Abstract
Recent advances in applied physics and chemistry have led to the development of novel microfluidic systems. Microfluidic systems allow minute amounts of reagents to be processed using μm-scale channels and offer several advantages over conventional analytical devices for use in biological sciences: faster, more accurate and more reproducible analytical performance, reduced cell and reagent consumption, portability, and integration of functional components in a single chip. In this review, we introduce how microfluidics has been applied to biological sciences. We first present an overview of the fabrication of microfluidic systems and describe the distinct technologies available for biological research. We then present examples of microsystems used in biological sciences, focusing on applications in molecular and cellular biology.
Keywords: cellular biology, human-on-a-chip, microfluidics, molecular biology, organ-on-a-chip, worm-on-a-chip
INTRODUCTION
In less than a century, the field of biology has experienced several revolutions that have brought new knowledge and new technologies. With the latest development of “omics” technologies such as genomics, proteomics, and metabolomics, the amount of biological information available has increased dramatically, leading to a need for sensitive high-throughput experimentations (Betz et al., 2005; Hopkins and Groom, 2002; Page et al., 1999; Sleno and Emili, 2008). However, conventional biological tools suffer from large reagent consumption, low throughput, and errors related to reagent transfer during experiments.
During the past decade, microfluidics has emerged as a potential platform for conducting cellular and molecular biological studies. By reducing conventional macroscale systems to microscale systems, microfluidics provides solutions to overcome the limitations of conventional experiments. From an experimental point of view, microfluidic technology requires lesser amounts of reagents, cells, and space than conventional methods. Moreover, microfluidics enables rapid analysis, high-throughput experimentation, and automation. From a biological point of view, microfluidics can partly reproduce in vivo-like molecular and physical microenvironments of cells and the 3D structure and organization of tissues or organs. From a technical point of view, the versatility of microfluidic systems allows distinct tools to be integrated on a single chip to reduce the errors related to reagent manipulation or to obtain multiple data points from a single experiment.
In this review, we first describe the technologies used to fabricate polydimethylsiloxane (PDMS) microfluidic chips and then present a succinct description of the existing microfluidic devices used in molecular and cellular biology.
BASIS OF MICROFLUIDICS
Soft-lithography-fabrication
In the 1980s and 1990s, microfluidics emerged as a promising tool for molecular biology (Regnier et al., 1999). During that period, microfluidic chips were fabricated using mainly silicon, glass, or quartz substrates, which required trained technicians working in cleanroom facilities and therefore limited the application of this technology to chromatography and electrophoresis on chips. In the late 1990s, the introduction of polymer-based soft-lithography freed the microfabrication process from the cleanroom (Duffy et al., 1998), facilitating the development of inexpensive microfluidic devices by almost any laboratory equipped with a traditional chemical fumehood. Currently, the most popular material used for fabricating microfluidic chips for biological applications is polydimethylsiloxane (PDMS), an elastomer that can be readily molded into microstructures and microchannels (Duffy et al., 1998). The mechanical and physical properties of this polymer are highly advantageous for fabricating microfluidic devices: PDMS is transparent, biocompatible, and gas-permeable, which make it suitable for molecular and cellular biological studies (Belanger and Marois, 2001; Piruska et al., 2005).
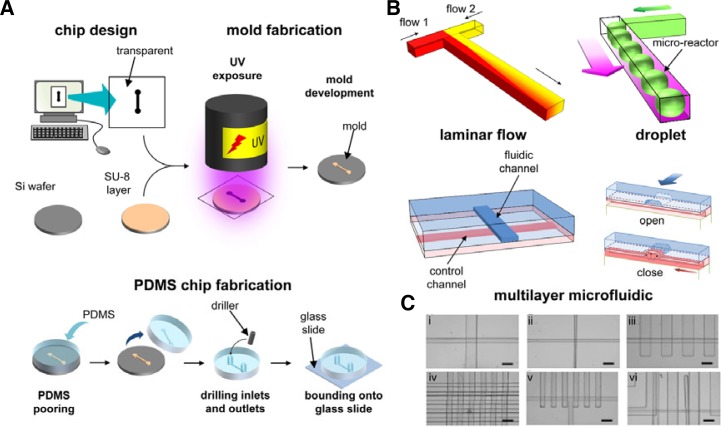
PDMS microfluidic devices are generally fabricated using molding methods (Fig. 1). A microfluidic chip is first drawn using computer-aided design (CAD) software and the drawing is printed at high resolution (20,000–40,000 dpi) on a transparent sheet, which serves as a mask for fabricating the mold (also known as “master”). To fabricate the mold, a thin layer of photoactive polymer is first spin-coated on the surface of a clean hard wafer. The photoactive materials used typically are epoxy-based polymers such as SU-8, and the wafers are made of silicone or glass. The photoactive polymer is then covered with the mask and exposed to UV light, which selectively crosslinks the polymer. The microstructures and microchannels are then revealed by removing the un-crosslinked polymer from the wafer by using solvent-based development. The surface of the mask is treated with silanizing agents (e.g., tridecafluoro-1,1,2,2-tetrahydrooctyl trichlorosilane) before use to facilitate the release of the cured PDMS from the master. PDMS linear polymers and crosslinking agents are mixed, degassed, and poured onto the mold and left to cure. After polymerization, the PDMS is peeled off the master and cut to desired dimensions, after which holes for inlets and outlets are drilled using a biopsy puncher or a syringe needle. The cured PDMS is then bounded and sealed onto a hard surface (such as a glass slide) or another cured PDMS layer to form the microfluidic channels.
Fig. 1.
Basis of microfluidics. (A) Step-by-step process of a PDMS microfluidic chip. (B) Schematique representation of different microfluidic techniques: laminar flow, droplet-based microfluidic, and multilayer-based microfluidics. (C) Pictures of valves in multilayer-based microfluidics (Unger et al., 2000).
Techniques of microfluidics
Continuous-flow microfluidics
Continuous-flow microfluidics involves continuous liquid flow through microchannels. Two common methods are used to actuate fluid flow in microchannels: pressure-driven flow, and electro-osmotic flow. In pressure-driven flow, the fluid is controlled by external pressure sources, external mechanical pumps, or integrated mechanical micropumps. In electro-osmotic flow, the fluid is controlled by combinations of capillary and electrokinetic forces (Chang and Yeo, 2010). Continuous-flow microfluidics is the main microfluidic method that has been used in many well-defined and simple biological applications including cell culture (Tourovskaia et al., 2004), cell sorting (Cho et al., 2003), and gradient formation (Li Jeon et al., 2002). However, the simplicity of the system limits its use to tasks that require a high degree of flexibility or the absence of stress (e.g., shear stress) induced by fluid flow, such as in cell cultures.
Droplet-based microfluidics
One subcategory of microfluidics is droplet-based microfluidics, which is also called “digital microfluidics” in reference to digital microelectronics to emphasize the use of discrete and distinct volumes of fluids and to contrast this with the use of continuous flow of fluids in the continuous-flow microfluidic system. In digital microfluidics, droplets with diameters in the nm-to-μm range are created by using immiscible phases and manipulated inside microdevices (Belder, 2005; Jensen and Lee, 2004). Unlike in continuous-flow systems, the independent control of each droplet in digital microfluidics allows discrete microreactors to be generated and individually transported, stored, mixed, reacted, and analyzed (Fair, 2007; Link et al., 2006) at rates up to 20,000/s (Kobayashi et al., 2007). This particular feature enables multiple identical microreactor units to be generated rapidly. Consequently, this system is especially well-suited for parallel processing and experimentation and facilitates large data sets to be acquired in a single experiment without increasing device size or complexity. In recent years, droplet-based microfluidics has been used in diverse applications including in protein crystallization (Chayen and Saridakis, 2008), drug development (Kintses et al., 2010), and biochemical testing (Jambovane et al., 2011).
Droplet-based technology offers various advantages including the physical and chemical isolation of droplets that eliminates the risk of cross-contamination, the small amount of reagent used (1 pl to 10 nl), the fast and efficient mixing of reagents inside the droplets, the ability to manipulate droplets digitally at extremely high throughputs, and the ability to incubate stable droplets off-chip and later reintroduce them into the microfluidic environment for further processing and analysis.
Multilayer soft-lithography for fabricating valve-based microfluidic systems
Multilayer soft-lithography refers to the methodology developed by Quake’s group and used to construct multilayer structures from multiple monolayers of PDMS, or any elastomer, each of which is cast separately from a specific mold (Unger et al., 2000). As introduced previously, the PDMS pre-polymer is made of 2 components: the linear polymer and the curing agent. To fabricate multilayer devices, the bottom layer is prepared with an excess of one of the components (for example, the linear polymer), whereas the upper layer is prepared with an excess of the other component (in this example, the curing agent). The layers are cured separately and the upper layer is peeled off from its mold and placed atop the lower layer. Further curing causes the excess reactive molecules at the interface between the 2 layers to react and irreversibly bond the 2 layers. By repeating this process, supplementary layers can be easily added (Unger et al., 2000). This is an easier method for fabricating multilayer devices than conventional micromachining.
Multilayer soft-lithography can be used to fabricate active valves and pumps. Valve are fabricated using a “crossed-channel architecture”, in which the “control channel” that contains the valve structures is in one layer and crosses the “fluidic channel” that is in the other layer. The PDMS membrane between the 2 channels is engineered to be thin (typically 30-μm thick), and thus when pressure is applied to the control channel, the membrane deflects and closes the fluidic channel. To prevent incomplete closing of the valve, the shape of the flow channel is critical: round flow channels close completely whereas rectangular ones do not. Moreover, the width of the control channels can be varied without affecting the efficiency of the control valve, which depends on membrane dimensions. Thus, control channels can pass over multiple fluidic channels and actuate only the desired channels. Furthermore, each control channel can actuate multiple valves concurrently, and distinct control channels with dedicated pressure controllers can independently actuate distinct valves in the same device (Unger et al., 2000). The valves control fluid flow in the channels and can be used to stop the flow, isolate a part of the device (to create microreactors), direct the flow, meter specific amounts of reagent, and create an integrated peristaltic pump (Fig. 1).
MICROFLUIDICS FOR MOLECULAR BIOLOGY
Microfluidics for genomics and transcriptomics
DNA sequencing
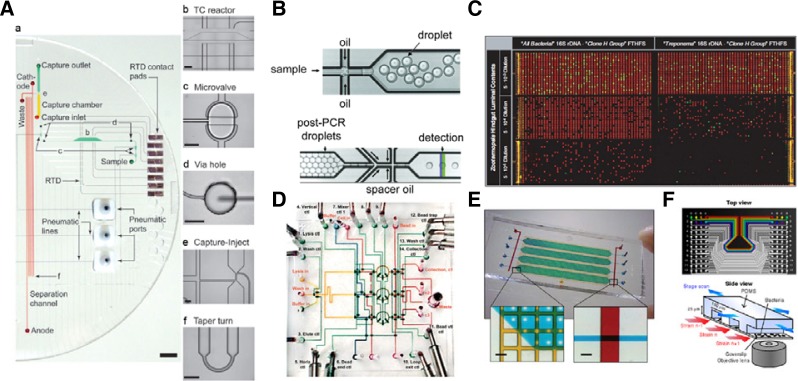
DNA sequencing is a crucial step in genomics. Although the Human Genome Project was completed almost 10 years ago (International Human Genome Sequencing Consortium, 2004), DNA sequencing remains a key process in studies aimed at understanding numerous diseases and identifying potential drug targets (Kubinyi, 2003). Various microfluidic devices for DNA sequencing have been successfully developed (Blazej et al., 2006; 2007; Fredlake et al., 2008; Kartalov and Quake, 2004), including ones that use microfluidic approaches for Sanger sequencing (Aborn et al., 2005; Blazej et al., 2006; 2007; Fredlake et al., 2008; Kumagai et al., 2008), which remains the most convenient technique for conventional de novo sequencing of genomes. These devices integrate the 3 steps of the conventional sequencing protocol on a single chip: thermal cycling, sample purification, and capillary electrophoresis (CE). For example, Blazej et al. (2006) have developed a microfluidic bioprocessor for integrated nl-scale Sanger sequencing (Fig. 2A). The chip is composed of 250-nl reactors integrated with CE channels that capture and purify DNA. This chip enables complete Sanger sequencing of 556 continuous bases with 99% accuracy from merely 1 fmol of DNA template. When integrated with an inline-injection system, this chip allows the sequencing sample to be purified and the sample plug to be defined narrowly, which eliminates the excess amounts of sample required previously for cross-injected CE separations and thus facilitates microchip-based Sanger sequencing of 365 bases with 99% of accuracy from only 30 nl of sample containing just 100 amol of template (Blazej et al., 2007). Although the sequencing length must be increased to reach the conventional Sanger sequencing level, these devices offer a proof-of-concept that integrated microfluidic systems can be developed for DNA sequencing for future applications such as low-cost personal sequencing (Liu and Mathies, 2009) or for single-cell genome analysis (Kalisky and Quake, 2011).
Fig. 2.
Microfluidic system for molecular biology. (A) Bioprocessor for nanoliter-scale Sanger DNA sequencing. (a) photograph of one of the two systems. (b–f) Close-up of the different components. (Blazej et al., 2006). (B) Device for high-throughput droplet digital PCR system. Samples are encapsulated in droplet (top) where PCR will be performed. After reaction, droplets are introduced into a microfluidic droplet reader (bottom) for analysis (Hindson et al., 2011). (C) Multiplex digital PCR platform for single cell analysis in environmental sample for amplification of “All bacteria” 16S rDNA and termite cluster 16S rDNA (red fluorescence) and clone H FTHFS gene (green fluorescence) (Ottesen et al., 2006). (D) Picture of a nanofluidic system containing three simultaneous parallel processors for DNA recovery (Hong et al., 2004). (E) Microfluidic device for parallel gene synthesis. Photograph of the 4 parallel reactors. Left inlet, Close-up of the gene synthesis chamber (blue and green) and water jacket (yellow). Right inlet, Close up of a control channel (red) crossing fluidic channel (blue) (Kong et al., 2007). (F) Top, Picture of a microfluidic system for proteomics. Bottom, Schematics of the experiments representing 3 channels of the device containing 3 different bacteria strains (Taniguchi et al., 2010).
Nucleic acid amplification on a chip
Nucleic acid amplification techniques, such as polymerase chain reaction (PCR) and the recent isothermal amplifications, are essential in every biology-related field ranging from basic biology to drug discovery and food science (Diaz-Sanchez et al., 2013; Gill and Ghaemi, 2008; Stals et al., 2012). For nucleic acid amplification, microfluidics offers numerous advantages when compared to conventional methods: reduced reagent consumption, lowered amplification times, increased analytical throughput, minimized risk of contamination, increased sensitivity, and integration. To design an optimal sample-in-answer-out gene analysis system, microfluidic PCR systems (or microPCR) have been developed using continuous-low (Kopp et al., 1998; Li et al., 2009) and droplet-based microreactors (Zhu et al., 2012) and valve-actuated PCR microchambers (Ottesen et al., 2006).
In 1998, Kopp et al. (1998) developed the first chip-based continuous-flow microPCR. The chip was composed of a 40-μm deep and 90-μm wide channel (etched in a Corning 0211 glass chip) that had a total length of 2.2 m. The single channel was passed repeatedly through 3 well-defined temperature zones that were maintained at 95°C, 77°C, and 60°C using thermostated copper blocks. The pattern defined the number of cycles performed per run through the chip. In this case, the device was designed to generate 20 cycles, each with a melting:annealing:extension time ratio of 4:4:9, and thus had a theoretical DNA-amplification factor of 220. Using this chip, a 176-bp DNA fragment was amplified at flow rates ranging from 5.8–72.9 nl/s, which correspond to a PCR time of 18.7 min to 1.5 min.
More recently, chip-based PCR has been developed using droplet-based microfluidics, allowing millions of discrete amplification reactions to be performed within a few minutes from a single-copy of the template DNA (Zhu et al., 2012). The use of droplet-based microfluidics prevents the channel walls from interacting with the polymerase and template DNA and thus eliminates the local binding of DNA or enzyme that leads to false results, improves reaction yield, and prevents cross contamination of samples. For example, Hindson et al. developed a high-throughput droplet digital PCR (ddPCR) system for quantifying DNA (Fig. 2B) (Hindson et al., 2011). The system was able to process, concurrently, 20,000 PCR reactions from approximately 20 μl of sample/reagent mixture. The partitioning of the sample/reagent mixture provides orders of magnitude greater precision and sensitivity than real-time PCR, and thereby enables the accurate measurement of distinct copy number variations (CNVs) implicated in human diseases, the detection of rare alleles with a capacity to identify mutant DNA in the presence of a 100,000-fold excess of wild-type DNA, and the absolute quantitation of circulating fetal and maternal DNA in cell-free plasma. However, despite such advantages, this technique may not be widely applied in routine experiments because the droplets have to be harvested from a droplet-generating cartridge and transferred to 96-well PCR plates for amplification, and then reintroduced into another microfluidic droplet-reader for analysis.
To overcome the limitations of conventional PCR in detecting the presence of a single pathogen, microfluidics has been integrated with PCR to increase sensitivity dramatically. An example of this technique was introduced by Ottesen et al. (2006), who developed a microfluidic digital PCR platform for single-cell detection (Fig. 2C) (Ottesen et al., 2006). The platform was composed of 1,176 distinct valve-actuated 6.25-nl reaction chambers that allowed 1,176 discrete PCR reactions to be performed. Unlike in the droplet-based method described previously, here the PCR reactions occurred in the chip placed on a conventional flat-block thermocycler, and 5′-nuclease probes were used to generate a fluorescent signal that was detected using a modified microarray scanner to monitor nucleic acid amplification. To design a sample-in-answer-out gene-analysis system, Cheong et al. (2008) developed a one-step real-time PCR method that integrated cell lysis and PCR on a single chip for detecting bacterial cells. In this device, Au nanorods were used to lyse the pathogens in the chip and then DNA was extracted from the cell lysate and amplified in the PCR chamber. Because reagents are not changed or removed during this entire process in such devises, the overall efficiency of the experiments is improved dramatically compared to conventional methods. Other lysis techniques have been integrated in microfluidic PCR systems using thermal (Privorotskaya et al., 2010), chemical (Grabski, 2009), physical (Di Carlo et al., 2003; Huh et al., 2007), and electrical methods (Grahl and Markl, 1996).
