mesendodermal markers). Expression domains of trb3 are partially overlapped with those of cas and ntl (Figs. 3A and 3B) but had more overlap with those of gata5 and bon (Figs. 3C and 3D), confirming its expression in the mesendodermal cells.
Fig. 3.
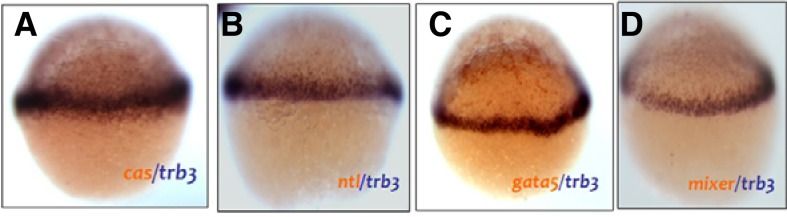
Localization of trb3 transcripts in the blastodermal marginal cells at late blastula. Two color WISH was performed with DIG labeled trb3 anti-sense probe and fluorescent labeled DIG antisense probes of cas (A), ntl (B), gata5 (C) and mixer (D) 30% epiboly (4.7 hpf). (A–C) lateral views. Control embryo at 30% epiboly (E).
Trb3 is a component of TGF-β signaling
It is known that Nodals, members of the TGF-β pathway, play a critical role in LR axis patterning (Schier, 2009) whereas lefty1/antivin works as an antagonist to Nodal signaling in zebrafish embryogenesis (Bisgrove et al., 1999). To test the functional relationship between TGF-β signaling and Trb3, we initially overexpressed TARAM-A* which structurally resembles Alk4, a receptor of TGF-β related ligands in the Xenopus embryos (Chen et al., 2004) that constitutively induces TGF-β signals (Peyrieras et al., 1998; Renucci et al., 1996). We then measured the level of trb3 transcripts in the TARAM-A* overexpressed embryos at 30% epiboly, and found that its level was significantly elevated while at the same time extending out to the blastodermal cells (Figs. 4A and 4B). In contrast to the TRARAM-A* effect, overexpression of antivin/lefty1, a Nodal inhibitor, clearly reduces the level of trb3 transcripts (Figs. 4A and 4C). It is thereby evident that TGF-β signaling positively regulates the transcription of trb3.
Fig. 4.
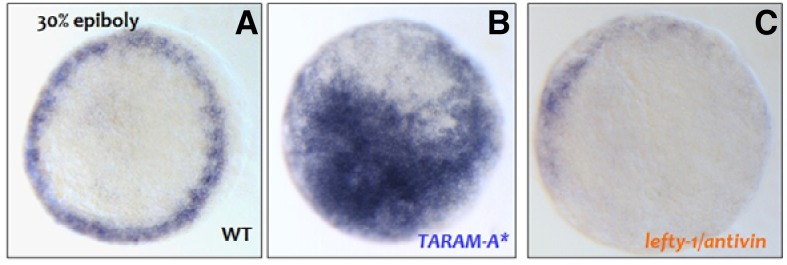
Embryo injected with 100 pg of TARAM-A* at two cell stage (B). Embryo injected with 50 pg of lefty-1 at two cell stage (C).
Forced expression of trb3 ventralizes the embryos
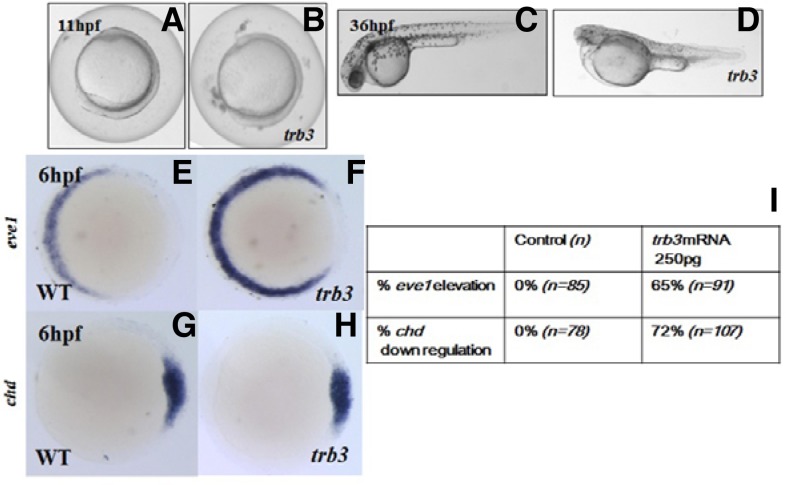
To assess embryological roles of Trb3 in embryogenesis, we over-expressed trb3 by microinjecting 250 pg of 5′-capped trb3 mRNAs (Figs. 5A–5H). Over-expression of trb3 induces ventralization in 65% (n =1 10) of trb3 mRNA injected embryos, giving rise to a shortened axis at 11 hpf (Figs. 5A and 5B), small eyes, elaborated somites, cardiac edema, and a thickened yolk plug at 36 hpf (Figs. 5C and 5D).
