Abstract
MicroRNAs are short 21–22 nucleotide single strand RNAs that are involved in post-transcriptional regulation of gene expression. Most microRNAs are first transcribed as long primary microRNAs and then undergo a two step-wise sequential processing to yield single-stranded mature microRNAs. It has been suggested that the loop region of primary microRNAs plays an important role in regulating microRNA biogenesis and target recognition. However, despite the fact that several single nucleotide polymorphisms have been identified in mature microRNA sequences and are related to human diseases, it remains unclear whether and how the single nucleotide polymorphisms in the loop regions of primary microRNAs would affect the biogenesis and function of microRNAs. Herein, we provide evidence that primary microRNAs loop nucleotides control the accuracy and efficiency of microRNA processing. Accordingly, we identified 32 single nucleotide polymorphisms in the loop regions of human primary microRNAs using bioinformatics, and further validated three loss-of-function and one gain-of-function single nucleotide polymorphisms using dual-luciferase assays. Thus, these results reveal a critical regulatory role encoded in the loop nucleotides of primary microRNAs for microRNA processing and function.
Keywords: let-7, microRNA processing, post-transcriptional regulation, SNP, terminal loop
INTRODUCTION
MicroRNAs (miRNAs) are an abundant class of 21–22-nt-long small endogenous RNAs that primarily downregulate gene expression at the post-transcriptional level (Ambros, 2004; Bartel, 2004). MiRNAs are reported to be involved in a wide range of biological activities, such as immunity (Chen et al., 2004; Li et al., 2007a), cancer (O’Donnell et al., 2005; Zhang et al., 2013), apoptosis (Xu et al., 2003), stem cell maintenance (Arnold et al., 2011; Gangaraju and Lin, 2009) and neurological diseases (Cheng et al., 2009; Ouyang et al., 2012a). MiRNAs are initially transcribed as part of a long primary transcript or primary-miRNA (pri-miRNA) in the nucleus and are further processed by a RNase III Drosha/DGCR8 complex to generate 60-70-nt-long hairpin precursor miRNAs (pre-miRNA) (Lee et al., 2003). The pre-miRNAs are then exported to the cytoplasm by Exportin5 (Yi et al., 2003) and further processed into a 21-22-nt miRNA duplex by another RNase III-Dicer complex (Hutvagner et al., 2001). One strand of the miRNA duplex, the mature miRNA, is subsequently loaded into the RNA-induced silencing complex (RISC) (Hammond et al., 2000) and directs the RNAi regulatory function by targeting their cognate sites in the 3′ UTR region of target mRNAs (Lee et al., 1993; Lewis et al., 2003; Wightman et al., 1993). Currently, it is estimated that one miRNA can regulate hundreds of target genes, and more than half of human protein coding genes are regulated by miRNAs (Bartel, 2009).
MiRNA biogenesis can be regulated at either the transcriptional or the post-transcriptional level (Kim et al., 2009). Recently, extensive research has focused on identifying miRNA regulatory proteins and further investigating their post-transcriptional regulatory functions during miRNA biogenesis. Lin-28, a pluri-potent protein, was first reported to interact with the let-7 terminal loop region and block the processing of both primary and precursor microRNAs (Heo et al., 2008; Newman et al., 2008; Viswanathan et al., 2008). The crystal structure of mouse Lin-28 revealed that two folded domains of Lin-28 recognize two distinct regions of the miRNA terminal loop regions and are sufficient to inhibit let-7 biogenesis in vivo (Nam et al., 2011). TUT4, a non-canonical polyA polymerase, can be recruited by Lin-28 to precursor let-7 to block Dicer processing through pre-miRNA uridylation (Heo et al., 2009). Heteronuclear ribonuc-leoprotein A1 (hnRNP A1), a negative regulator of let-7a, binds the conserved terminal loop of pri-let-7a-1 and inhibits its processing by Drosha (Michlewski and Caceres, 2010). Several other studies showed a specific role of the pri/pre-miRNA hairpin loop nucleotides in miRNA target recognition and regulation (Liu et al., 2008; Trujillo et al., 2010; Yue et al., 2011). Except for the miRNA loop region, the pri-miRNA tertiary structure of the miR-17-92 cluster has been demonstrated to modulate differential expression of constituent miRNAs (Chaulk et al., 2011). Emerging evidence strongly implies that the loop region of miRNAs plays a central role in the biogenesis of miRNAs.