Because the PCR microfluidic chip uses small volumes, particular attention must be devoted to preventing reagent evaporation during the thermocycling step. One alternative is to use isothermal techniques for amplifying DNA/RNA samples, which do not require either high temperature or large temperature variations (Asiello and Baeumner, 2011; Craw and Balachandran, 2012). Isothermal amplification techniques are inexpensive and not so labor-intensive as to prevent their use in routine experiments, making isothermal amplification techniques an excellent choice for detecting nucleic acids using microfluidics. Recently, we have developed a microfluidic chip in our laboratory for detecting pathogenic bacteria by using the isothermal real-time helicase-dependent amplification (HDA) technique (unpublished data). The chip is composed of a 16 × 24 array of nL-scale microchambers interconnected by channels and actuated with valves. Real-time HDA for single cell analysis can be performed in each discrete microchamber.
Sample preparation
Sample preparation is a critical step in achieving high sensitivity and specificity in PCR. Hong et al. (2004) have developed 2 distinct microfluidic nL-scale nucleic acid processors for DNA and mRNA purification. Each chip integrated 3 separate compartments to isolate and to lyse cells and to purify DNA or mRNA from lysates without pre- or post-treatment of the samples. For DNA purification, the chip was composed of 3 independent processors. In each processor, lysis buffer, dilution buffer, and Escherichia coli in culture medium were introduced into a metering section and then injected into a microfluidic mixer for the cells to be lysed. The lysate was then flushed over the DNA-affinity column, and the genomic DNA was recovered from the chip for PCR amplification. This methodology allows DNA recovery from a minute number of bacteria (< 28 bacteria), making this process 3–4 orders of magnitude more sensitive than conventional methods. For mRNA purification, magnetic beads bearing oligo-dT polymers were introduced into the separation chamber to form an affinity column; after cell lysis, the lysates were flushed over this affinity column and mRNA was recovered from the column for further analysis or amplification (Fig. 2D).
Nucleic acid hybridization
Recently, various microfluidic DNA-based probes have been used with diverse measurement techniques including surface plasmon resonance imaging (Malic et al., 2011), conductance impedance (Javanmard and Davis, 2011), and fluorescence (Chen et al., 2008a). Ben-Yoav et al. (2012) have developed a microfluidics-based electrochemical biochip containing an array of individually addressable 25-nl reaction chambers, which was fabricated using micro-electromechanical systems (MEMS) technology. Each chamber contains a grid of 3 × 3 sensors and each row of 3 sensors also contains a counter and a reference electrode to complete the 3-electrode system. Three unique single-stranded DNA (ssDNA, 30-mers) probes were functionalized onto patterned electrodes of the chip to detect cDNA hybridization events using electrochemical impedance spectroscopy analysis. This impedimetric biosensor-based technique is similar to whole-cell detection methods but uses ssDNA as bio-recognition elements. This biosensor can detect ssDNA targets on the nM scale and with low cross-reactivity (13%).
DNA synthesis
The ability to produce synthetic genes offers a powerful tool for biological research fields such as genomics and transcriptomics and for small-molecule production. However, high costs currently hamper de novo synthesis of DNA constructs. Microfluidic oligonucleotide synthesis presents a less expensive alternative to conventional methods, requiring lower amounts of reagents than ml-scale macro-experiments.
A few proofs-of-concept for microfluidic DNA synthesis have been presented. For example, Kong et al. introduced a multi-chamber microfluidic system for synthesizing a 1-kb-long gene that uses low concentrations of oligonucleotides (as low as 10–25 nM) and 2 orders of magnitude less reagents than conventional approaches (Fig. 2E) (Kong et al., 2007). More recently, a microfluidic device has been developed for synthesizing 16 oligonucleotides that can be assembled into a 200-bp long DNA construct (Lee et al., 2010). The device is a valve-integrated microfluidic system that enables individual sample manipulation and product collection. Moreover, the microfluidic system provides similar results as conventional methods at costs that are 2 orders of magnitude lower. Finally, by integrating selective amplification of oligonucleotide pools, optimized gene assembly, and enzymatic error correction for highly parallel gene synthesis, Kosuri et al. were able to synthesize 35 kb of DNA encoding 47 genes from a complex background containing 13,000 oligonucleotides (Kosuri et al., 2010).
Proteomics
Identifying all cellular proteins is of major interest to researchers in numerous biology-related fields such as drug discovery or pathogen detection (Hopkins and Groom, 2002; Yan, 2010). However, proteins are often present in extremely low copy numbers in the cell, which makes detecting the proteins in single-cell analysis challenging (Blake et al., 2003; Cai et al., 2006; Elowitz et al., 2002; Rosenfeld et al., 2005). Microfluidic reactors have been developed to analyze proteins from multiple or single cells (Hellmich et al., 2005; Hong et al., 2004; Ottesen et al., 2006; Thorsen et al., 2002). For example, Taniguchi et al. have developed an automated imaging platform based on a microfluidic system to quantify the E. coli proteome and transcriptome with single-molecule sensitivity in single cells. (Taniguchi et al., 2010) The PDMS chip, shown in Fig. 2F, was composed of 96 independent channels designed to hold 96 distinct cell samples in parallel channels, each channel measuring 150 μm (width) × 10 mm (length) × 25 μm (height). The channels were pre-coated with poly-L-lysine to immobilize bacteria within the microchannels (Huang et al., 2007). Cells were then injected into the channels and incubated to ensure stable binding to the channel surface as a monolayer of cells. The test used 1018 strains of an E. coli yellow fluorescent protein fusion bank, and revealed concurrently, in single cells, the spatial (nucleus, cytoplasm, membrane) and quantitative expressions of specific proteins or mRNAs at a single-molecule level.
Flow cytometry is another technique for measuring protein expression that is commonly used to count microscopic elements (Nolan and Sklar, 1998). Recently, microfluidic devices have been developed for flow cytometry (Schrum et al., 1999) to measure antibody staining and transfection efficiency of green fluorescent protein and to characterize cells (Chan et al., 2003; Preckel, 2002). For example, a commercial lab-on-a-chip device (2100 Agilent Bioanalyzer, Agilent Technologies GmbH) has been used to analyze protein expression in primary cells (Chan et al., 2003). In this device made of glass and plastic, a buffer channel is connected to a sample channel to align cells in a stream for analysis in the detection area, which can detect 2 distinct fluorescent colors. Protein profiling and apoptosis measurement can be performed with this chip on mammalian cells, both non-adherent (lymphocytes) and adherent (human umbilical vein endothelial cells (HUVECs) and normal human dermal fibroblasts).
Metabolomics
Metabolomics has attracted considerable attention of researchers recently (Lindon et al., 2007; Morris and Watkins, 2005). Metabolomics is concerned primarily with characterizing small-molecule metabolites, which are intermediates or products of metabolism, and focuses on their physiological effects (Wishart, 2008) such as the regulation of gene expression, cell signaling, cell-cell communication, and cellular differentiation. Conventionally, the metabolome is studied using spectroscopy-based techniques such as nuclear magnetic resonance (NMR) spectroscopy (Griffin, 2003) and mass spectrometry (MS) (Werner et al., 2008a; 2008b). These techniques are coupled to high-resolution separation-chromatography techniques that can detect many distinct metabolites in a single sample (Kraly et al., 2009), such as liquid chromatography (LC) (Porter et al., 2006) and gas chromatography (GC) (Phelps et al., 2002). Quantifying metabolites in vivo under physiological and dynamic conditions is extremely challenging because of the low concentration of the metabolites, their high turn-over rates, and their chemical diversity. Microfluidics can help overcome these challenges because in microfluidics samples are treated and analyzed at high spatial and temporal resolution. Many microfluidic studies or devices have been developed for metabolomics, but they still remain at the proof-of-concept level and have been rarely applied to metabolomic analysis. To date, devices have been developed to perform NMR (Lee et al., 2008) and high-performance LC (HPLC) (Liu et al., 2009) coupled to MS (Bai et al., 2010), but no integrated microfluidic system exists that allows the culture of cells and the analysis of their metabolism on a single chip.
MICROFLUIDICS FOR CELLULAR BIOLOGY
Cell culture
In 1907, Ross Harrison presented his findings on the growth of nerve fibers in vitro and introduced the first reproducible technique for culturing cells for experimentation (Harrison, 1906). From Harrison’s simple experiments, cell culture evolved to become the principal tool for research in cell biology and associated fields including drug discovery, bioengineering, and biomedical engineering. In conventional cultures, cells are maintained in a static environment with defined physicochemical properties (temperature, pH, O2 and CO2 concentration, surface treatment, and culture medium composition). However, conventional culture technologies may be insufficient for overcoming various challenges in cell biology such as high-throughput screening, controlling cellular microenvironment, and single-cell analysis. Diverse cell-culture technologies have been developed to establish high-throughput platforms, to control the cellular microenvironment better, and for cellular analysis (Du et al., 2006; Fan et al., 1995; Hung et al., 2005; Khetani and Bhatia, 2008; Kostov et al., 2001; Maffia et al., 1999; Sorrell et al., 2007; Sundberg, 2000). In the development of these technologies, substantial research effort has been devoted to designing microfluidic cell-culture systems (Bauer et al., 2010; Gomez-Sjoberg et al., 2007; Meyvantsson and Beebe, 2008; Park et al., 2010; Whitesides, 2006; Zhang and Noort, 2011).
For culturing cells in a microfluidic environment, principles from multiple fields including biology, physics, and engineering must be understood. The design of a microchamber for cell culture in a microfluidic system must consider parameters ranging from the choice of material for device fabrication to the geometry of the culture region.
Material
The most popular material used in microfluidics is PDMS because of its numerous physical and practical properties. PDMS is inexpensive and easy to mold, making it ideal for rapid prototyping of microfluidic designs, and PDMS is biocompatible, gas permeable, and transparent, and has low autofluorescence, which make it particularly suitable for use in cell cultures. Although a few recent reports have revealed certain unfavorable characteristic of PDMS (such as sequestration of small hydrophilic molecules and binding of cell membranes by un-cross-linked polymer released from devices) (Regehr et al., 2009) that can affect cell growth and metabolism (Paguirigan and Beebe, 2009), PDMS is likely to remain an affordable rapid-prototyping option for most research laboratories, and will likely be used in association with other materials such as polystyrene, glass, or hydrogels.
Geometry
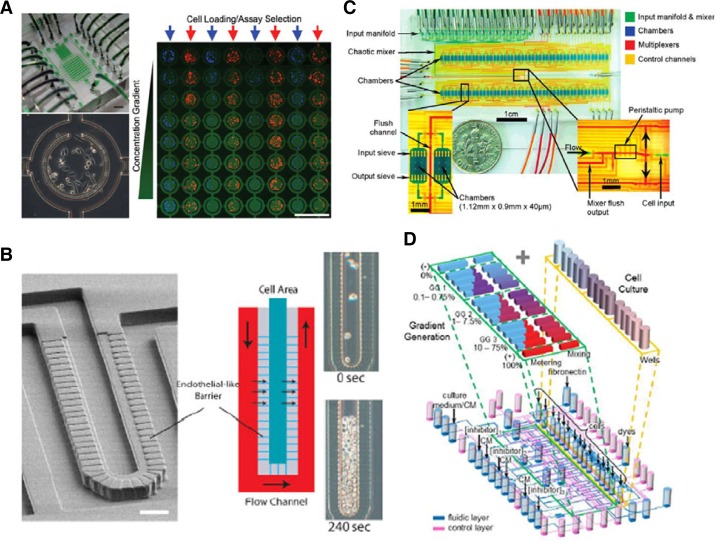
Microchamber geometry is a key factor to consider during chip design. The geometry will determine the amount of culture medium available for the cell culture (the effective culture volume) and the time between each culture-medium exchange (the effective culture time). The simplest culture-chamber design is the microfluidic channel, in which cells adhere to the surface and grow as a monolayer at the bottom of the channel. In this system, culture medium is provided continuously to the cell, constantly renewing the nutrients and removing cellular wastes. However, the stream of fluidic flow generates mechanical stress that can detach cells or influence their behavior (Tilles et al., 2001). To protect cells against the detrimental effects of hydrodynamic shear, walls or barriers have been integrated between the flow and the cells (Hung et al., 2005). For example, Lee et al. (2006) introduced a microchamber C-shaped structure that trapped cells and protected them from the flow of the culture medium (Fig. 3A) while nutrients were provided to the cells through a 2-μm opening at the bottom of the structure. Lee et al. (2007) have developed a microporous perfusion barrier that consists of a grid of microchannels 5-μm wide and 2-μm tall to separate cells from the stream of culture medium while providing nutrients to the cells (Fig. 3B). Shear-sensitive hepatocytes (Ledezma et al., 1999; Yuki et al., 2006) were introduced into this system and they remained viable and functional for the 1-wk culture time. Fabricating micro-wells integrated in a microchannel is another method for shielding cells from direct convective flow (Lecault et al., 2011; Powers et al., 2002). In this configuration, the flow velocity is reduced, with minimal values being near the bottom of the chambers, as a result of the volume expansion from the flow channels to the culture chambers (Lecault et al., 2011). To accurately control the flow velocity of the culture medium delivered to the cell, Gomez-Sjoberg et al. (2007) have integrated an on-chip peristaltic pump at the root of their cell-culture chip. The micropump is composed of 3 valves in series and can inject precise doses of culture media or other reagents at a controlled flow speed into each of the 96 culture chambers; the number of actuation cycles applied to the pump valves controls the dosage, and the frequency at which the valves are switched controls the flow speed. Although all these systems have been applied successfully to cell cultures in microfluidic environments, they required complex designs, microfabrication processes, or substantial amounts of culture medium (Fig. 3C). Recently, we have introduced a simple cell-culture system composed of open-well microchambers, which is represented in Fig. 3D (Hamon et al., 2013). The microchambers were designed to provide sufficient culture medium to hundreds of cells for several days of experiments without renewing culture medium, and thus without subjecting cells to flow stream. Furthermore, we designed the microchamber to introduce cells directly from the top of the open micro-wells, which makes our system accessible to biologists without specific training in microfluidics.
Fig. 3.
Microfluidic systems for cell culture. (A) C-ring microchamber for cell culture. Left top, Photograph of the 64-cell culture unit chip. Left bottom, Close-up of the microchamber with the C-ring that trap and protect cell. Right, Fluorescence image of cell assay done in a array of 64 cell culture units (Lee et al., 2006). (B) Artificial liver sinusoid with microfluidic endothelial-like barrier. Left, SEM of the microchip. Center schematic representation of the fluidic flow in the device. Right. Hepatocytes loaded into the chip at t = 0 s (top) and 240 s (bottom). (Lee et al., 2007). (C) Photograph of a 96 cell chamber chip. Channels are filled with colored water to indicate different parts of the device. Left inset, closer view of two culture chambers. Right inset, Root of the input multiplexer, with the peristaltic pump, a waste output for flushing the mixer, and the cell input line (Gomez-Sjoberg et al., 2007). (D) Three-dimensional schematic representation of the open well microchamber system. Fluidic channels and wells are in blue, control channels are in pink. Inlets for culture medium (CM), inhibitors at different concentrations (1, 2, and 3), fibronectin, and cells are indicated (Hamon et al., 2013).
Cellular environment
Cellular phenotype is governed by 4 distinct factors that direct cell fate, differentiation, and function: chemical factors (e.g., glucose, growth factors, toxins, drugs), physical factors (e.g., temperature, 3D organization, shear stress), cell-cell interactions (e.g., heterotypic or homotypic interactions, cell-cell contact, paracrine communication), and cell-extracellular matrix (ECM) interaction (e.g., collagen type, RDG peptide). In conventional cultures, these parameters are applied to entire cell populations but cannot be applied, observed, and analyzed at the single-cell level. Microfluidics can control the environment both spatially and temporally at the cell level and has been developed successfully to study various cellular behaviors and phenotypes including cell growth (Gomez-Sjoberg et al., 2007), differentiation (Tourovskaia et al., 2004), signal transduction (Liedert et al., 2006), protein secretion and transport (Huang et al., 2004), gene expression (Toriello et al., 2008), cell and ECM behaviors (Tan and Desai, 2003), and cytoskeletal dynamics (Fletcher and Mullins, 2010). In this section, we describe various devices used currently to control the microenvironment of the cell.
Chemical factors
Controlling the chemical microenvironment is one of the main applications of microfluidics, which can be used to reproduce chemical gradients that are involved in vivo in various biological processes such as cellular chemotaxis (Abhyankar et al., 2006; Li Jeon et al., 2002; Lin et al., 2005; Mao et al., 2003; Wang et al., 2004) and migration (Barkefors et al., 2008; Irimia et al., 2006b), differentiation (Chung et al., 2005), gene regulation (Paliwal et al., 2007), and pathology (Walker et al., 2004). Diverse devices have been developed for generating gradients in microfluidic chips, including T-sensor-based devices, premixer gradient generators, and “Universal” gradient generators.
A simple method for generating gradients in a microfluidic system is to use the diffusion properties of 2 parallel laminar flows. At a constant flow rate, the shape concentration gradient will remain constant in the channel. T-sensor devices, which have the simplest design among gradient generators, are composed of 2 or more channels that join in a T-shaped configuration, into a single channel where the gradient is formed (Kamholz et al., 1999).