Fig. 5.
Gain-of-function studies of trb3. Forced expression of trb3 induces ventralization of the embryos (A–H). Control embryos (A, C), injected embryos (B, D) are at 11 hpf and 36 hpf, respectively. (E–H) Molecular marker studies with eve1 (E, F) and chd (G, H); control embryos (E, G). Quantification of molecular marker study of trb3 overexpressed embryos (I).
Embryos overexpressing trb3 are further assayed with the dorsal marker, chordin (chd) encoding a BMP4 inhibitor (Sasai et al., 1995) and the ventral marker, even-skipped-like1 (eve1) (Joly et al., 1993). At 60% epiboly, eve1 transcripts are increased in the ventral marginal cells of the embryos as to those of the control embryos (Figs. 5E and 5F). The dorsal marker, chd expression domains are markedly reduced in the dorsal side cells of the trb3 injected embryos in comparison with the control embryos at 6 hpf (Figs. 5G and 5H). All these observations support that Trb3 is an essential regulator for the ventral patterning during the late gastrula stage and thereafter until it reaches 36 hpf (Figs. 5A–5D).
Trb3 is required for left-right (LR) axis orientation of the organs
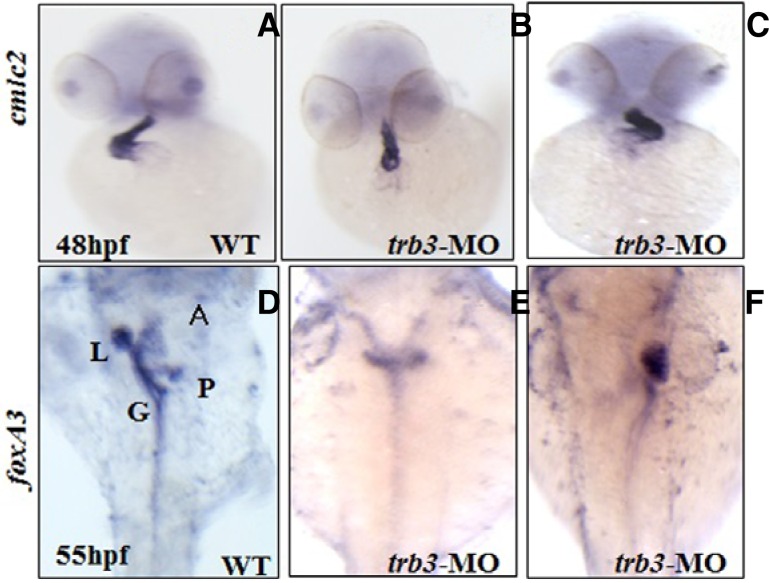
Specific expression of trb3 in the mesendodermal cells (Figs. 3A–3D) raises the possibility that Trb3 is of critical importance to the development of organs derived from the mesendodermal cells. To test this hypothesis we performed loss-of-function studies of trb3 by injecting trb3 specific translation blocking morpholino into the embryos at cell stage one. To assess molecular events associated with the loss-of-function of Trb3 in the embryos at 48 hpf we performed WISH with the LR axis patterning marker for heart development, cardiac myosin light chain-2 (cmlc2). In contrast to the control embryos (n = 75), the molecular labeling found that morphological defects in the cardiac looping and positioning, reversed looping (D-looping) and caused failure in the remaining ventricle looping to the midline (39%; n = 108) (Figs. 6A–6C). trb3 morphants at 55 hpf were further assayed with forkhead box A3 (foxa3), an endoderm marker, which specifies visceral organs, such as the liver and the pancreatic bud (Field et al., 2003). Spatiotemporal expression patterns of foxa3 detect heterotaxy in the gut looping as well as positional changes of the liver and pancreatic bud. Morphants show randomized positioning of the liver and pancreatic bud as well as organ duplication (Figs. 6D–6F). In order to conform to the specificity of trb3 and its morpholino we performed rescue experiments by coinjection of trb3 mRNA and trb3 MO, which showed phenotype with rescued heart laterality defects (Table 1). These findings strongly suggest that Trb3 is an essential element in regulating LR axis orientation of the heart, gut, pancreas and liver in organogenesis.