From T-sensor-based devices, more complex systems have been developed to generate more complex gradients (Cooksey et al., 2009; Dertinger et al., 2001; Irimia et al., 2006a). These systems, referred to as premixer gradient generators, have been used in studies of cell chemotaxis (Li Jeon et al., 2002; Lin et al., 2005; Mao et al., 2003; Wang et al., 2004), migration (Barkefors et al., 2008), and differentiation (Chung et al., 2005). For example, Chung et al. (2005) described a gradient-generating microfluidic platform for optimizing the proliferation and differentiation of cells in a laminar-flow-based microfluidic system; the device contains a gradient generator that generates a gradient of growth factors (GFs) in a cell-culture chamber. Human neural stem cells (hNSCs) were cultured for more than 1 wk in the microfluidic device, and the cells were exposed constantly to a continuous gradient of a GF mixture composed of epidermal growth factor (EGF), fibroblast growth factor 2 (FGF2), and platelet-derived growth factor (PDGF). During the culture, hNSCs proliferated and differentiated into astrocytes in proportions that varied with the concentrations of the GFs. These results were similar to results obtained in parallel conventional cultures prepared in 6-well plates. More recently, Sugiura et al. (2010) presented a premixer gradient generator-based system that generated a logarithmic-scale concentration gradient over 6 orders of magnitudes. The chip was composed of a serial-dilution microfluidic network for generating the concentration gradient, which was connected to an array of cell-culture microchambers, where cell assays were performed.
The “Universal” gradient generator (Irimia et al., 2006a) is another device derived from the T-sensor. In the mixing channel, a series of physical walls controls the orthogonal diffusion of chemicals between adjacently flows. By controlling the location of the dividing walls, gradients with distinct profiles can be generated (Irimia et al., 2006) to apply to cells.
The continuous perfusion of culture medium in the cell chamber is a great advantage because it renews the culture medium constantly, providing fresh nutrients to the cells while flushing away waste products. However, the flow generates shear stress that can change cellular behavior (Diao et al., 2006; Keenan and Folch, 2008) and remove autocrine and paracrine factors secreted by cells (Keenan and Folch, 2008).
One solution for limiting the influence of fluidic flow on cells and the cellular microenvironment is to separate the cells from the flow by using a barrier that allows passive diffusion of bio-molecules. For this purpose, various systems have been developed that include barriers made from hydrogels (Mosadegh et al., 2007; Saadi et al., 2007; Wu et al., 2006), nanoporous membranes (Abhyankar et al., 2006), and microchannels (Saadi et al., 2007). These systems can prevent shear stress generated by flow, retain the autocrine/paracrine signals in the cellular microenvironment, and be integrated with the gradient generator (Hung et al., 2005). Furthermore, these systems use small amounts of reagents and facilitate experiments on non-adherent cells such as yeasts (Paliwal et al., 2007).
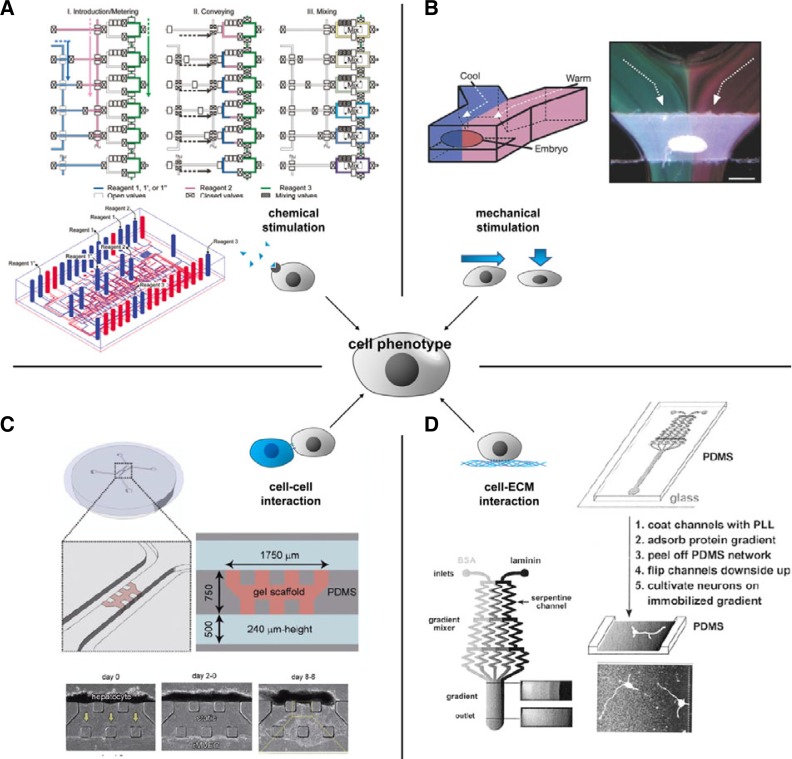
Droplet-based microfluidic approaches offer several advantages that can be exploited while testing for the influence of chemical factors on cells: high-throughput screening, low sample volumes (Jambovane et al., 2011), and single-cell analysis (He et al., 2005; Teh et al., 2008). In 2009, Brouzes et al. (2009) developed a droplet-based microfluidic system for encapsulating single mammalian cells and reagents and for high-throughput cytotoxicity screening. Three separate chips were used, one each for generating drug libraries, merging, and assaying. First, an optically labeled library of drugs was generated on the library-generation chip. Next, each drug-library droplet was combined with a cell-containing droplet on the merging chip and the combined droplets were incubated. After incubation, the combined droplets were introduced into the assay chip to determine whether the cells were alive or dead. The treated-cell droplets were merged with fluorescent-dye droplets by using a fusion module. Inside each droplet, the dye was mixed with the cells while passing through a mixing module. The droplet passed through a 15-min delay line for optimizing the incubation of the cells with the live/dead-marking fluorescent dyes. Finally, fluorescent signals were detected in a detection module. The advantages of using valve-based microfluidics to study the effects of chemical factors on cells are the following: prevention of cross-contamination (by isolating each cell chamber from the rest of the system), rapid mixing, and controlled formation of accurate, non-linear, and wide concentration gradients. In conventional experimentation, concentration gradients are generated using a series of dilutions usually by pipetting, which may affect the precision of each step of the dilution process. Addition of each small imprecision may lead to results that do not accurately represent the true potential of the chemical factors being tested (Behringer, 1989). To develop an error-free system, Hong’s group recently introduced a microfluidic system that can generate accurate logarithmic-scale concentration gradients (Fig. 4A) (Yun et al., 2011) and integrated this system with open-well microchambers for cell culture and dose-response assays (Hamon et al., 2013). The chip comprised 3 gradient generators (GG). Each GG was composed of 4 microreactors, and each microreactor contained a metering section, a mixing section, and an open-well microchamber. In the metering section, valves from the control layer were used to control the mixing ratio of the 2 reagents. The valves were positioned along channels at specific distances, which allowed a precise volume of liquid to be contained in the channels. By controlling the placement of 3 valves along a channel, it was possible to recreate a mixing ratio (Jambovane et al., 2009; Yun et al., 2011). In each GG, the metering section was designed to obtain final concentrations that were 75%, 50%, 25%, and 10% of the concentration of the solution introduced originally. For example, by introducing 10× inhibitor and 1× working buffer into a GG, concentrations that were 1×, 2.5×, 5×, and 7.5× of the original could be obtained. Similarly, by introducing 10× inhibitor into the first GG, 100× into the second GG, and 1,000× into the third GG, effective concentrations could be obtained that were 1×, 2.5×, 5×, 7.5×, 10×, 25×, 50×, 75×, 100×, 250×, 500×, and 750× of the original and included “0” for the negative control and 1,000× for the positive control. In the mixing section, valves were used to create a peristaltic pump. Three valves were positioned closely, in parallel, along the mixing chamber. By opening and closing the valves sequentially, a flow was generated in the channel and the 2 reagents were mixed. The mixed chemicals were then injected into the micro-well chambers, resulting in further dilution of the solutions. Each microchamber was isolated from the rest of the system by 4 distinct valves. Using this chip, the researchers demonstrated the possibility of creating a logarithmic concentration gradient and the possibility of conducting cell-based assays for determining the IC50 values of 3 cytotoxic agents. These IC50 values were comparable to those obtained using conventional methods.
Fig. 4.
Microfluidic system for cell environment studies. (A) Device for the study of soluble factor on cells. Step-by-step process (top) and schematic representation (bottom) of a microfluidic chip for wide range of concentration gradient of soluble factor. Blue line represent the fluidic channels, red lines represent the control channels (Yun et al., 2011). (B) Laminar flow-based device for mechanical stimulation of cells. Schematic representation (left) and photograph (righ) of a Drosophilia embryo submitted to different temperature (Lucchetta et al., 2005). (C) Device for cell-cell interaction studies. The gel scaffold separate endothelial cells and hepatocytes. Photographs shows the migration of endothelial cells through the gel (bottom) (Sudo et al., 2009). (D) Device for the formation of gradient of laminin and the study of neurons laminin-based orientation (Dertinger et al., 2002).
Physical factors
Cellular phenotypes have been shown to be directly related to the mechanical environment of cells. Confinement, shear stress, and temperature are physical factors that influence cellular behaviors including growth, differentiation, gene expression, metabolism, and death. Microfluidic systems can help elucidate the physical factors that affect the behavior of cells: the flexibility of the fabrication process and device actuation allows physical parameters such as channel size, flow speed, and stream temperature to be controlled.
Shear stress
Shear stress is a critical factor that affects the physiology and pathophysiology of vascularized tissues (Cunningham and Gotlieb, 2005). Understanding the influence of shear stress on cells is crucial for both in vivo studies and in vitro bioengineering (Imberti et al., 2002; Jain et al., 2005). Conventionally, cell responses to flow streams are studied in systems derived from the parallel-plate chamber first developed by Frangos et al. (1988). These systems were successfully used to study the influence of fluid flow on endothelial cell (EC) functions in vitro (Malek Am, 1999; Resnick et al., 2003). However, the systems consume large amounts of reagent and do not entirely mimic the in vivo environment because they do not present the geometry of in vivo microvascular structures. Microfluidics is an attractive alternative to conventional methods because it consumes lesser amounts of reagents and can control fluid flow in the cellular microenvironment (Gu et al., 2004; Unger et al., 2000). Various devices have been developed to control the shear stress applied on cells to mimic the in vivo microenvironment or to study the influence of shear stress on cellular behavior (Khan and Sefton, 2011; Mack et al., 2009; Vickerman et al., 2008). For example, Mack et al. (2009) used a microfluidic chip to demonstrate that collateral flow regulates EC gene-expression profiles and endothelium-derived paracrine signaling that affect vascular events critical for adaptive remodeling, particularly the migration of smooth muscle cells (SMCs); these findings revealed the control by biomechanical forces of cellular phenotype and cell-cell interactions. The chip had 2 independent microchannels separated by a 3D region filled with a hydrogel scaffold. First, a type I collagen scaffold was injected into the 3D region, and then SMCs and ECs were introduced into their respective channels (smooth-muscle channel and endothelial channel). Next, when distinct collateral waveforms were applied to ECs, dissimilar gene-expression profiles and phenotypes were observed. Factors generated by the ECs were transported by passive diffusion across the scaffold, where they stimulated SMC migration. The configuration of this chip (i.e., 2 independent microchannels separated by a gel/scaffold compartment) has been used often to control (Vickerman et al., 2008) or study (Mack et al., 2009) the influence of shear stress on cultured cells, with microfluidics allowing the flow to be controlled precisely in the channels as well as across the gel compartment (Song and Munn, 2011).
Confinement
The cellular microenvironment in vivo is a confined space where cells live, proliferate, and move. This confined environment is challenging to recreate in conventional cell cultures and is often limited to studies on cells in 3D collagen gels. By contrast, artificial confinement can be achieved readily using microfluidic systems to study the impact of confinement on cell structure (Minc et al., 2009; Takeuchi et al., 2005) and behavior (Irimia and Toner, 2009; Tse et al., 2012). Moreover, microchannels can be used to reproduce the physical confinement encountered by cells in vivo during migration through the extracellular matrix (Friedl and Alexander, 2011; Huang et al., 2013). Microfluidic platforms for studying cell motility have been reported in several studies (Faure-Andre et al., 2008; Friedl and Alexander, 2011; Heuzé et al., 2011; Irimia and Toner, 2009; Tse et al., 2012). For example, Faure-Andre et al. designed a simple microfluidic chip to study the regulation of dendritic cell (DC) migration (Faure-Andre et al., 2008). The researchers observed the migration patterns of wild-type DCs and CD74-deficient DCs in 4-μm width parallel, fibronectin-coated microchannels and demonstrated the importance of CD74 in controlling the cell-migration mode. Irimia and Toner (2009) have developed microfluidic devices that mechanically constrain migrating cancer cells inside microchannels that have a cross-section comparable to the cell size. Interestingly, cell migration was observed in the absence of external chemical gradients, suggesting that mechanical confinement regulated the control of cell motility.
Temperature
Temperature is a key parameter that influences many aspects of cell biology. Because microfluidic systems use small volumes, these systems have enormous potential for application in temperature-based biological studies. We previously described technologies in which integrated thermostated copper electrodes were used for spatiotemporal control of on-chip temperatures (Kopp et al., 1998). However, because the electrodes were not transparent, this technology is not suitable for cell-related studies. One simple solution is to perfuse temperature-controlled liquid into the microfluidic device. Two distinct laminar-flow-based systems have been developed to control temperature in microchannels. The first system uses microfluidic laminar flow to create temperature disparities by flowing 2 converging aqueous streams, each at a controlled temperature, in the microchannel (Lucchetta et al., 2005; Pearce et al., 2005). Lucchetta et al. (2005) applied this approach to investigate Drosophila embryologic development. The researchers trapped a live embryo in the cross-section of a T-sensor-based microfluidic device and submitted the 2 sides of the embryo to distinct temperatures (Fig. 4B); the results obtained demonstrated that the developmental rate of each half of the embryo depended on the temperature. The second system uses a 2-layer microfluidic device. The temperature on the chip is controlled by flowing water through a 0.3-mm-deep and 1.4-mm-wide channel in the top layer, above the array of channels and cell chambers. Velve Casquillas et al. (2010) used a similar system in which temperature could be switched reversibly between 5°C and 45°C in less than 10 s and sinusoidal shifts between the 2 temperatures could be generated at a rate of 1 shift/min. By controlling the temperature within the cell chamber, the researchers could control cytoskeletal dynamics and activate or deactivate temperature-sensitive gene in yeast (Velve Casquillas et al., 2010).
Cell-cell interaction
Intercellular communications across heterotypic cell-cell junctions or through paracrine signaling mechanisms are essential for responding to stimuli and determining and adjusting cellular phenotypes. Numerous studies have demonstrated the key roles played by intercellular communications in applications of cellular biology such as tissue engineering, cancer therapy, and regenerative medicine using stem cells. Despite the importance of intercellular communications, these complex processes remain incompletely understood because conventional cocultures in dishes and artificially manipulated cell patterns only provide general information on cell populations and cannot isolate specific signals among mixtures of multiple, time-variable signals. Microfluidics offers the possibility to downsize the cell population and thus focus on cellular interactions at the single-cell level. The ability to perform high-throughput screening on a single chip is also critical for obtaining a clear understanding of genotypic and phenotypic variation among similar cell type. Several microfluidic devices have been developed to study cellular communication (Hong et al., 2013; Huang et al., 2009; Mao et al., 2013; Okuyama et al., 2010; van der Meer et al., 2013), but only a few have focused on specific homotypic or heterotypic cell-cell interactions (i.e., direct interactions between cell surfaces) (Huang et al., 2009; Okuyama et al., 2010; van der Meer et al., 2013). Most of the microfluidic systems are composed of 2 channels, one for each cell type, separated by a gel channel or intervening gel, usually filled with type-1 collagen gel (Chung et al., 2009; Huang et al., 2009; Sudo et al., 2009). Paracrine communication generally initiates the migration of one of the cell types through the gel channel to reach and interact with the other cell type in the second channel. Sudo et al. applied this system to liver cell cultures (Sudo et al., 2009). When cultured under interstitial flow, hepatocytes formed 3D tissue-like structures, with cell-cell cohesion apparently being enhanced by the interstitial flow. When hepatocytes and ECs were cocultured in the microfluidic chip, the ECs formed 3D capillary-like structures that migrated across the gel to the hepatocyte compartment. However, although heterotypic cell-cell interactions occurred in this device, robust outgrowths of hepatocytes toward the ECs were not observed. Nevertheless, these results demonstrated the possibility of using microfluidics to study homotypic and heterotypic cell-cell interactions (Fig. 4C).
Cell-extracellular matrix interaction
Elucidating the properties of the ECM surrounding the cell is critical for understanding the role of the microenvironment on cell fate (Flaim et al., 2005) because ECM composition and physical features directly influence a range of cellular processses including cell life/death, differentiation, shape, polarization, and motility (Chen et al., 1997; Discher et al., 2005; Engler et al., 2006; McBeath et al., 2004; Pelham and Wang, 1997; Prager-Khoutorsky et al., 2011; Selhuber-Unkel et al., 2010; Trappmann et al., 2012; Watt et al., 1988).