Fig. 6.
Knock down of trb3 alters LR axis orientation of organs. Morphant embryos are analyzed with organ specific markers, heart (A–C), liver, gut and pancreatic bud (D–F); heart position was identified with cardiac specific marker, cmlc2 while the positions of liver, gut, pancreatic bud with foxa3.
Table 1.
Phenotype rescue of heart laterality by coinjecting trb3 mRNAs and trb3 morpholino
| Trb3 MO (ng) | mRNA (pg) | n | Heart laterality defects (%) |
|---|---|---|---|
| 10 | - | 108 | 39 |
| 10 | 250 | 94 | 6 |
Over-expression of trb3 reduces level of smurf1 transcripts
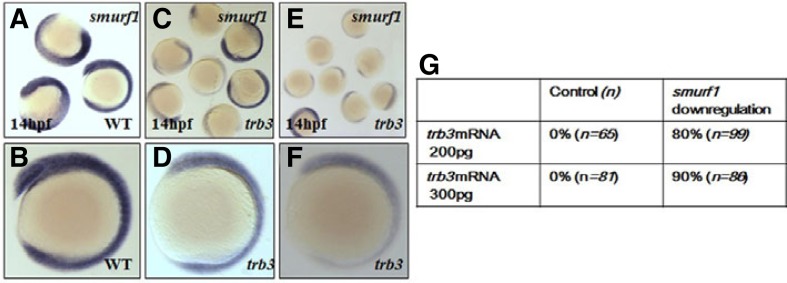
Studies in the pulmonary artery smooth muscle cells (PASMCs) show that TRB3 regulates Smurf1 in association with BMP signaling components (Chan et al., 2007). We thus examined the putative correlation between Smurf1 and Trb3 in zebrafish embryogenesis. We first measured endogenous level of smurf1 transcripts in the embryos at 14 hpf, in which trb3 transcripts were microinjected at one cell stage for over-expression. Over-expression of trb3 remarkably decreases the level of smurf1 transcripts in the embryos (Figs. 7A–7F). However, during embryonic development expression domains of trb3 (Figs. 2F and 2H) are largely overlapped with the expression domains of smurf1 (Figs. 7A and 2B). Two different amounts (200 pg or 300 pg per embryo) of trb3 mRNA reduce the transcriptional level of smurf1 in a concentration-dependent manner (Figs. 7C–7F) in the 80% (n = 98) of trb3 mRNA injected embryos, evidently demonstrating that Trb3 is a critical regulator of Smurf1 to modulate TGF-β signaling in embryos.
Fig. 7.
Forced expression of trb3 alters level of smurf1 transcripts. Level of smurf1 transcripts are measured in trb3 overexpressed embryos (A–F). (A, B) Control embryos. (C, D) 200 pg trb3 mRNA injected embryos. (E, F) 300 pg trb3 mRNA injected embryos. Quantification of the number of embryos with reduced smurf1 levels in the trb3 overexpressed embryos (G).