Conventional cell-ECM interaction studies involve absorbing purified matrix proteins on cell-culture substrates. However, determining the role of the ECM and ECM concentration (McCarthy and Furcht, 1984; McCarthy et al., 1983; 1985) on cellular behavior, such as in haptotaxis studies, through conventional methods is hindered by the large amount of molecules required for each experiment, the insufficient availability of certain ECM molecules, and the difficulty to generate consistent gradients of ECM molecules over distances required for biological studies (a few 100 μm) (Dertinger et al., 2001; 2002; Flaim et al., 2005). Laminar-flow microfluidics can be employed to easily generate a linear gradient using small amounts of molecules, and thus this approach appears to be an attractive alternative to conventional methods for conducting studies on specific cell-ECM interactions (Dertinger et al., 2002; Gunawan et al., 2006). Dertinger et al. (2002) were the first to use micro-fluidics for haptotaxis studies. The PDMS device shown in the Fig. 4D was designed to generate and coat concentration gradients of laminin and bovine serum albumin (BSA) in a single channel. The linear-gradient generator was based on a previous design from the Whiteside group: a T-sensor-based network with 2 inlets (one for BSA, the other for laminin) was used. Between each dilution step, 55-μm-long serpentine channels mixed all streams at a flow rate of 1 mm/s, which was sufficient to ensure complete diffusive mixing of ECM proteins. The 5 streams, each carrying distinct concentrations of laminin and BSA, were combined in a single channel to form a gradient perpendicular to the direction of the flow. After absorbing the gradients on the channel walls, the microfluidic device was sealed, inverted, and placed in a culture dish, and cells were then cultured on the floor of the channel in the region where the gradient had formed. Rat hippocampal neurons were cultured on these substrate-bound gradients, and analysis showed that the surface density of laminin oriented axon specification. More recently, Hsu et al. (2005) have developed a simple microfluidic device to investigate the interactions between haptotaxis and shear stress during EC migration. A microfluidic chip was patterned with type-1 collagen and, after patterning, the device was removed and the entire substrate was coated with collagen, resulting in the formation of step changes in collagen density. Because of haptotaxis, ECs migrated into the area with a high density of collagen. To study the influence of shear stress, a parallel flow chamber was used to generate a flow perpendicular to the collagen strips, and various levels of fluid shear stress were applied on the ECs. The results obtained using this system suggested that shear stress beyond a certain threshold can overcome the influence of haptotaxis and predominantly direct EC migration.
Cell separation and detection
The isolation of pure populations of cells is critical in diverse biological applications such as biological research (Choi et al., 2009; El-Ali et al., 2006; Roda et al., 2009), diagnostics (Li et al., 2002; Toner and Irimia, 2005; Villanueva et al., 2006), pathogen detection (Cheng et al., 2007; Gagnon and Chang, 2005; Tan et al., 2011; Varshney et al., 2007), and therapeutics (Sethu et al., 2006). Flow cytometry, fluorescence-activated cell sorting (FACS), magnetic-activated cell sorting (MACS), and centrifugation are common conventional methods used for detecting cell separation. However, these techniques are expensive, require trained personnel, and can alter cell functions (Kumar and Bhardwaj, 2008). Microfluidics presents several characteristics that make it a suitable technique for isolating cells of interest: small sample volumes, low cost, integration with analytical techniques, and portability (Boedicker et al., 2008; Boehm et al., 2007; Petersen et al., 2002; Qiu et al., 2009; Wu et al., 2005; Yi et al., 2006). Several concepts based on microfluidics have been developed over the past 10 years for cell isolation, separation, and detection (Autebert et al., 2012). These systems can be classified arbitrarily into 2 categories: systems based on the biomolecular properties of cells (presence of specific surface antigens), or affinity-based techniques; and systems on the physical properties of cells (such as size, deformability, and density), or label-free techniques.
Affinity-based techniques
A common approach used to isolate specific cells is based on recognizing specific cell-surface markers. This approach is similar to the one used to develop conventional, non-microfluidic techniques such as flow cytometry-based techniques (FACS) (Crosland-Taylor, 1953) or MACS (Molday et al., 1977).
Fluorescence-based cell detection
Fluorescence-based cell detection methods such as flow cytometry present several advantages when compared with other techniques, including sensitivity, multicolor detection for concurrent detection of multiple cell types, stability, low hazard, availability, and low cost (Van Rood et al., 1976; Zhao et al., 1992). Recently, Yamaguchi et al. (2011) developed a simplified and rapid on-chip method in which a microfluidic cytometry device was used for rapidly quantifying bacteria such as E. coli in water. The chip had 4 parts: a T-shaped introduction section, a mixing channel, an alignment part, and a detecting part. Samples and fluorescently labeled antibodies against E. coli O157:H7 were injected into the T-shaped introduction section; bacterial cells from the sample were stained with the fluorescent antibodies in the mixing channel; the cells were then aligned by injecting a sheath fluid (PBS) into the main stream through 2 microchannels; and the fluorescently stained cells were detected in the detecting part. The researchers demonstrated that this simple fluorescence-based technique is capable of detecting a small number of bacterial cells (104 cells/ml). Although this system’s sensitivity does not differ substantially from that of conventional epifluorescence microscopy (EFM), using microfluidics presents several advantages over EFM: the microfluidics method is simpler because pretreatment apparatus is not required; the method is rapid and results can be obtained in 1 h; and the method can be integrated with small light sources such as light-emitting diodes (LEDs) for on-site analysis.
Impedimetric biosensor
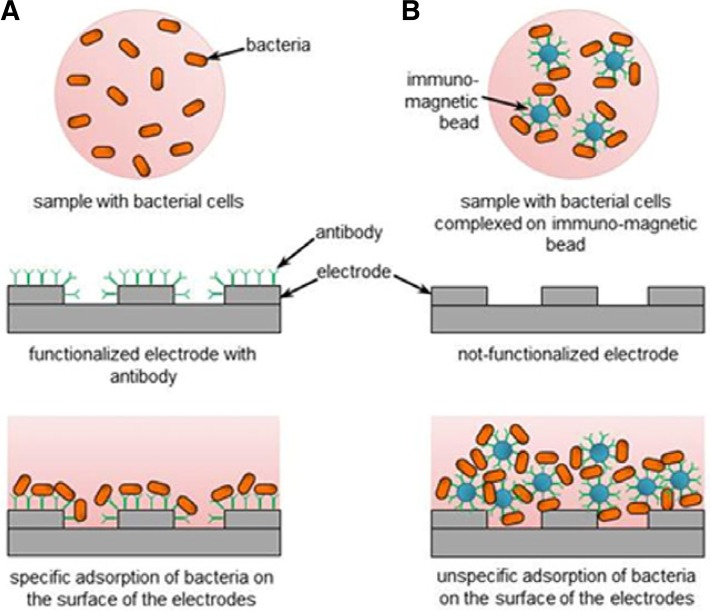
Impedimetric biosensors have been used for concentrating and detecting microorganisms present in food and water. These biosensors rely on the insulating properties of bacterial cell membranes. Impedimetric biosensors are composed of a solid electrode to which cells attach through specific or non-specific adsorption (Fig. 5) (Varshney and Li, 2009). After attaching to the electrode, the natural capacitance and resistance of the cell membrane (Pethig, 1979) affect the current that reaches the electrode. The adsorption of bacteria on the electrodes is thus revealed through detecting changes in the electrical properties of the electrode (Berggren et al., 1998; Mirsky et al., 1998; Sadik et al., 2009; Towe and Pizziconi, 1997). To improve the sensitivity these biosensors, microelectrodes have been developed that allow low Ohmic drop, quick establishment of steady-state, rapid reaction kinetics, increased signal-to-noise ratio (Amatore et al., 1983; Ciszkowska and Stojek, 1999; Maruyama et al., 2006), and integration into microfluidic systems (Bayoudh et al., 2008; Boehm et al., 2007; Gottschamel et al., 2009; Liu et al., 2007; Mernier et al., 2010; Richter et al., 2007; Yu et al., 2009; Zaytseva et al., 2005; Zhu et al., 2010). Integration in microfluidic systems offers various advantages over impedance macrosystems (Deng et al., 1996): increased detection sensitivity, quicker response, and lower reagent consumption (Tan et al., 2011; Varshney and Li, 2007).
Fig. 5.
Working principle of impedance detection technique with (A) or without (B) the use of bio-recognition elements for bacterial cell adherence on the electrodes (Varshney and Li, 2009).
For absorbing specific cells on the surface of the electrodes, the electrode surface must be functionalized with bio-recognition elements (or bioreceptors), usually antibodies specific to pathogenic bacteria. When bacteria attach to the antibodies, the insulating properties of the cell membrane induce measurable electrical changes in the electrode (Radke and Alocilja, 2005a; Radke and Alocilja, 2004; 2005a; 2005b; Yang and Bashir, 2008). Recently, Tan et al. (2011) integrated an electrochemical impedance spectroscopy (EIS)-based immunosensor, which was composed of an alumina nanoporous membrane with immobilized antibodies specific to foodborne pathogens, with a PDMS microfluidic system to detect E. coli and Staphylococcus aureus rapidly. This microfluidic immunosensor based on the impedance spectrum of the nanoporous membrane detected bacteria within 2 h with a sensitivity of 100 cells/ml in pure cultures.
In the “non-specific adsorption” method, cells are first separated, re-suspended, and concentrated with the help of magnetic nanoparticle-antibody conjugates (MNACs), functionalized with antibodies (against E. coli in the following example), and then spread uniformly on the surface of an electrode network. In 2007, Varshney et al. (2007) developed an impedimetric biosensor based on the non-specific absorption of bio-recognition elements for detecting E. coli O157:H7 in food samples. The impedance sensor detected a minimum of 7.4 × 104 and 8.0 × 105 cells/mL of E. coli O157:H7 in pure cultures and ground beef samples, respectively (Varshney and Li, 2009). The integration of this technology within a microfluidic system increased detection sensitivity to 1.6 × 102 and 1.2 × 103 cells/ ml of E. coli O157:H7 in pure cultures and ground beef samples, respectively, in 60-nl samples (Varshney et al., 2007).
Label-free techniques
An intuitive way to separate target cells from a circulating flow is to use the cells’ intrinsic and specific physical properties such as size, density, and deformability. These methods free cell detection from the laborious steps required for labeling cells. Systems have been developed to separate cells using mechanical properties of microfluidic flows, microfabrication, and integrated systems.
Hydrodynamic sorting
In a microfluidic system, at a low Reynolds number, fluid streams follow a laminar flow. When particles such as cells circulate in a microchannel, the center of the particle follows a stream line. Hydrodynamic sorting relies on this behavior. Based on this principle, sorting systems have been developed using microchannel bifurcation or pinched-flow fractionation. In the bifurcation-based system (Fig. 6A), several perpendicular channels branch out from the main channel, and at each intersection, a fraction of the fluid flows down the perpendicular channels, dragging cells closer to the walls. As the cells travel downstream, they leave the main channel and enter the branched channels, with smaller cells leaving the main channel earlier. Yamada et al. (2007) for example, have used this system to separate hepatocytes and non-parenchymal cells from a pool of liver cells.
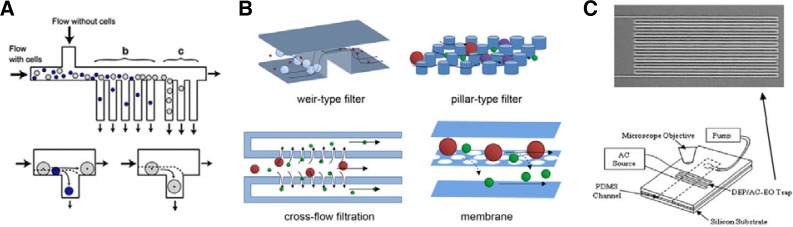
Fig. 6.
Microfluidics for cell sorting. (A) Hydrodynamic-based cell sorting using microchannel bifurcation (Yamada et al., 2007). (B) Schematic representation of four types of filters for size-based cell sorting (Ji et al., 2008). (C) Dielectrophoresis system for cell sorting a wire of platinum (top) for trapping bacteria (Gagnon and Chang, 2005).
Physical filters
The size of a cell is the cellular phenotype observed most easily. Thus, cell size is a common criterion used for cell separation. Cells can be separated based on their size most simply by using physical filters. Four types of filtration have been reported in microfluidic systems: weir-type, pillar, cross-flow, and membrane (Fig. 6B) (Ji et al., 2008). In weir-type filtration, a barrier is fabricated to constrict the height of the channel, with a space maintained between the barrier and the ceiling of the channel that allows fluids and small particles to pass through while retaining large particles. In pillar-type filtration, micro-pillars are spaced apart in an array along the micro-channel to filter out particles. Cross-flow filtration is based on the same principle, but the flow is perpendicular to the micro-pillar array. Membrane filters contain well-defined pores that separate larger cells from the fluid and smaller particles. Size-based filtrations face challenges such as clogging and fouling. However, designing micro-pillars (Tan et al., 2009) or using cross-flow systems (Chen et al., 2008b) can prevent cells from obstructing flow.
Dielectrophoresis
One main limitation of whole-cell detection using microfluidic systems is the difficulty in detecting small numbers of cells in nl-volume samples. Among the methods used for whole-cell detection, dielectrophoresis techniques can concentrate the target cells within a specific area and thus amplify the signal for efficient detection (Cheng et al., 2007; Gagnon and Chang, 2005). For example, Gagnon and Chang (2005) have developed a microfluidic device (Fig. 6C) composed of microchannels aligned atop a thin continuous serpentine micro-wire of platinum. When alternating current passes through the wire, it generates an electro-osmotic flow that traps and concentrates cells in the micro-wire area and directs the cells towards a designated point on the device, while suspended particles are swept toward the outlet with the fluidic flow (Gagnon and Chang, 2005). This system requires less than 100 μl of solution and can concentrate 103 particles/ml in a few seconds.
OTHER BIOLOGICAL PERSPECTIVE: ORGAN-ON-A-CHIP AND ORGANISM-ON-A-CHIP
Organ-on-a-chip
Microfluidic systems offer the unique opportunity to fabricate cell culture systems, called organ-on-a-chip (Fig. 7), that can simulate the biological activity, mechanical properties, and physiological and pathophysiological responses of whole organs and organ systems (Baker, 2011; van der Meer and van den Berg, 2012). Examples of these include the lungs (Huh et al., 2010), intestines (Kim et al., 2012), or vasculature systems (Korin et al., 2012). Two main laboratory techniques have been developed to fabricate organ-on-a-chip.
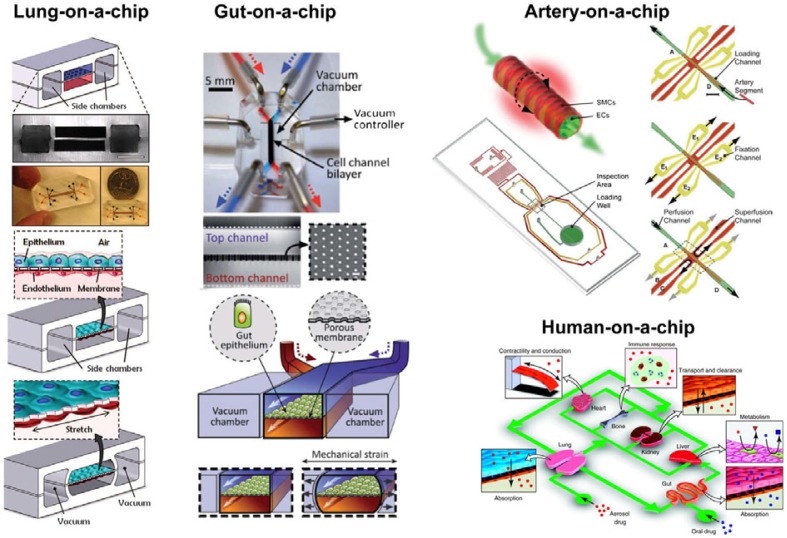
Fig. 7.
Biomimicking organs-on-a-chip. Lung-on-a-chip: Top, Schematic representation and photograph of the microfluidic device. Bottom, Representation of the working chip. Epithelial and endothelial are culture on both side of a porous PDMS membrane. Vacuum is made in the side chamber to mimic breathing motions (Huh et al., 2010). Gut-on-a-chip: Top, Photographic images of the gut-on-a-chip PDMS device. Blue and red dyes have been introduced into the upper and lower microchannels, respectively. A cross-sectional view of the top and bottom channels (150 mm high) of the chip; square inset shows a top view of the porous PDMS membrane (10 mm pores). Bottom, A schematic of the gut-on-a-chip device showing the culture of gut epithelial cells on the flexible porous PDMS membrane in the central microchannel, and two vacuum chambers on both sides (Kim et al., 2012). Artery-on-a-chip: Top, Schematic representation of an artery segment for the measurement of the transendothelial resistance (top) and of the chip (bottom) with the microchannel network, the loading well and the inspection area. Right, Representation of the reversible procedures for artery segment loading, fixation and inspection (Gunther et al., 2010). Human-on-a-chip, Schematic representation of a human-on-a-chip concept in which different organs are integrated into a single microdevice and linked by a microfluidic circulatory system in a physiologically relevant manner (Esch et al., 2011).
In the first, cells are cultured in a microfluidic device that mimics the organ microenvironment to direct cell organization and functions. For example, Huh et al. (2010) have developed a biomimetic microsystem that reproduces the functional alveolar-capillary interface of the human lung. In this embodiment, a PDMS microfluidic chip containing 3 parallel channels was constructed. The middle channel is composed of 2 opposing microchannels separated by a thin, porous PDMS membrane. Cells were cultured on each side of the membrane - alveolar epithelial cells on one side, and pulmonary microvascular endothelial cells on the other side. The 2 side channels were connected to a vacuum pump. A decrease of the pressure in the side channels pulls the sides of the middle channel, allowing a pressure-driven stretching of the porous membrane and, subsequently, of the cells, mimicking the cyclic mechanical strain of the alveolar-capillary complex observed during respiration in vivo. To validate the biological accuracy of their device, the researchers simulated diverse pathophysiological environments including inflammatory responses, absorption and toxicity responses to airborne nanoparticulates, and bacterial infection.
In the second technique for organ-on-a-chip fabrication, tissue samples are taken from a living animal and integrated to a microfluidic chip. One example is the recently developed artery-on-a-chip, a primary blood vessel on a chip that can assess functions and structures of intact blood vessels. In this system, shown in Fig. 7, animal blood vessels are manually isolated, introduced into the microfluidic chip, and reversibly fixed for perfusion and studies (Gunther et al., 2010).
Ultimately, human physiology results from the interaction of many different organs. To reproduce the organ-organ interactions observed in vivo, platforms to culture thick sections of whole living organs are usually used (Blake et al., 2010; Huh et al., 2011; van Midwoud et al., 2011a; 2011b). In an alternative approach, researchers are working toward the development of a “human-on-a-chip”, which integrates multiple compartmentalized microchambers in which cells are cultured to mimic multiple organs in the body (Fig. 7) (Esch et al., 2011). Microchambers are interconnected by microchannels that are arranged to permit the recirculation and exchange of metabolites in a physiologically-relevant manner. However, early platforms lacked tissue-specific function due to a poorly relevant organ microenvironment in the microchambers. More recently, the integration of biomaterial and microfluidics has allowed the formation of a 3D microenvironment with heterotypic cell cultures that provide a more organ-relevant culture microenvironment for better physiological response of the systems (Sung and Shuler, 2009; Sung et al., 2010).
Organism-on-a-chip
Caenorhabditis Elegans, Danio rerio (or zebrafish), and Drosophila melanogaster are model animals that have drawn great scientific attention because of their relative systemic simplicity, their similarity with mammals, their easy handling, and/or their relative transparency during their embryonic (D. rerio), larval (D. melanogaster) or entire life (C. elegans). To address specific biological questions, animal models must be studied in a well-controlled environment where a precise stimulus can be applied, and a specific behavior or phenotype studied. Despite great potential, conventional methods are facing several issues, particularly in the animal immobilization approach. Conventional immobilization methods, which include glue (Hilliard et al., 2005; Suzuki et al., 2003), anesthetic (Fuger et al., 2007; Schmid et al., 2008) or dissection (Gunawardena et al., 2003; Pilling et al., 2006), perturb the animal, leading to a misrepresentation of its behavior or phenotype. Microfluidics platforms have been developed as an alternative to immobilize animal with limited impact while providing a controlled environment in a high-throughput manner. Here, we will briefly review some of the devices developed for on chip study of worms, flies, and fish.
Fly-on-a-chip
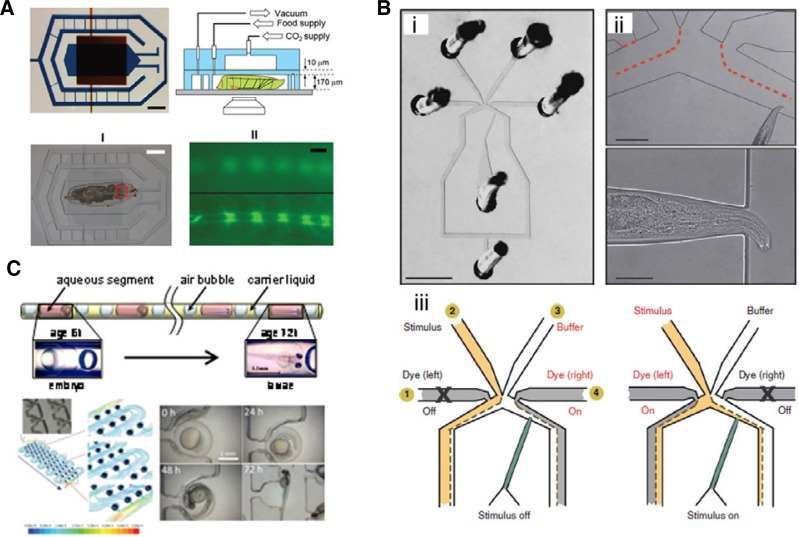
Ghannad-Rezaie et al. (2012) reported a microfluidic- based immobilization methodology for time-lapse imaging in Drosophila larvae (Fig. 8A). To overcome the inherent issue of organism immobilization, they regulated mechanical forces and the carbon dioxide supply to temporarily immobilize larva. The microfluidic chip is composed of an immobilization chamber, feeder channels and a CO2 chamber on the top of the immobilization chamber, separated by a 10 μm thick PDMS membrane. A larva is introduced into the immobilization chamber and is anesthetized with CO2 passing through the PDMS membrane. This ‘larva chip’ has been used successfully in the characterization of several subcellular responses to axotomy, including waves of calcium induction and changes in intracellular transport of vesicles that occur within seconds after axotomy, and axonal regeneration, which is initiated after a 7-h period of dormancy. While this work focused upon cellular responses to targeted injury of neurons, the microfluidic chips could be used for in vivo imaging of many different other pro-cesses in Drosophila larvae.
Fig. 8.
(A) Fly-on-a-chip, Top, Top view and side view representation of the two-layer chip. The larvae is immobilized in the lower microchamber (blue), which is connected to two microfluidic channels to supply food to the larva head. A second PDMS chamber (red) is above the first PDMS layer to deliver CO2. Bottom, (I) Bright-field image of a 3rd instar larva immobilized in the chip. (II) Fluorescent images of the larva body before (top image) and after (bottom image) immobilization (Ghannad-Rezaie et al., 2012). (B) Worm-on-a-chip, (i) Photographic image of the ‘olfactory’ chip. (ii) Higher magnification photographs of the chip with a trapped worm, which nose is exposed to the buffer stream (dotted lines). (iii) Schematic representation of how the stimulus and buffer streams are directed by the side flows (gray). In the ‘off’ state (left), the channel 4 is open and pushes the stimulus stream to the exit. In the ‘on’ state (right), the channel 1 is open, and the worm is exposed to stimulus (Chronis, 2007). (C) Fish-on-a-chip, Microfluidic chips for fish embryology study. Top, Schematic representation of micro fluid segment technique for screening and development studies on fish embryos. Bottom, Photo and 3D streamline of flow obtained by computational fluid dynamic simulations across the fully loaded device. Time-lapse images of developing fish embryos collected every 24 h (Akagi et al., 2011; Fun-fak et al., 2007, respectively).
Worm-on-a-chip
Interestingly, the first microfluidic system developed to study worm behavior was aimed to study C. elegans in space (Lange et al., 2005). Since then, microfluidics has been largely used for the study of C. elegans biology and physiology on earth (Ben-Yakar et al., 2009; Chronis et al., 2007; Pulak, 2006; Wlodkowic et al., 2011; Zhang et al., 2005). Chronis et al. (2007) have developed an ‘olfactory chip’ to study the olfactory sense of the worm. The chip is composed of a worm trap that immobilizes the worm and presents it to a chemical delivery system composed of four laminar flow microchannels (Fig. 8B). The system delivers olfactory stimuli to the nose of the worm and neural responses to the stimuli are registered. Using this system, Chronis and coworkers have demonstrated that specific G-cAMP-expressing ASH neurons responded to the presentation of high osmotic-strength stimuli and to the removal of the high osmolarity solution.
Fish-on-a-chip
Under conventional conditions, analysis and imaging of fish embryos and juveniles in a high-throughput and high-content manner remains challenging (Giacomotto and Segalat, 2010; Lammer et al., 2009; Westerfield, 1993; Wheeler and Brandli, 2009; Wlodkowic et al., 2011). These limitations may be overcome with miniaturized lab-on-a-chip technologies that provide automated platforms for the manipulation and immobilization of micron-size organisms (Chung et al., 2008; Hulme et al., 2010; Khoshmanesh et al., 2011; Lucchetta et al., 2006; Wlodkowic et al., 2011) as well as high-throughput screening (Sundberg, 2000). Recently, several microfluidic-based methodologies have been developed involving droplet-based microfluidic systems (Funfak et al., 2007; Son and Garrell, 2009) or continuous- flow systems (Akagi et al., 2011; Choudhury et al., 2012; Pardo-Martin et al., 2010; Wielhouwer et al., 2011; Yang et al., 2011). Droplet-based microfluidic systems have been the first technology to be used to study fish embryogenesis. In 2006, Funfak et al. (2007) introduced a microfluidic system composed of a Teflon® microtube perfused with immiscible perfluoromethyldecalin (PP9) in which fish eggs in aqueous solution are introduced, resulting in the formation of droplets containing a single embryo each (Fig. 8C). The development process of the fish embryos is observed over a time period of up to 80 h. After 5 days, the fish larvae may be collected from the droplets and transferred into breeding reservoirs for further experiments. More recently, Akagi et al. (2011) introduced a miniaturized embryo array that automatically traps, immobilizes, and perfuses fish embryos. The device, shown in Fig. 8C, is composed of a serpentine channels that contain embryo traps (where embryos are trapped and immobilized), suction channels (to draw embryos into the trap), and hydrodynamic deflectors (to enhance embryo trapping). Embryos are loaded in the chip one-by-one, once every 5 s. After entering the serpentine channel, embryos roll on the bottom surface of the main channels under the influence of drag force. An embryo approaching an empty trap is affected by the flow passing through the suction channel and directed toward the trap, which is designed to assure single embryo occupancy, and the unobstructed passage of other embryos in the main channel. The succeeding embryo rolls freely along the serpentine channel toward the next available trap. This process is repeated until all traps are occupied. After loading, the chip is perfused for long term culture (up to 72 h) and the normal and uniform development of all embryos may be observed.
CONCLUSION
In this review, we have presented a brief overview of how microfluidic systems may be applied to biological sciences from the molecular up to the multicellular organism level. Most of the systems described here remain proofs-of-concept. To be integrated fully into biological labs, further interdisciplinary collaborations are required to develop useful tools for routine experimentation. Microfluidics brings new tools for biology, and biology brings new application challenges to microfluidics. It is certain that applying microfluidic systems to the biological sciences will become routine in the near future, thus offering a wide choice of inexpensive, easy-to-use, and personalized tools for diverse types of research.
Acknowledgments
This work was supported by the National Institutes of Health (NIH Grant R01 008392) and the National Science Foundation (NSF CBET Grant 1063536) of the USA.
REFERENCES
- Abhyankar VV, Lokuta MA, Huttenlocher A, Beebe DJ. Characterization of a membrane-based gradient generator for use in cell-signaling studies. Lab Chip. 2006;6:389–393. doi: 10.1039/b514133h. [DOI] [PubMed] [Google Scholar]
- Aborn JH, El-Difrawy SA, Novotny M, Gismondi EA, Lam R, Matsudaira P, McKenna BK, O’Neil T, Streechon P, Ehrlich DJ. A 768-lane microfabricated system for high-throughput DNA sequencing. Lab Chip. 2005;5:669–674. doi: 10.1039/b501104c. [DOI] [PubMed] [Google Scholar]
- Akagi J, Khoshmanesh K, Evans B, Hall CJ, Crosier KE, Cooper JM, Crosier PS, Wlodkowic D. Miniaturized embryo array for automated trapping, immobilization and microperfusion of zebrafish embryos. PLoS One. 2011;7:e36630. doi: 10.1371/journal.pone.0036630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatore C, Saveant JM, Tessier D. Kinetics of electron-transfer to organic-molecules at solid electrodes in organic media. J Electroanal Chem. 1983;146:37–45. [Google Scholar]
- Asiello PJ, Baeumner AJ. Miniaturized isothermal nucleic acid amplification, a review. Lab Chip. 2011;11:1420–1430. doi: 10.1039/c0lc00666a. [DOI] [PubMed] [Google Scholar]
- Autebert J, Coudert B, Bidard FC, Pierga JY, Descroix S, Malaquin L, Viovy JL. Microfluidic: an innovative tool for efficient cell sorting. Methods. 2012;57:297–307. doi: 10.1016/j.ymeth.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Bai HY, Lin SL, Chan SA, Fuh MR. Characterization and evaluation of two-dimensional microfluidic chip-HPLC coupled to tandem mass spectrometry for quantitative analysis of 7-aminoflunitrazepam in human urine. Analyst. 2010;135:2737–2742. doi: 10.1039/c0an00355g. [DOI] [PubMed] [Google Scholar]
- Baker M. Tissue models: a living system on a chip. Nature. 2011;471:661–665. doi: 10.1038/471661a. [DOI] [PubMed] [Google Scholar]
- Barkefors I, Le Jan S, Jakobsson L, Hejll E, Carlson G, Johansson H, Jarvius J, Park JW, Li Jeon N, Kreuger J. Endothelial cell migration in stable gradients of vascular endothelial growth factor A and fibroblast growth factor 2: effects on chemotaxis and chemokinesis. J Biol Chem. 2008;283:13905–13912. doi: 10.1074/jbc.M704917200. [DOI] [PubMed] [Google Scholar]
- Bauer M, Su G, Beebe DJ, Friedl A. 3D microchannel co-culture: method and biological validation. Integr Biol. 2010;2:371–378. doi: 10.1039/c0ib00001a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayoudh S, Othmane A, Ponsonnet L, Ben Ouada H. Electrical detection and characterization of bacterial adhesion using electrochemical impedance spectroscopy-based flow chamber. Colloids Surf. A Physicochem. Eng. Asp. 2008;318:291–300. [Google Scholar]
- Behringer MP. Techniques and Materials in Biology: Care and Use of Living Animals Plants and Microorganisms. 2nd ed. Krieger Pub Co; 1989. [Google Scholar]
- Belanger MC, Marois Y. Hemocompatibility, biocompatibility, inflammatory and in vivo studies of primary reference materials low-density polyethylene and polydimethylsiloxane: a review. J Biomed Mater Res. 2001;58:467–477. doi: 10.1002/jbm.1043. [DOI] [PubMed] [Google Scholar]
- Belder D. Microfluidics with droplets. Angew Chem Int Ed Engl. 2005;44:3521–3522. doi: 10.1002/anie.200500620. [DOI] [PubMed] [Google Scholar]
- Ben-Yakar A, Chronis N, Lu H. Microfluidics for the analysis of behavior, nerve regeneration, and neural cell biology in C. elegans. Curr Opin Neurobiol. 2009;19:561–567. doi: 10.1016/j.conb.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yoav H, Dykstra PH, Bentley WE, Ghodssi R. A microfluidic-based electrochemical biochip for label-free diffusion-restricted DNA hybridization analysis. Biosens Bioelectron. 2012;38:114–120. doi: 10.1016/j.bios.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Berggren C, Bjarnason B, Johansson G. An immunological interleukin-6 capacitive biosensor using perturbation with a potentiostatic step. Biosens Bioelectron. 1998;13:1061–1068. doi: 10.1016/s0956-5663(98)00058-x. [DOI] [PubMed] [Google Scholar]
- Betz UAK, Farquhar R, Ziegelbauer K. Genomics: success or failure to deliver drug targets? Curr Opin Chem Biol. 2005;9:387–391. doi: 10.1016/j.cbpa.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Blake WJ, Kaern M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- Blake AJ, Rodgers FC, Bassuener A, Hippensteel JA, Pearce TM, Pearce TR, Zarnowska ED, Pearce RA, Williams JC. A microfluidic brain slice perfusion chamber for multisite recording using penetrating electrodes. J. Neurosci. Methods. 2010;189:5–13. doi: 10.1016/j.jneumeth.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazej RG, Kumaresan P, Mathies RA. Microfabricated bioprocessor for integrated nanoliter-scale Sanger DNA sequencing. Proc. Natl. Acad. Sci. USA. 2006;103:7240–7245. doi: 10.1073/pnas.0602476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazej RG, Kumaresan P, Cronier SA, Mathies RA. Inline injection microdevice for attomole-scale sanger DNA sequencing. Anal Chem. 2007;79:4499–4506. doi: 10.1021/ac070126f. [DOI] [PubMed] [Google Scholar]
- Boedicker JQ, Li L, Kline TR, Ismagilov RF. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip. 2008;8:1265–1272. doi: 10.1039/b804911d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm DA, Gottlieb PA, Hua SZ. On-chip microfluidic biosensor for bacterial detection and identification. Sensors and Actuators B: Chemical. 2007;126:508–514. [Google Scholar]
- Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl. Acad. Sci. USA. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- Chan SD, Luedke G, Valer M, Buhlmann C, Preckel T. Cytometric analysis of protein expression and apoptosis in human primary cells with a novel microfluidic chip-based system. Cytometry A. 2003;55:119–125. doi: 10.1002/cyto.a.10070. [DOI] [PubMed] [Google Scholar]
- Chang HC, Yeo LY. Electrokinetically Driven Microfluidics and Nanofluidics. Cambridge, New York: Cambridge University Press; 2010. [Google Scholar]
- Chayen NE, Saridakis E. Protein crystallization: from purified protein to diffraction-quality crystal. Nat. Methods. 2008;5:147–153. doi: 10.1038/nmeth.f.203. [DOI] [PubMed] [Google Scholar]
- Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- Chen L, Lee S, Lee M, Lim C, Choo J, Park JY, Lee S, Joo SW, Lee KH, Choi YW. DNA hybridization detection in a microfluidic channel using two fluorescently labelled nucleic acid probes. Biosens Bioelectron. 2008a;23:1878–1882. doi: 10.1016/j.bios.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Chen X, Cui DF, Liu CC, Li H. Microfluidic chip for blood cell separation and collection based on crossflow filtration. Sensors and Actuators B: Chemical. 2008b;130:216–221. [Google Scholar]
- Cheng IF, Chang HC, Hou D. An integrated dielectrophoretic chip for continuous bioparticle filtering, focusing, sorting, trapping, and detecting. Biomicrofluidics. 2007;1:21503. doi: 10.1063/1.2723669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong KH, Yi DK, Lee JG, Park JM, Kim MJ, Edel JB, Ko C. Gold nanoparticles for one step DNA extraction and real-time PCR of pathogens in a single chamber. Lab Chip. 2008;8:810–813. doi: 10.1039/b717382b. [DOI] [PubMed] [Google Scholar]