DISCUSSION
Pseudokinases are functionally important in various cellular signaling processes although they are a functionally inactive in phosphorylation (Reviewd by Boudeau et al., 2006). Tribbles proteins are initially identified in Drosophila and are critical for cell division and mitosis (Rorth et al., 2000). Three isoforms of TRB are reported with unique function in Drosophila while their functional roles in developing embryos remain unclear. In this report we aimed to analyze the functional roles of a pseudokinase, tribbles3 in a vertebrate animal model zebrafish (Danio rerio) embryogenesis. We identified tribbles3 in the zebrafish genome and isolated zebrafish trb3 which is an orthologue of human TRB3. We found that trb3 is expressed throughout embryogenesis. In particular, trb3 transcripts are restricted to mesendodermal cells, the precursor cells for the heart, liver, gut and pancreatic bud. Transforming growth factor (TGF-β) signaling has been demonstrated as a pivotal regulator for the induction of mesendoderm, a tissue specific to the Spemann organizer (Rodaway and Patient, 2001). Overexpression of the TGF-β activator (TARAM-A*) and the antagonist, Lefty1 confirmed that TGF-β signaling is required for trb3 expression in the embryos.
Studies have shown that Trb3 physically interacts with the BMP type II receptor in mammalian cells and thereby regulates BMP signaling (Chan et al., 2007). BMPs and their receptors belong to the TGF-β super family and are critical for ventral patterning in vertebrates (von Bubnoff and Cho, 2001). Over-expression of trb3 induced ventralized phenotypes, which were further confirmed by molecular marker studies. Existing evidence from animal models revealed that the TGF-β super family contributes to the left-right asymmetry (LR) process, notably Nodal, Pitx2, Lefty 1 and Lefty2 (Meno et al., 1998). Bone morphogenic protein (BMP) signaling also governs LR patterning; it subdues nodal expression in the left lateral plate mesoderm (LPM) to regulate heart laterality and dorsoventral fate in Xenopus and zebrafish embryos (Monterio et al., 2008; Ramsdell, 1998; Tucker et al., 2008). Secreted BMPs belonging to the TGF-β super-family transduce the signals by interacting with BMP type I and II receptors (Chen at al., 2004; Hua et al., 2011). Considering these combined features of the signaling pathways including Trb3, trb3 expression in the mesendodermal cells (Figs. 2C and 4), and the altered expression pattern of cmlc2 and foxa3 in the trb3 MO (Fig. 6), it is conceivable that Trb3 plays a pivotal role in controlling LR axis patterning.
The evolutionarily conserved LR patterning process is tightly regulated during embryogenesis and involves various elements such as ubiquitin proteasome system (UPS) components. Ubiquitin E3 ligases, Smad ubiquitination-related factor 1 (Smurf1) and Smurf2 control TGF-β signaling by regulating cellular levels of Smad molecules (Izzi and Attisano, 2004). SMADs are vital regulators of the TGF-β signaling with numerous interactions in the cytoplasm and nucleus (Rebagliati et al., 1998) and SMADs levels are modulated by Smad ubiquitination regulatory factor 1 (Smurf1) and Smurf2. Smurfs are E3 ubiquitin ligases and master regulators of SMADs (Moustakas et al., 2001). Over-expression of trb3 reduces the level of endogenous Smurf1 levels in the embryos (Fig. 7). Expression and functional studies on trb3 in the zebrafish embryos demonstrated that trb3 is expressed in mesendodermal cells during the late blastula stage and is a vital component of TGF-β signaling (Figs. 2 and 4). Over-expression of trb3 induces ventralization and reduces the smurf1 transcripts (Fig. 7) whereas the knock-down of trb3 using trb3-specific morpholino alters the positions of the heart, liver, pancreatic bud, and gut loop (Fig. 6). It is most likely that Trb3 exerts its biological roles via Smurf in developing vertebrate embryos. It would be of great interest to identify target molecules of Trb3 using proteomic approaches for extension of the Trb3 network during vertebrate embryonic development.
Supplementary Material
Acknowledgments
We would like to thank Dr. Shunji Jia, Tsinghua University, Beijing for providing the smurf1-gfp vector and Korea Zebrafish Organogenesis Mutant Bank (ZOMB) for providing wild type zebrafish. This work was supported by the National Foundation of Korea Grants funded by the Korean Government (2009-0074496).