- Cho BS, Schuster TG, Zhu X, Chang D, Smith GD, Takayama S. Passively driven integrated microfluidic system for separation of motile sperm. Anal Chem. 2003;75:1671–1675. doi: 10.1021/ac020579e. [DOI] [PubMed] [Google Scholar]
- Choi S, Song S, Choi C, Park JK. Microfluidic self-sorting of mammalian cells to achieve cell cycle synchrony by hydrophoresis. Anal Chem. 2009;81:1964–1968. doi: 10.1021/ac8024575. [DOI] [PubMed] [Google Scholar]
- Choudhury D, van Noort D, Iliescu C, Zheng B, Poon KL, Korzh S, Korzh V, Yu H. Fish and chips: a microfluidic perfusion platform for monitoring zebrafish development. Lab Chip. 2012;12:892–900. doi: 10.1039/c1lc20351g. [DOI] [PubMed] [Google Scholar]
- Chronis N, Zimmer M, Bargmann CI. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat. Methods. 2007;4:727–731. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]
- Chung BG, Flanagan LA, Rhee SW, Schwartz PH, Lee AP, Monuki ES, Jeon NL. Human neural stem cell growth and differentiation in a gradient-generating microfluidic device. Lab Chip. 2005;5:401–406. doi: 10.1039/b417651k. [DOI] [PubMed] [Google Scholar]
- Chung K, Crane MM, Lu H. Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nat. Methods. 2008;5:637–643. doi: 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- Chung S, Sudo R, Mack PJ, Wan CR, Vickerman V, Kamm RD. Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab Chip. 2009;9:269–275. doi: 10.1039/b807585a. [DOI] [PubMed] [Google Scholar]
- Ciszkowska M, Stojek Z. Voltammetry in solutions of low ionic strength Electrochemical and analytical aspects. J Electroanal Chem. 1999;466:129–143. [Google Scholar]
- Cooksey GA, Sip CG, Folch A. A multi-purpose microfluidic perfusion system with combinatorial choice of inputs, mixtures, gradient patterns, and flow rates. Lab Chip. 2009;9:417–426. doi: 10.1039/b806803h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craw P, Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12:2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- Crosland-Taylor PJ. A device for counting small particles suspended in a fluid through a tube. Nature. 1953;171:37–38. doi: 10.1038/171037b0. [DOI] [PubMed] [Google Scholar]
- Cunningham KS, Gotlieb AI. The role of shear stress in the pathogenesis of atherosclerosis. Lab Invest. 2005;85:9–23. doi: 10.1038/labinvest.3700215. [DOI] [PubMed] [Google Scholar]
- Deng L, Bao LL, Yang ZY, Nie LH, Yao SZ. In situ continuous detection of bacteria on the surface of solid medium with a bulk acoustic wave-impedance sensor. J. Microbiol. Methods. 1996;26:197–203. [Google Scholar]
- Dertinger SKW, Chiu DT, Jeon NL, Whitesides GM. Generation of gradients having complex shapes using microfluidic networks. Anal Chem. 2001;73:1240–1246. [Google Scholar]
- Dertinger SK, Jiang X, Li Z, Murthy VN, Whitesides GM. Gradients of substrate-bound laminin orient axonal specification of neurons. Proc. Natl. Acad. Sci. USA. 2002;99:12542–12547. doi: 10.1073/pnas.192457199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo D, Jeong KH, Lee LP. Reagentless mechanical cell lysis by nanoscale barbs in microchannels for sample preparation. Lab Chip. 2003;3:287–291. doi: 10.1039/b305162e. [DOI] [PubMed] [Google Scholar]
- Diao J, Young L, Kim S, Fogarty EA, Heilman SM, Zhou P, Shuler ML, Wu M, DeLisa MP. A three-channel microfluidic device for generating static linear gradients and its application to the quantitative analysis of bacterial chemotaxis. Lab Chip. 2006;6:381–388. doi: 10.1039/b511958h. [DOI] [PubMed] [Google Scholar]
- Diaz-Sanchez S, Hanning I, Pendleton S, D’Souza D. Next-generation sequencing the future of molecular genetics in poultry production and food safety. Poult Sci. 2013;92:562–572. doi: 10.3382/ps.2012-02741. [DOI] [PubMed] [Google Scholar]
- Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Du Y, Chia S.-m, Han R, Chang S, Tang H, Yu H. 3D hepatocyte monolayer on hybrid RGD/galactose substratum. Biomaterials. 2006;27:5669–5680. doi: 10.1016/j.biomaterials.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Duffy DC, McDonald JC, Schueller OJ, Whitesides GM. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane) Anal Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- El-Ali J, Sorger PK, Jensen KF. Cells on chips. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Esch MB, King TL, Shuler ML. The role of body-on-a-chip devices in drug and toxicity studies. Annu Rev Biomed Eng. 2011;13:55–72. doi: 10.1146/annurev-bioeng-071910-124629. [DOI] [PubMed] [Google Scholar]
- Fair RB. Digital microfluidics: is a true lab-on-a-chip possible? Microfluid. Nanofluidics. 2007;3:245–281. [Google Scholar]
- Fan Y.-Y, Chapkin R, Ramos K. A macrophage-smooth muscle cell co-culture model. Applications in the study of atherogenesis. In Vitro Cell Dev Biol Anim. 1995;31:492–493. doi: 10.1007/BF02634024. [DOI] [PubMed] [Google Scholar]
- Faure-Andre G, Vargas P, Yuseff MI, Heuze M, Diaz J, Lankar D, Steri V, Manry J, Hugues S, Vascotto F, et al. Regulation of dendritic cell migration by CD74, the MHC class II-associated invariant chain. Science. 2008;322:1705–1710. doi: 10.1126/science.1159894. [DOI] [PubMed] [Google Scholar]
- Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat. Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463:485–492. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangos JA, McIntire LV, Eskin SG. Shear stress induced stimulation of mammalian cell metabolism. Biotechnol Bioeng. 1988;32:1053–1060. doi: 10.1002/bit.260320812. [DOI] [PubMed] [Google Scholar]
- Fredlake CP, Hert DG, Kan CW, Chiesl TN, Root BE, Forster RE, Barron AE. Ultrafast DNA sequencing on a microchip by a hybrid separation mechanism that gives 600 bases in 6.5 minutes. Proc. Natl. Acad. Sci. USA. 2008;105:476–481. doi: 10.1073/pnas.0705093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Fuger P, Behrends LB, Mertel S, Sigrist SJ, Rasse TM. Live imaging of synapse development and measuring protein dynamics using two-color fluorescence recovery after photo-bleaching at Drosophila synapses. Nat Protoc. 2007;2:3285–3298. doi: 10.1038/nprot.2007.472. [DOI] [PubMed] [Google Scholar]
- Funfak A, Brosing A, Brand M, Kohler JM. Micro fluid segment technique for screening and development studies on Danio rerio embryos. Lab Chip. 2007;7:1132–1138. doi: 10.1039/b701116d. [DOI] [PubMed] [Google Scholar]
- Gagnon Z, Chang HC. Aligning fast alternating current electroosmotic flow fields and characteristic frequencies with dielectrophoretic traps to achieve rapid bacteria detection. Electrophoresis. 2005;26:3725–3737. doi: 10.1002/elps.200500129. [DOI] [PubMed] [Google Scholar]
- Ghannad-Rezaie M, Wang X, Mishra B, Collins C, Chronis N. Microfluidic chips for in vivo imaging of cellular responses to neural injury in Drosophila larvae. PLoS One. 2012;7:e29869. doi: 10.1371/journal.pone.0029869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomotto J, Segalat L. High-throughput screening and small animal models, where are we? Br J Pharmacol. 2010;160:204–216. doi: 10.1111/j.1476-5381.2010.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill P, Ghaemi A. Nucleic acid isothermal amplification technologies: a review. Nucleosides Nucleotides Nucleic Acids. 2008;27:224–243. doi: 10.1080/15257770701845204. [DOI] [PubMed] [Google Scholar]
- Gomez-Sjoberg R, Leyrat AA, Pirone DM, Chen CS, Quake SR. Versatile, fully automated, microfluidic cell culture system. Anal. Chem. 2007;79:8557–8563. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- Gottschamel J, Richter L, Mak A, Jungreuthmayer C, Birnbaumer G, Milnera M, Bruckl H, Ertl P. Development of a disposable microfluidic biochip for multiparameter cell population measurements. Anal Chem. 2009;81:8503–8512. doi: 10.1021/ac901420u. [DOI] [PubMed] [Google Scholar]
- Grabski AC. Advances in preparation of biological extracts for protein purification. Method Enzymol. 2009;463:285–303. doi: 10.1016/S0076-6879(09)63018-4. [DOI] [PubMed] [Google Scholar]
- Grahl T, Markl H. Killing of microorganisms by pulsed electric fields. Appl Microbiol Biotechnol. 1996;45:148–157. doi: 10.1007/s002530050663. [DOI] [PubMed] [Google Scholar]
- Griffin JL. Metabonomics: NMR spectroscopy and pattern recognition analysis of body fluids and tissues for characterisation of xenobiotic toxicity and disease diagnosis. Curr Opin Chem Biol. 2003;7:648–654. doi: 10.1016/j.cbpa.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Gu W, Zhu X, Futai N, Cho BS, Takayama S. Computerized microfluidic cell culture using elastomeric channels and Braille displays. Proc. Natl. Acad. Sci. USA. 2004;101:15861–15866. doi: 10.1073/pnas.0404353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawan RC, Silvestre J, Gaskins HR, Kenis PJA, Leckband DE. Cell migration and polarity on microfabricated gradients of extracellular matrix proteins. Langmuir. 2006;22:4250–4258. doi: 10.1021/la0531493. [DOI] [PubMed] [Google Scholar]
- Gunawardena S, Her LS, Brusch RG, Laymon RA, Niesman IR, Gordesky-Gold B, Sintasath L, Bonini NM, Goldstein LS. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 2003;40:25–40. doi: 10.1016/s0896-6273(03)00594-4. [DOI] [PubMed] [Google Scholar]
- Gunther A, Yasotharan S, Vagaon A, Lochovsky C, Pinto S, Yang J, Lau C, Voigtlaender-Bolz J, Bolz S.-S. A microfluidic platform for probing small artery structure and function. Lab Chip. 2010;10:2341–2349. doi: 10.1039/c004675b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon M, Jambovane S, Bradley L, Khademhosseini A, Hong JW. Cell-based dose responses from open-well microchambers. Anal Chem. 2013;85:5249–5254. doi: 10.1021/ac400743w. [DOI] [PubMed] [Google Scholar]
- Harrison RG. Observations on the living developing nerve fiber. Paper presented at: Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine; New York, NY. 1906. (Royal Society of Medicine) [Google Scholar]
- He M, Edgar JS, Jeffries GD, Lorenz RM, Shelby JP, Chiu DT. Selective encapsulation of single cells and subcellular organelles into picoliter- and femtoliter-volume droplets. Anal Chem. 2005;77:1539–1544. doi: 10.1021/ac0480850. [DOI] [PubMed] [Google Scholar]
- Hellmich W, Pelargus C, Leffhalm K, Ros A, Anselmetti D. Single cell manipulation, analytics, and label-free protein detection in microfluidic devices for systems nanobiology. Electrophoresis. 2005;26:3689–3696. doi: 10.1002/elps.200500185. [DOI] [PubMed] [Google Scholar]
- Heuzé M, Collin O, Terriac E, Lennon-Duménil A.-M, Piel M. Cell migration in confinement: a micro-channel-based assay. In: Wells CM, Parsons M, editors. Cell Migration. Humana Press; 2011. pp. 415–434. [DOI] [PubMed] [Google Scholar]
- Hilliard MA, Apicella AJ, Kerr R, Suzuki H, Bazzicalupo P, Schafer WR. In vivo imaging of C. elegans ASH neurons cellular response and adaptation to chemical repellents. EMBO J. 2005;24:63–72. doi: 10.1038/sj.emboj.7600493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JW, Studer V, Hang G, Anderson WF, Quake SR. A nanoliter-scale nucleic acid processor with parallel architecture. Nat Biotechnol. 2004;22:435–439. doi: 10.1038/nbt951. [DOI] [PubMed] [Google Scholar]
- Hong JW, Song S, Shin JH. A novel microfluidic co-culture system for investigation of bacterial cancer targeting. Lab Chip. 2013;13:3033–3040. doi: 10.1039/c3lc50163a. [DOI] [PubMed] [Google Scholar]
- Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Hsu S, Thakar R, Liepmann D, Li S. Effects of shear stress on endothelial cell haptotaxis on micropatterned surfaces. Biochem Biophys Res Commun. 2005;337:401–409. doi: 10.1016/j.bbrc.2005.08.272. [DOI] [PubMed] [Google Scholar]
- Huang W.-H, Cheng W, Zhang Z, Pang D.-W, Wang Z.-L, Cheng J.-K, Cui D.-F. Transport, location, and quantal release monitoring of single cells on a microfluidic device. Anal Chem. 2004;76:483–488. doi: 10.1021/ac035026y. [DOI] [PubMed] [Google Scholar]
- Huang J, Yamaji H, Fukuda H. Immobilization of Escherichia coli cells using porous support particles coated with cationic polymers. J Biosci Bioeng. 2007;104:98–103. doi: 10.1263/jbb.104.98. [DOI] [PubMed] [Google Scholar]
- Huang CP, Lu J, Seon H, Lee AP, Flanagan LA, Kim HY, Putnam AJ, Jeon NL. Engineering microscale cellular niches for three-dimensional multicellular co-cultures. Lab Chip. 2009;9:1740–1748. doi: 10.1039/b818401a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-J, Samorajski J, Kreimer R, Searson PC. The influence of electric field and confinement on cell motility. PLoS One. 2013;8:e59447. doi: 10.1371/journal.pone.0059447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh YS, Choi JH, Park TJ, Hong YK, Hong WH, Lee SY. Microfluidic cell disruption system employing a magnetically actuated diaphragm. Electrophoresis. 2007;28:4748–4757. doi: 10.1002/elps.200700366. [DOI] [PubMed] [Google Scholar]
- Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs-on-chips Trends. Cell Biol. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme SE, Shevkoplyas SS, McGuigan AP, Apfeld J, Fontana W, Whitesides GM. Lifespan-on-a-chip: microfluidic chambers for performing lifelong observation of C. elegans. Lab Chip. 2010;10:589–597. doi: 10.1039/b919265d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung PJ, Lee PJ, Sabounchi P, Lin R, Lee LP. Continuous perfusion microfluidic cell culture array for high-throughput cell-based assays. Biotechnol Bioeng. 2005;89:1–8. doi: 10.1002/bit.20289. [DOI] [PubMed] [Google Scholar]
- Imberti B, Seliktar D, Nerem RM, Remuzzi A. The response of endothelial cells to fluid shear stress using a co-culture model of the arterial wall. Endothelium. 2002;9:11–23. doi: 10.1080/10623320210714. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Irimia D, Toner M. Spontaneous migration of cancer cells under conditions of mechanical confinement. Integr Biol. 2009;1:506–512. doi: 10.1039/b908595e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia D, Geba DA, Toner M. Universal microfluidic gradient generator. Anal Chem. 2006a;78:3472–3477. doi: 10.1021/ac0518710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia D, Liu SY, Tharp WG, Samadani A, Toner M, Poznansky MC. Microfluidic system for measuring neutrophil migratory responses to fast switches of chemical gradients. Lab Chip. 2006b;6:191–198. doi: 10.1039/b511877h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RK, Au P, Tam J, Duda DG, Fukumura D. Engineering vascularized tissue. Nat Biotechnol. 2005;23:821–823. doi: 10.1038/nbt0705-821. [DOI] [PubMed] [Google Scholar]
- Jambovane S, Duin EC, Kim SK, Hong JW. Determination of kinetic parameters, Km and kcat, with a single experiment on a chip. Anal Chem. 2009;81:3239–3245. doi: 10.1021/ac8020938. [DOI] [PubMed] [Google Scholar]
- Jambovane S, Kim DJ, Duin EC, Kim S.-K, Hong JW. Creation of stepwise concentration gradient in picoliter droplets for parallel reactions of matrix metalloproteinase II and IX. Anal Chem. 2011;83:3358–3364. doi: 10.1021/ac103217p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanmard M, Davis RW. A microfluidic platform for electrical detection of DNA hybridization. Sensor Actuat B-Chem. 2011;154:22–27. doi: 10.1016/j.snb.2010.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K, Lee A. The science & applications of droplets in microfluidic devices. Lab Chip. 2004;4:31N–32N. [Google Scholar]
- Ji HM, Samper V, Chen Y, Heng CK, Lim TM, Yobas L. Silicon-based microfilters for whole blood cell separation. Biomed. Microdevices. 2008;10:251–257. doi: 10.1007/s10544-007-9131-x. [DOI] [PubMed] [Google Scholar]
- Kalisky T, Quake SR. Single-cell genomics. Nat. Methods. 2011;8:311–314. doi: 10.1038/nmeth0411-311. [DOI] [PubMed] [Google Scholar]
- Kamholz AE, Weigl BH, Finlayson BA, Yager P. Quantitative analysis of molecular interaction in a microfluidic channel: the T-sensor. Anal Chem. 1999;71:5340–5347. doi: 10.1021/ac990504j. [DOI] [PubMed] [Google Scholar]
- Kartalov EP, Quake SR. Microfluidic device reads up to four consecutive base pairs in DNA sequencing-by-synthesis. Nucleic Acids Res. 2004;32:2873–2879. doi: 10.1093/nar/gkh613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan TM, Folch A. Biomolecular gradients in cell culture systems. Lab Chip. 2008;8:34–57. doi: 10.1039/b711887b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan OF, Sefton MV. Endothelial cell behaviour within a microfluidic mimic of the flow channels of a modular tissue engineered construct. Biomed. Microdevices. 2011;13:69–87. doi: 10.1007/s10544-010-9472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotech. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- Khoshmanesh K, Kiss N, Nahavandi S, Evans CW, Cooper JM, Williams DE, Wlodkowic D. Trapping and imaging of micron-sized embryos using dielectrophoresis. Electrophoresis. 2011;32:3129–3132. doi: 10.1002/elps.201100160. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- Kintses B, van Vliet LD, Devenish SR, Hollfelder F. Microfluidic droplets new integrated workflows for biological experiments. Curr Opin Chem Biol. 2010;14:548–555. doi: 10.1016/j.cbpa.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Kobayashi I, Uemura K, Nakajima M. Formulation of monodisperse emulsions using submicron-channel arrays. Colloids Surf. A Physicochem. Eng. Asp. 2007;296:285–289. [Google Scholar]
- Kong DS, Carr PA, Chen L, Zhang S, Jacobson JM. Parallel gene synthesis in a microfluidic device. Nucleic Acids Res. 2007;35:e61. doi: 10.1093/nar/gkm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp MU, Mello AJ, Manz A. Chemical amplification: continuous-flow PCR on a chip. Science. 1998;280:1046–1048. doi: 10.1126/science.280.5366.1046. [DOI] [PubMed] [Google Scholar]
- Korin N, Kanapathipillai M, Matthews BD, Crescente M, Brill A, Mammoto T, Ghosh K, Jurek S, Bencherif SA, Bhatta D, et al. Shear-activated nanotherapeutics for drug targeting to obstructed blood vessels. Science. 2012;337:738–742. doi: 10.1126/science.1217815. [DOI] [PubMed] [Google Scholar]
- Kostov Y, Harms P, Randers-Eichhorn L, Rao G. Low-cost microbioreactor for high-throughput bioprocessing. Biotechnol Bioeng. 2001;72:346–352. doi: 10.1002/1097-0290(20010205)72:3<346::aid-bit12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Kosuri S, Eroshenko N, Leproust EM, Super M, Way J, Li JB, Church GM. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat Biotechnol. 2010;28:1295–1299. doi: 10.1038/nbt.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraly JR, Holcomb RE, Guan Q, Henry CS. Review: Microfluidic applications in metabolomics and metabolic profiling. Anal. Chim. Acta. 2009;653:23–35. doi: 10.1016/j.aca.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubinyi H. Drug research: myths, hype and reality. Nat Rev Drug Discov. 2003;2:665–668. doi: 10.1038/nrd1156. [DOI] [PubMed] [Google Scholar]
- Kumagai H, Utsunomiya S, Nakamura S, Yamamoto R, Harada A, Kaji T, Hazama M, Ohashi T, Inami A, Ikegami T, et al. Large-scale microfabricated channel plates for high-throughput, fully automated DNA sequencing. Electrophoresis. 2008;29:4723–4732. doi: 10.1002/elps.200800301. [DOI] [PubMed] [Google Scholar]
- Kumar A, Bhardwaj A. Methods in cell separation for biomedical application: cryogels as a new tool. Biomed Mater. 2008;3:034008. doi: 10.1088/1748-6041/3/3/034008. [DOI] [PubMed] [Google Scholar]
- Lammer E, Kamp HG, Hisgen V, Koch M, Reinhard D, Salinas ER, Wendler K, Zok S, Braunbeck T. Development of a flow-through system for the fish embryo toxicity test (FET) with the zebrafish (Danio rerio) Toxicol. In Vitro. 2009;23:1436–1442. doi: 10.1016/j.tiv.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Lange D, Storment CW, Conley CA, Kovacs GTA. A microfluidic shadow imaging system for the study of the nematode Caenorhabditis elegans in space. Sensors and Actuators B: Chemical. 2005;107:904–914. [Google Scholar]
- Lecault V, VanInsberghe M, Sekulovic S, Knapp DJHF, Wohrer S, Bowden W, Viel F, McLaughlin T, Jarandehei A, Miller M, et al. High-throughput analysis of single hematopoietic stem cell proliferation in microfluidic cell culture arrays. Nat. Methods. 2011;8:581–586. doi: 10.1038/nmeth.1614. [DOI] [PubMed] [Google Scholar]
- Ledezma GA, Folch A, Bhatia SN, Balis UJ, Yarmush ML, Toner M. Numerical model of fluid flow and oxygen transport in a radial-flow microchannel containing hepatocytes. J Biomech Eng. 1999;121:58–64. doi: 10.1115/1.2798043. [DOI] [PubMed] [Google Scholar]
- Lee PJ, Hung PJ, Rao VM, Lee LP. Nanoliter scale microbioreactor array for quantitative cell biology. Biotechnol Bioeng. 2006;94:5–14. doi: 10.1002/bit.20745. [DOI] [PubMed] [Google Scholar]
- Lee PJ, Hung PJ, Lee LP. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol Bioeng. 2007;97:1340–1346. doi: 10.1002/bit.21360. [DOI] [PubMed] [Google Scholar]
- Lee H, Sun E, Ham D, Weissleder R. Chip-NMR biosensor for detection and molecular analysis of cells. Nat Med. 2008;14:869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-C, Snyder TM, Quake SR. A microfluidic oligonucleotide synthesizer. Nucleic Acids Res. 2010;38:2514–2521. doi: 10.1093/nar/gkq092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48:1296–1304. [PubMed] [Google Scholar]
- Li YY, Xing D, Zhang CS. Rapid detection of genetically modified organisms on a continuous-flow polymerase chain reaction microfluidics. Anal Biochem. 2009;385:42–49. doi: 10.1016/j.ab.2008.10.028. [DOI] [PubMed] [Google Scholar]
- Li Jeon N, Baskaran H, Dertinger SKW, Whitesides GM, Van De Water L, Toner M. Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device. Nat. Biotech. 2002;20:826–830. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- Liedert A, Kaspar D, Blakytny R, Claes L, Ignatius A. Signal transduction pathways involved in mechanotransduction in bone cells. Biochem Biophys Res Commun. 2006;349:1–5. doi: 10.1016/j.bbrc.2006.07.214. [DOI] [PubMed] [Google Scholar]
- Lin F, Nguyen CM, Wang SJ, Saadi W, Gross SP, Jeon NL. Neutrophil migration in opposing chemoattractant gradients using microfluidic chemotaxis devices. Ann Biomed Eng. 2005;33:475–482. doi: 10.1007/s10439-005-2503-6. [DOI] [PubMed] [Google Scholar]
- Lindon JC, Holmes E, Nicholson JK. Metabonomics in pharmaceutical R & D. FEBS J. 2007;274:1140–1151. doi: 10.1111/j.1742-4658.2007.05673.x. [DOI] [PubMed] [Google Scholar]
- Link DR, Grasland-Mongrain E, Duri A, Sarrazin F, Cheng Z, Cristobal G, Marquez M, Weitz DA. Electric control of droplets in microfluidic devices. Angew Chem Int Ed Engl. 2006;45:2556–2560. doi: 10.1002/anie.200503540. [DOI] [PubMed] [Google Scholar]
- Liu P, Mathies RA. Integrated microfluidic systems for high-performance genetic analysis. Trends Biotechnol. 2009;27:572–581. doi: 10.1016/j.tibtech.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Liu Y.-S, Walter TM, Chang W.-J, Lim K.-S, Yang L, Lee SW, Aronson A, Bashir R. Electrical detection of germination of viable model Bacillus anthracis spores in microfluidic biochips. Lab Chip. 2007;7:603–610. doi: 10.1039/b702408h. [DOI] [PubMed] [Google Scholar]
- Liu JK, Chen CF, Tsao CW, Chang CC, Chu CC, Devoe DL. Polymer microchips integrating solid-phase extraction and high-performance liquid chromatography using reversed-phase polymethacrylate monoliths. Anal Chem. 2009;81:2545–2554. doi: 10.1021/ac802359e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta EM, Lee JH, Fu LA, Patel NH, Ismagilov RF. Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature. 2005;434:1134–1138. doi: 10.1038/nature03509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta EM, Munson MS, Ismagilov RF. Characterization of the local temperature in space and time around a developing Drosophila embryo in a microfluidic device. Lab Chip. 2006;6:185–190. doi: 10.1039/b516119c. [DOI] [PubMed] [Google Scholar]
- Mack PJ, Zhang Y, Chung S, Vickerman V, Kamm RD, Garcia-Cardena G. Biomechanical regulation of endothelium-dependent events critical for adaptive remodeling. J Biol Chem. 2009;284:8412–8420. doi: 10.1074/jbc.M804524200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffia AM, Kariv I, Oldenburg KR. Miniaturization of a mammalian cell-based assay: luciferase reporter gene readout in a 3 microliter 1536-well plate. J Biomol Screen. 1999;4:137–142. doi: 10.1177/108705719900400307. [DOI] [PubMed] [Google Scholar]
- Malek AM, Alper SL, Izumo S. HEmodynamic shear stress and its role in atherosclerosis. JAMA. 1999;282:2035–2042. doi: 10.1001/jama.282.21.2035. [DOI] [PubMed] [Google Scholar]
- Malic L, Sandros MG, Tabrizian M. Designed biointerface using near-infrared quantum dots for ultrasensitive surface plasmon resonance imaging biosensors. Anal Chem. 2011;83:5222–5229. doi: 10.1021/ac200465m. [DOI] [PubMed] [Google Scholar]
- Mao H, Cremer PS, Manson MD. A sensitive, versatile microfluidic assay for bacterial chemotaxis. Proc. Natl. Acad. Sci. USA. 2003;100:5449–5454. doi: 10.1073/pnas.0931258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S, Zhang J, Li H, Lin JM. Strategy for signaling molecule detection by using an integrated microfluidic device coupled with mass spectrometry to study cell-to-cell communication. Anal Chem. 2013;85:868–876. doi: 10.1021/ac303164b. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Ohkawa H, Ogawa S, Ueda A, Niwa O, Suzuki K. Fabrication and characterization of a nanometer-sized optical fiber electrode based on selective chemical etching for scanning electrochemical/optical microscopy. Anal Chem. 2006;78:1904–1912. doi: 10.1021/ac0502549. [DOI] [PubMed] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- McCarthy JB, Furcht LT. Laminin and fibronectin promote the haptotactic migration of B16 mouse melanoma cells in vitro. J Cell Biol. 1984;98:1474–1480. doi: 10.1083/jcb.98.4.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JB, Palm SL, Furcht LT. Migration by haptotaxis of a Schwann cell tumor line to the basement membrane glycoprotein laminin. J Cell Biol. 1983;97:772–777. doi: 10.1083/jcb.97.3.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JB, Basara ML, Palm SL, Sas DF, Furcht LT. The role of cell adhesion proteins--laminin and fibronectin--in the movement of malignant and metastatic cells. Cancer Metastasis Rev. 1985;4:125–152. doi: 10.1007/BF00050692. [DOI] [PubMed] [Google Scholar]
- Mernier G, Hasenkamp W, Piacentini N, Renaud P. Multiple-frequency impedance measurements in continuous flow for the evaluation of electrical lysis of yeast cells. Procedia Eng. 2010;5:37–40. [Google Scholar]
- Meyvantsson I, Beebe DJ. Cell culture models in microfluidic systems. Annu Rev Anal Chem. 2008;1:423–449. doi: 10.1146/annurev.anchem.1.031207.113042. [DOI] [PubMed] [Google Scholar]
- Minc N, Bratman SV, Basu R, Chang F. Establishing new sites of polarization by microtubules. Curr Biol. 2009;19:83–94. doi: 10.1016/j.cub.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsky VM, Mass M, Krause C, Wolfbeis OS. Capacitive approach to determine phospholipase A(2) activity toward artificial and natural substrates. Anal Chem. 1998;70:3674–3678. doi: 10.1021/ac980102w. [DOI] [PubMed] [Google Scholar]
- Molday RS, Yen SP, Rembaum A. Application of magnetic microspheres in labelling and separation of cells. Nature. 1977;268:437–438. doi: 10.1038/268437a0. [DOI] [PubMed] [Google Scholar]
- Morris M, Watkins SM. Focused metabolomic profiling in the drug development process: advances from lipid profiling. Curr Opin Chem Biol. 2005;9:407–412. doi: 10.1016/j.cbpa.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Mosadegh B, Huang C, Park JW, Shin HS, Chung BG, Hwang SK, Lee KH, Kim HJ, Brody J, Jeon NL. Generation of stable complex gradients across two-dimensional surfaces and three-dimensional gels. Langmuir. 2007;23:10910–10912. doi: 10.1021/la7026835. [DOI] [PubMed] [Google Scholar]
- Nolan JP, Sklar LA. The emergence of flow cytometry for sensitive, real-time measurements of molecular interactions. Nat Biotechnol. 1998;16:633–638. doi: 10.1038/nbt0798-633. [DOI] [PubMed] [Google Scholar]
- Okuyama T, Yamazoe H, Mochizuki N, Khademhosseini A, Suzuki H, Fukuda J. Preparation of arrays of cell spheroids and spheroid-monolayer cocultures within a microfluidic device. J Biosci Bioeng. 2010;110:572–576. doi: 10.1016/j.jbiosc.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science. 2006;314:1464–1467. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- Page MJ, Amess B, Rohlff C, Stubberfield C, Parekh R. Proteomics: a major new technology for the drug discovery process. Drug Discov. Today. 1999;4:55–62. doi: 10.1016/s1359-6446(98)01291-4. [DOI] [PubMed] [Google Scholar]
- Paguirigan AL, Beebe DJ. From the cellular perspective: exploring differences in the cellular baseline in macroscale and microfluidic cultures. Integr Biol. 2009;1:182–195. doi: 10.1039/b814565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliwal S, Iglesias PA, Campbell K, Hilioti Z, Groisman A, Levchenko A. MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature. 2007;446:46–51. doi: 10.1038/nature05561. [DOI] [PubMed] [Google Scholar]
- Pardo-Martin C, Chang TY, Koo BK, Gilleland CL, Wasserman SC, Yanik MF. High-throughput in vivo vertebrate screening. Nat. Methods. 2010;7:634–636. doi: 10.1038/nmeth.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Takayama S, Lee S.-H. Regulating microenvironmental stimuli for stem cells and cancer cells using microsystems. Integr Biol. 2010;2:229–240. doi: 10.1039/c000442a. [DOI] [PubMed] [Google Scholar]
- Pearce TM, Wilson JA, Oakes SG, Chiu SY, Williams JC. Integrated microelectrode array and microfluidics for temperature clamp of sensory neurons in culture. Lab Chip. 2005;5:97–101. doi: 10.1039/b407871c. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen NJ, Mogensen KB, Kutter P., Jr Performance of an in-plane detection cell with integrated wave-guides for UV/Vis absorbance measurements on microfluidic separation devices. Electrophoresis. 2002;23:3528–3536. doi: 10.1002/1522-2683(200210)23:20<3528::AID-ELPS3528>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Pethig R. Dielectric and Electronic Properties of Biological Materials. John Wiley & Sons; 1979. [Google Scholar]
- Phelps TJ, Palumbo AV, Beliaev AS. Metabolomics and microarrays for improved understanding of phenotypic characteristics controlled by both genomics and environmental constraints. Curr Opin Biotech. 2002;13:20–24. doi: 10.1016/s0958-1669(02)00279-3. [DOI] [PubMed] [Google Scholar]
- Pilling AD, Horiuchi D, Lively CM, Saxton WM. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell. 2006;17:2057–2068. doi: 10.1091/mbc.E05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piruska A, Nikcevic I, Lee SH, Ahn C, Heineman WR, Limbach PA, Seliskar CJ. The autofluorescence of plastic materials and chips measured under laser irradiation. Lab Chip. 2005;5:1348–1354. doi: 10.1039/b508288a. [DOI] [PubMed] [Google Scholar]
- Porter SEG, Stoll DR, Rutan SC, Carr PW, Cohen JD. Analysis of four-way two-dimensional liquid chromatography-diode array data: application to metabolomics. Anal Chem. 2006;78:5559–5569. doi: 10.1021/ac0606195. [DOI] [PubMed] [Google Scholar]
- Powers MJ, Janigian DM, Wack KE, Baker CS, Beer Stolz D., Griffith LG. Functional behavior of primary rat liver cells in a three-dimensional perfused microarray bioreactor. Tissue Eng. 2002;8:499–513. doi: 10.1089/107632702760184745. [DOI] [PubMed] [Google Scholar]
- Prager-Khoutorsky M, Lichtenstein A, Krishnan R, Rajendran K, Mayo A, Kam Z, Geiger B, Bershadsky AD. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat Cell Biol. 2011;13:1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- Preckel T, Luedke G, Chan S, Wang B, Dubrow R, Buhlmann C. Detection of cellular parameters using a microfluidic chip-based system. JALA. 2002;7:5. [Google Scholar]
- Privorotskaya N, Liu YS, Lee JC, Zeng HJ, Carlisle JA, Radadia A, Millet L, Bashir R, King WP. Rapid thermal lysis of cells using silicon-diamond microcantilever heaters. Lab Chip. 2010;10:1135–1141. doi: 10.1039/b923791g. [DOI] [PubMed] [Google Scholar]
- Pulak R. Techniques for analysis, sorting, and dispensing of C. elegans on the COPAS flow-sorting system. Methods Mol Biol. 2006;351:275–286. doi: 10.1385/1-59745-151-7:275. [DOI] [PubMed] [Google Scholar]
- Qiu J, Zhou Y, Chen H, Lin J.-M. Immunomagnetic separation and rapid detection of bacteria using bioluminescence and microfluidics. Talanta. 2009;79:787–795. doi: 10.1016/j.talanta.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Radke SM, Alocilja EC. Design and fabrication of a microimpedance biosensor for bacterial detection. IEEE Sensor J. 2004;4:434–440. [Google Scholar]
- Radke SA, Alocilja EC. A high density microelectrode array biosensor for detection of E-coli O157 : H7. Biosens Bioelectron. 2005a;20:1662–1667. doi: 10.1016/j.bios.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Radke SM, Alocilja EC. A microfabricated biosensor for detecting foodborne bioterrorism agents. IEEE Sensor J. 2005b;5:744–750. [Google Scholar]
- Regnier FE, He B, Lin S, Busse J. Chromatography and electrophoresis on chips: critical elements of future integrated, microfluidic analytical systems for life science. Trends Biotechnol. 1999;17:101–106. doi: 10.1016/s0167-7799(98)01294-3. [DOI] [PubMed] [Google Scholar]