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Avery J, Etzion S, DeBosch BJ, Jin X, Lupu TS, Beitinjaneh B, Grand J, Kovacs A, Sambandam N, Muslin AJ. TRB3 function in cardiac endoplasmic reticulum stress. Circ Res. 2010;106:1516–1523. doi: 10.1161/CIRCRESAHA.109.211920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove BW, Essner JJ, Yost HJ. Regulation of midline development by antagonism of lefty and nodal signaling. Development. 1999;126:3253–3262. doi: 10.1242/dev.126.14.3253. [DOI] [PubMed] [Google Scholar]
- Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Bowers AJ, Scully S, Boylan JF. SKIP3, a novel Drosophila tribbles ortholog, is overexpressed in human tumors and is regulated by hypoxia. Oncogene. 2003;22:2823–2835. doi: 10.1038/sj.onc.1206367. [DOI] [PubMed] [Google Scholar]
- Chan MC, Nguyen PH, Davis BN, Ohoka N, Hayashi H, Du K, Lagna G, Hata A. A novel regulatory mechanism of the bone morphogenetic protein (BMP) signaling pathway involving the carboxyl-terminal tail domain of BMP type II receptor. Mol Cell Biol. 2007;27:5776–5789. doi: 10.1128/MCB.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Mironova E, Whitaker LL, Edwards L, Yost HJ, Ramsdell AF. ALK4 functions as a receptor for multiple TGF beta-related ligands to regulate left-right axis determination and mesoderm induction in Xenopus. Dev Biol. 2004;268:280–294. doi: 10.1016/j.ydbio.2003.12.035. [DOI] [PubMed] [Google Scholar]
- Du K, Herzig S, Kulkarni RN, Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–1577. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- Hua F, Mu R, Liu J, Xue J, Wang Z, Lin H, Yang H, Chen X, Hu Z. TRB3 interacts with SMAD3 promoting tumor cell migration and invasion. J Cell Sci. 2011;124:3235–3246. doi: 10.1242/jcs.082875. [DOI] [PubMed] [Google Scholar]
- Izzi L, Attisano L. Regulation of the TGF-beta signaling pathway by ubiquitin-mediated degradation. Oncogene. 2004;23:2071–2078. doi: 10.1038/sj.onc.1207412. [DOI] [PubMed] [Google Scholar]
- Joly JS, Joly C, Schulte-Merker S, Boulekbache H, Condamine H. The ventral and posterior expression of the zebrafish homeobox gene eve1 is perturbed in dorsalized and mutant embryos. Development. 1993;119:1261–1275. doi: 10.1242/dev.119.4.1261. [DOI] [PubMed] [Google Scholar]
- Jousse C, Deval C, Maurin AC, Parry L, Chérasse Y, Chaveroux C, Lefloch R, Lenormand P, Bruhat A, Fafournoux P. TRB3 inhibits the transcriptional activation of stress-regulated genes by a negative feedback on the ATF4 pathway. J Biol Chem. 2007;282:15851–15861. doi: 10.1074/jbc.M611723200. [DOI] [PubMed] [Google Scholar]
- Kato S, Du K. TRB3 modulates C2C12 differentiation by interfering with Akt activation. Biochem Biophys Res Commun. 2007;353:933–993. doi: 10.1016/j.bbrc.2006.12.161. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Ro H, Huh TL, Lee CJ, Choi J, Rhee M. A novel Kinesin-like protein, Surhe is associated with dorsalization in the zebrafish embryos. Animal Cells Syst. 2008;12:219–230. [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Lee SW, Seong MW, Jeon YJ, Chung CH. Ubiquitin E3 ligases controlling p53 stability. Animal Cells and Syst. 2012;16:173–182. [Google Scholar]
- Maddirevula S, Anuppalle M, Huh TL, Kim SH, Rhee M. Nrdp1 governs differentiation of the melanocyte lineage via Erbb3b signaling in the zebrafish embryogenesis. Biochem Biophys Res Commun. 2011;409:454–458. doi: 10.1016/j.bbrc.2011.05.025. [DOI] [PubMed] [Google Scholar]
- Mata J, Curado S, Ephrussi A, Rørth P. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell. 2000;101:511–522. doi: 10.1016/s0092-8674(00)80861-2. [DOI] [PubMed] [Google Scholar]
- Mayumi-Matsuda K, Kojima S, Suzuki H, Sakata T. Identification of a novel kinase-like gene induced during neuronal cell death. Biochem Biophys Res Commun. 1999;258:260–264. doi: 10.1006/bbrc.1999.0576. [DOI] [PubMed] [Google Scholar]