- Regehr KJ, Domenech M, Koepsel JT, Carver KC, Ellison-Zelski SJ, Murphy WL, Schuler LA, Alarid ET, Beebe DJ. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip. 2009;9:2132–2139. doi: 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick N, Yahav H, Shay-Salit A, Shushy M, Schubert S, Zilberman LC, Wofovitz E. Fluid shear stress and the vascular endothelium: for better and for worse. Prog Biophys Mol Biol. 2003;81:177–199. doi: 10.1016/s0079-6107(02)00052-4. [DOI] [PubMed] [Google Scholar]
- Richter L, Stepper C, Mak A, Reinthaler A, Heer R, Kast M, Bruckl H, Ertl P. Development of a microfluidic biochip for online monitoring of fungal biofilm dynamics. Lab Chip. 2007;7:1723–1731. doi: 10.1039/b708236c. [DOI] [PubMed] [Google Scholar]
- Roda B, Reschiglian P, Zattoni A, Alviano F, Lanzoni G, Costa R, Di Carlo A, Marchionni C, Franchina M, Bonsi L, et al. A tag-less method of sorting stem cells from clinical specimens and separating mesenchymal from epithelial progenitor cells. Cytometry B Clin Cytom. 2009;76:285–290. doi: 10.1002/cyto.b.20472. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- Saadi W, Rhee SW, Lin F, Vahidi B, Chung BG, Jeon NL. Generation of stable concentration gradients in 2D and 3D environments using a microfluidic ladder chamber. Biomed Microdevices. 2007;9:627–635. doi: 10.1007/s10544-007-9051-9. [DOI] [PubMed] [Google Scholar]
- Sadik OA, Aluoch AO, Zhou A. Status of biomolecular recognition using electrochemical techniques. Biosensors Bioelectronics. 2009;24:2749–2765. doi: 10.1016/j.bios.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Schmid A, Hallermann S, Kittel RJ, Khorramshahi O, Frolich AM, Quentin C, Rasse TM, Mertel S, Heckmann M, Sigrist SJ. Activity-dependent site-specific changes of glutamate receptor composition in vivo. Nat Neurosci. 2008;11:659–666. doi: 10.1038/nn.2122. [DOI] [PubMed] [Google Scholar]
- Schrum DP, Culbertson CT, Jacobson SC, Ramsey JM. Microchip flow cytometry using electrokinetic focusing. Anal Chem. 1999;71:4173–4177. doi: 10.1021/ac990372u. [DOI] [PubMed] [Google Scholar]
- Selhuber-Unkel C, Erdmann T, Lopez-Garcia M, Kessler H, Schwarz US, Spatz JP. Cell adhesion strength is controlled by intermolecular spacing of adhesion receptors. Biophys J. 2010;98:543–551. doi: 10.1016/j.bpj.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethu P, Sin A, Toner M. Microfluidic diffusive filter for apheresis (leukapheresis) Lab Chip. 2006;6:83–89. doi: 10.1039/b512049g. [DOI] [PubMed] [Google Scholar]
- Sleno L, Emili A. Proteomic methods for drug target discovery. Curr Opin Chem Biol. 2008;12:46–54. doi: 10.1016/j.cbpa.2008.01.022. [DOI] [PubMed] [Google Scholar]
- Son SU, Garrell RL. Transport of live yeast and zebrafish embryo on a droplet digital microfluidic platform. Lab Chip. 2009;9:2398–2401. doi: 10.1039/b906257b. [DOI] [PubMed] [Google Scholar]
- Song JW, Munn LL. Fluid forces control endothelial sprouting. Proc. Natl. Acad. Sci. USA. 2011;108:15342–15347. doi: 10.1073/pnas.1105316108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell JM, Baber MA, Caplan AI. A self-assembled fibroblast-endothelial cell co-culture system that supports in vitro vasculogenesis by both human umbilical vein endothelial cells and human dermal microvascular endothelial cells. Cells Tissues Organs. 2007;186:157–168. doi: 10.1159/000106670. [DOI] [PubMed] [Google Scholar]
- Stals A, Mathijs E, Baert L, Botteldoorn N, Denayer S, Mauroy A, Scipioni A, Daube G, Dierick K, Herman L, et al. Molecular detection and genotyping of noroviruses. Food Environ Virol. 2012;4:153–167. doi: 10.1007/s12560-012-9092-y. [DOI] [PubMed] [Google Scholar]
- Sudo R, Chung S, Zervantonakis IK, Vickerman V, Toshimitsu Y, Griffith LG, Kamm RD. Transport-mediated angiogenesis in 3D epithelial coculture. FASEB J. 2009;23:2155–2164. doi: 10.1096/fj.08-122820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura S, Hattori K, Kanamori T. Microfluidic serial dilution cell-based assay for analyzing drug dose response over a wide concentration range. Anal Chem. 2010;82:8278–8282. doi: 10.1021/ac1017666. [DOI] [PubMed] [Google Scholar]
- Sundberg SA. High-throughput and ultra-high-throughput screening: solution- and cell-based approaches. Curr Opin Biotech. 2000;11:47–53. doi: 10.1016/s0958-1669(99)00051-8. [DOI] [PubMed] [Google Scholar]
- Sung JH, Shuler ML. A micro cell culture analog (microCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab Chip. 2009;9:1385–1394. doi: 10.1039/b901377f. [DOI] [PubMed] [Google Scholar]
- Sung JH, Kam C, Shuler ML. A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip. Lab Chip. 2010;10:446–455. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Kerr R, Bianchi L, Frokjaer-Jensen C, Slone D, Xue J, Gerstbrein B, Driscoll M, Schafer WR. In vivo imaging of C. elegans mechanosensory neurons demonstrates a specific role for the MEC-4 channel in the process of gentle touch sensation. Neuron. 2003;39:1005–1017. doi: 10.1016/j.neuron.2003.08.015. [DOI] [PubMed] [Google Scholar]
- Takeuchi S, DiLuzio WR, Weibel DB, Whitesides GM. Controlling the shape of filamentous cells of Escherichia coli. Nano Lett. 2005;5:1819–1823. doi: 10.1021/nl0507360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W, Desai TA. Microfluidic patterning of cells in extracellular matrix biopolymers: effects of channel size, cell type, and matrix composition on pattern integrity. Tissue Eng. 2003;9:255–267. doi: 10.1089/107632703764664729. [DOI] [PubMed] [Google Scholar]
- Tan SJ, Yobas L, Lee GY, Ong CN, Lim CT. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed. Microdevices. 2009;11:883–892. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- Tan F, Leung PHM, Liu Z-b, Zhang Y, Xiao L, Ye W, Zhang X, Yi L, Yang M. A PDMS microfluidic impedance immunosensor for E. coli O157:H7 and Staphylococcus aureus detection via antibody-immobilized nanoporous membrane. Sensors and Actuators B: Chemical. 2011;159:328–335. [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh SY, Lin R, Hung LH, Lee AP. Droplet microfluidics. Lab Chip. 2008;8:198–220. doi: 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- Tilles AW, Baskaran H, Roy P, Yarmush ML, Toner M. Effects of oxygenation and flow on the viability and function of rat hepatocytes cocultured in a microchannel flat-plate bioreactor. Biotechnol Bioeng. 2001;73:379–389. doi: 10.1002/bit.1071. [DOI] [PubMed] [Google Scholar]
- Toner M, Irimia D. Blood-on-a-chip. Annu Rev Biomed Eng. 2005;7:77–103. doi: 10.1146/annurev.bioeng.7.011205.135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriello NM, Douglas ES, Thaitrong N, Hsiao SC, Francis MB, Bertozzi CR, Mathies RA. Integrated microfluidic bioprocessor for single-cell gene expression analysis. Proc. Natl. Acad. Sci. USA. 2008;105:20173–20178. doi: 10.1073/pnas.0806355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourovskaia A, Figueroa-Masot X, Folch A. Differentiation-on-a-chip: a microfluidic platform for long-term cell culture studies. Lab Chip. 2004;5:14–19. doi: 10.1039/b405719h. [DOI] [PubMed] [Google Scholar]
- Towe BC, Pizziconi VB. A microflow amperometric glucose biosensor. Biosens Bioelectron. 1997;12:893–899. doi: 10.1016/s0956-5663(97)00018-3. [DOI] [PubMed] [Google Scholar]
- Trappmann B, Gautrot JE, Connelly JT, Strange DGT, Li Y, Oyen ML, Cohen Stuart MA, Boehm H, Li B, Vogel V, et al. Extracellular-matrix tethering regulates stem-cell fate. Nat Mater. 2012;11:642–649. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- Tse HTK, Weaver WM, Di Carlo D. Increased asymmetric and multi-daughter cell division in mechanically confined microenvironments. PLoS One. 2012;7:e38986. doi: 10.1371/journal.pone.0038986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger MA, Chou H-P, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- van der Meer AD, van den Berg A. Organs-on-chips: breaking the in vitro impasse. Integr Biol. 2012;4:461–470. doi: 10.1039/c2ib00176d. [DOI] [PubMed] [Google Scholar]
- van der Meer AD, Orlova VV, Ten Dijke P, van den Berg A, Mummery CL. Three-dimensional co-cultures of human endothelial cells and embryonic stem cell-derived pericytes inside a microfluidic device. Lab Chip. 2013;13:3562–3568. doi: 10.1039/c3lc50435b. [DOI] [PubMed] [Google Scholar]
- van Midwoud PM, Janssen J, Merema MT, de Graaf IA, Groothuis GM, Verpoorte E. On-line HPLC analysis system for metabolism and inhibition studies in precision-cut liver slices. Anal Chem. 2011a;83:84–91. doi: 10.1021/ac1018638. [DOI] [PubMed] [Google Scholar]
- van Midwoud PM, Verpoorte E, Groothuis GMM. Microfluidic devices for in vitro studies on liver drug metabolism and toxicity. Integr Biol. 2011b;3:509–521. doi: 10.1039/c0ib00119h. [DOI] [PubMed] [Google Scholar]
- Van Rood JJ, Van Leeuwen A, Ploem JS. Simultaneous detection of two cell populations by two-colour fluorescence and application to the recognition of B-cell determinants. Nature. 1976;262:795–797. doi: 10.1038/262795a0. [DOI] [PubMed] [Google Scholar]
- Varshney M, Li YB. Interdigitated array microelectrode based impedance biosensor coupled with magnetic nanoparticle-antibody conjugates for detection of Escherichia coli O157 : H7 in food samples. Biosens Bioelectron. 2007;22:2408–2414. doi: 10.1016/j.bios.2006.08.030. [DOI] [PubMed] [Google Scholar]
- Varshney M, Li YB. Interdigitated array microelectrodes based impedance biosensors for detection of bacterial cells. Biosens Bioelectron. 2009;24:2951–2960. doi: 10.1016/j.bios.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Varshney M, Li YB, Srinivasan B, Tung S. A label-free, microfluidics and interdigitated array microelectrode-based impedance biosensor in combination with nanoparticles immunoseparation for detection of Escherichia coli O157 : H7 in food samples. Sensor Actuat. B-Chem. 2007;128:99–107. [Google Scholar]
- Velve-Casquillas G, Costa J, Carlier-Grynkorn F, Mayeux A, Tran PT. A fast microfluidic temperature control device for studying microtubule dynamics in fission yeast. Methods Cell Biol. 2010;97:185–201. doi: 10.1016/S0091-679X(10)97011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman V, Blundo J, Chung S, Kamm R. Design, fabrication and implementation of a novel multi-parameter control microfluidic platform for three-dimensional cell culture and real-time imaging. Lab Chip. 2008;8:1468–1477. doi: 10.1039/b802395f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J, Shaffer DR, Philip J, Chaparro CA, Erdjument-Bromage H, Olshen AB, Fleisher M, Lilja H, Brogi E, Boyd J, et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest. 2006;116:271–284. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GM, Ozers MS, Beebe DJ. Cell infection within a microfluidic device using virus gradients. Sensors and Actuators B: Chemical. 2004;98:347–355. [Google Scholar]
- Wang SJ, Saadi W, Lin F, Minh-Canh Nguyen C, Li Jeon N. Differential effects of EGF gradient profiles on MDA-MB-231 breast cancer cell chemotaxis. Exp Cell Res. 2004;300:180–189. doi: 10.1016/j.yexcr.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Watt FM, Jordan PW, O’Neill CH. Cell shape controls terminal differentiation of human epidermal keratinocytes. Proc. Natl. Acad. Sci. USA. 1988;85:5576–5580. doi: 10.1073/pnas.85.15.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner E, Croixmarie V, Umbdenstock T, Ezan E, Chaminade P, Tabet JC, Junot C. Mass spectrometry-based metabolomics: Accelerating the characterization of discriminating signals by combining statistical correlations and ultrahigh resolution. Anal Chem. 2008a;80:4918–4932. doi: 10.1021/ac800094p. [DOI] [PubMed] [Google Scholar]
- Werner E, Heilier JF, Ducruix C, Ezan E, Junot C, Tabet JC. Mass spectrometry for the identification of the discriminating signals from metabolomics: Current status and future trends. J. Chromatogr. B. 2008b;871:143–163. doi: 10.1016/j.jchromb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Westerfield M. 1993. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio Rerio) (M. Wester-field).
- Wheeler GN, Brandli AW. Simple vertebrate models for chemical genetics and drug discovery screens: lessons from zebrafish and Xenopus. Dev Dyn. 2009;238:1287–1308. doi: 10.1002/dvdy.21967. [DOI] [PubMed] [Google Scholar]
- Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- Wielhouwer EM, Ali S, Al-Afandi A, Blom MT, Riekerink MB, Poelma C, Westerweel J, Oonk J, Vrouwe EX, Buesink W, et al. Zebrafish embryo development in a microfluidic flow-through system. Lab Chip. 2011;11:1815–1824. doi: 10.1039/c0lc00443j. [DOI] [PubMed] [Google Scholar]
- Wishart DS. Applications of metabolomics in drug discovery and development. Drugs R D. 2008;9:307–322. doi: 10.2165/00126839-200809050-00002. [DOI] [PubMed] [Google Scholar]
- Wlodkowic D, Khoshmanesh K, Akagi J, Williams DE, Cooper JM. Wormometry-on-a-chip: Innovative technologies for in situ analysis of small multicellular organisms. Cytometry A. 2011;79:799–813. doi: 10.1002/cyto.a.21070. [DOI] [PubMed] [Google Scholar]
- Wu J, Ben Y, Chang H-C. Particle detection by electrical impedance spectroscopy with asymmetric-polarization AC electroosmotic trapping. Microfluid. Nanofluidics. 2005;1:161–167. [Google Scholar]
- Wu H, Huang B, Zare RN. Generation of complex, static solution gradients in microfluidic channels. J Am Chem Soc. 2006;128:4194–4195. doi: 10.1021/ja058530o. [DOI] [PubMed] [Google Scholar]
- Yamada M, Kano K, Tsuda Y, Kobayashi J, Yamato M, Seki M, Okano T. Microfluidic devices for size-dependent separation of liver cells. Biomed. Microdevices. 2007;9:637–645. doi: 10.1007/s10544-007-9055-5. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Torii M, Uebayashi Y, Nasu M. Rapid, semiautomated quantification of bacterial cells in freshwater by using a microfluidic device for on-chip staining and counting. Appl Environ Microb. 2011;77:1536–1539. doi: 10.1128/AEM.01765-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q. Systems Biology in Drug Discovery and Development (Springer Protocols) Humana Press; 2010. [Google Scholar]
- Yang L, Bashir R. Electrical/electrochemical impedance for rapid detection of foodborne pathogenic bacteria. Biotechnol Adv. 2008;26:135–150. doi: 10.1016/j.biotechadv.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Yang F, Chen Z, Pan J, Li X, Feng J, Yang H. An integrated microfluidic array system for evaluating toxicity and teratogenicity of drugs on embryonic zebrafish developmental dynamics. Biomicrofluidics. 2011;5:24115. doi: 10.1063/1.3605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Zhang Q, Li C-W, Yang J, Zhao J, Yang M. Optical and electrochemical detection techniques for cell-based microfluidic systems. Anal Bioanal Chem. 2006;384:1259–1268. doi: 10.1007/s00216-005-0252-x. [DOI] [PubMed] [Google Scholar]
- Yu J, Liu Z, Liu Q, Yuen KT, Mak AFT, Yang M, Leung P. A polyethylene glycol (PEG) microfluidic chip with nanostructures for bacteria rapid patterning and detection. Sensors Actuators A: Physical. 2009;154:288–294. [Google Scholar]
- Yuki T, Masayuki Y, Teruo O, Takehiko K, Kiichi S. Evaluation of effects of shear stress on hepatocytes by a microchip-based system. Meas Sci Technol. 2006;17:3167. [Google Scholar]
- Yun JY, Jambovane S, Kim SK, Cho SH, Duin EC, Hong JW. Log-scale dose response of inhibitors on a chip. Anal Chem. 2011;83:6148–6153. doi: 10.1021/ac201177g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytseva NV, Goral VN, Montagna RA, Baeumner AJ. Development of a microfluidic biosensor module for pathogen detection. Lab Chip. 2005;5:805–811. doi: 10.1039/b503856a. [DOI] [PubMed] [Google Scholar]
- Zhang C, Noort D. Cells in microfluidics. In: Lin B, editor. Microfluidics. Springer Berlin; Heidelberg: 2011. pp. 295–321. [Google Scholar]
- Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
- Zhao J-Y, Chen D-Y, Dovichi NJ. Low-cost laser-induced fluorescence detector for micellar capillary zone electrophoresis: detection at the zeptomol level of tetramethylrhodamine thiocarbamyl amino acid derivatives. J. Chromatography A. 1992;608:117–120. [Google Scholar]
- Zhu T, Pei Z, Huang J, Xiong C, Shi S, Fang J. Detection of bacterial cells by impedance spectra via fluidic electrodes in a microfluidic device. Lab Chip. 2010;10:1557–1560. doi: 10.1039/b925968f. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Jenkins G, Zhang W, Zhang M, Guan Z, Yang CJ. Single-molecule emulsion PCR in microfluidic droplets. Anal Bioanal Chem. 2012;403:2127–2143. doi: 10.1007/s00216-012-5914-x. [DOI] [PubMed] [Google Scholar]