- Meno C, Shimono A, Saijoh Y, Yashiro K, Mochida K, Ohishi S, Noji S, Kondoh H, Hamada H. lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell. 1998;94:287–297. doi: 10.1016/s0092-8674(00)81472-5. [DOI] [PubMed] [Google Scholar]
- Monteiro R, van Dinther M, Bakkers J, Wilkinson R, Patient P, ten Dijke R, Mummery C. Two novel type II receptors mediate BMP signaling and are required to establish left-right asymmetry in zebrafish. Dev Biol. 2008;315:55–71. doi: 10.1016/j.ydbio.2007.11.038. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Souchelnytskyi S, Heldin CH. Smad regulation in TGF-beta signal transduction. J Cell Sci. 2001;114:4359–4369. doi: 10.1242/jcs.114.24.4359. [DOI] [PubMed] [Google Scholar]
- Peyrieras N, Strahle U, Rosa F. Conversion of zebrafish blastomeres to an endodermal fate by TGF-beta-related signaling. Curr Biol. 1998;8:783–786. doi: 10.1016/s0960-9822(98)70303-3. [DOI] [PubMed] [Google Scholar]
- Qi L, Heredia JE, Altarejos JY, Screaton R, Goebel N, Niessen S, MacLeod IX, Liew CW, Kulkarni RN, Bain J, et al. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science. 2006;312:1763–1766. doi: 10.1126/science.1123374. [DOI] [PubMed] [Google Scholar]
- Ramsdell AF. Left-right asymmetry and congenital cardiac defects: getting to the heart of the matter in vertebrate left-right axis determination. Dev Biol. 2005;288:1–20. doi: 10.1016/j.ydbio.2005.07.038. [DOI] [PubMed] [Google Scholar]
- Rebagliati MR, Toyama R, Fricke C, Haffter P, Dawid IB. Zebrafish nodal-related genes are implicated in axial patterning and establishing left-right asymmetry. Dev Biol. 1998;199:261–272. doi: 10.1006/dbio.1998.8935. [DOI] [PubMed] [Google Scholar]
- Renucci A, Lemarchandel V, Rosa F. An activated form of type I serine/threonine kinase receptor TARAM-A reveals a specific signalling pathway involved in fish head organiser formation. Development. 1996;122:3735–3743. doi: 10.1242/dev.122.12.3735. [DOI] [PubMed] [Google Scholar]
- Rodaway A, Patient R. Mesendoderm an ancient germ layer? Cell. 2001;20:169–172. doi: 10.1016/s0092-8674(01)00307-5. [DOI] [PubMed] [Google Scholar]
- Rorth P, Szabo K, Texido G. The level of C/EBP protein is critical for cell migration during Drosophila oogenesis and is tightly controlled by regulated degradation. Mol Cell. 2000;6:23–30. doi: 10.1016/s1097-2765(05)00008-0. [DOI] [PubMed] [Google Scholar]
- Rzymski T, Paantjens A, Bod J, Harris AL. Multiple pathways are involved in the anoxia response of SKIP3 including HuR-regulated RNA stability, NF-κB and ATF4. Oncogene. 2008;27:4532–4543. doi: 10.1038/onc.2008.100. [DOI] [PubMed] [Google Scholar]
- Sasai Y, Lu B, Steinbeisser H, De Robertis EM. Regulation of neural induction by the Chd and Bmp-4 antagonistic patterning signals in Xenopus. Nature. 1995;376:333–336. doi: 10.1038/376333a0. [DOI] [PubMed] [Google Scholar]
- Schier AF. Nodal morphogens. Cold Spring Harb Perspect Biol. 2009;1:a003459. doi: 10.1101/cshperspect.a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Tucker JA, Mintzer KA, Mullins MC. The BMP signalling gradient patterns dorsoventral tissues in a temporally progressive manner along the anteroposterior axis. Dev Cell. 2008;14:108–119. doi: 10.1016/j.devcel.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Bubnoff A, Cho KW. Intracellular BMP signaling regulation in vertebrates: pathway or network? Dev Biol. 2001;239:1–14. doi: 10.1006/dbio.2001.0388. [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Nakamura T. Tribbles in disease: Signaling pathways important for cellular function and neoplastic transformation. Cancer Sci. 2011;102:1115–1122. doi: 10.1111/j.1349-7006.2011.01914.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.