To date, many miRNA-related single nucleotide polymorphisms (SNPs) have been identified in human miRNA seeds and mature regions, as well as their target sites in the 3′ UTR of mRNAs, suggesting a potential role of SNPs in regulating miR-NA target recognition through disrupting or creating new miR-NA target interactions (Gong et al., 2012; Ryan et al., 2010). A number of studies have demonstrated that SNPs in miRNA genes or target binding sites are associated with human diseases (Chin et al., 2008; Jazdzewski et al., 2009; Mencia et al., 2009; Sun et al., 2010). For example, an SNP in the seed region of human miR-96 is involved in non-syndromic progressive hearing loss (Mencia et al., 2009). A SNP in the 3′ UTR of KRAS is associated with increased risk of non-small cell lung cancer by weakening or abolishing its interaction with let-7 (Chin et al., 2008). However, there is little information about SNPs in the miRNA loop region and their roles in miRNA biogenesis and target regulation. In this study, we found that the miRNA terminal loop not only controls the efficiency of miRNA biogenesis, but also modulates Drosha/DGCR8 processing fidelity for pre-miRNAs. We also present evidence showing that SNPs in human pri-miRNA loop regions affect mature miRNA production and miRNA target regulation by modulating miRNA biogenesis.
MATERIALS AND METHODS
MiRNA and 3′ UTR reporter constructs
A bi-cistronic vector driven by murine 3-phosphoglycerate kinase (PGK) promoter (Trujillo et al., 2010) was used to co-express miRNAs and a GFP reporter. DNA fragments containing the pre-miRNA hairpin and ∼250–400 nt flanking sequence on each side were amplified from Caenorhabditis elegans or human genomic DNA and placed under the control of the PGK promoter. Loop mutants and SNP miRNA mutants were generated using an overlapping PCR strategy to introduce mutations into the loop regions of the miRNA genes. Full-length human CDKN1B, FBXW7, and 1000 nt flanking the predicted binding sites from the 3′ UTRs of the YAP and BCL-2 genes were cloned into the Renilla luciferase reporter vector phRL-TK (Pro-mega).
Cell culture and transfection
Adherent BOSC 23 cells were grown in DMEM, 10% FBS, and 1% of penicillin/streptomycin antibiotics, supplemented with glutamine. For Northern blotting and primer extension assays, BOSC 23 cells were plated at a density of 7.5 × 105 cells/well in a 6-well plate, 24 h before transfection. Cells were transfected with 2 μg of constructs expressing miRNAs, mutants and control vector using Fugene transfection reagents (Roche). After 48 h of transfection, FACS analysis showed that ∼70% cells expressed a GFP signal (data not shown).
Luciferase reporter assay
The luciferase reporter assay was performed as previously described (Ouyang et al., 2012b). BOSC23 cells were plated at a density of 2 × 104 cells/well in a 96-well plate, 24 h before transfection. Cells were co-transfected with 10 ng firefly luciferase control reporter plasmid, 5 ng Renilla luciferase target reporter, and 50 ng miRNA expression vector using Fugene (Roche), according to the manufacturer’s instructions. Cells were harvested 48 h post-transfection and assayed using the Dual-Luciferase system (E1960, Promega). Results were expressed as relative luciferase activity by first normalizing to the firefly luciferase transfection control, and then to the Renil-la/firefly value of the empty control vector.
Northern blot analyses and pre-miRNA processing efficiency analyses
Northern blot analyses were carried out as previously described (Liu et al., 2008). Total RNA was prepared from BOSC 23 cells transfected with equal amount of miRNA expression vectors. The U6 RNA level was used as a loading control. Phosphoimaging was used to determine band intensities and representative blots and averaged data of three replicates from cells transfected with independent DNA preparations are shown. To calculate pre-miRNA processing efficiency, truncated 20 nt or rescued 22 nt (LPAA/LPA1) mature cel-let-7 band signals were first subtracted from the endogenous human let-7 signal and then normalized to the truncated or rescued (LPAA/LPA1) pre-cel-let-7 signal, respectively. The value was further normalized to the value of WT cel-let-7 group (processing efficiency defined as 1). The same method was applied to human miRNA SNP mutants.
Primer extension analysis
Primer extension analyses were carried out as previously described (Trujillo et al., 2010). Pre-let-7 or mature let-7 RNA fractions were analyzed by primer extension analyses to determine the 5′ ends of pre-let-7 and mature let-7. Primer and DNA Ladder sequences are listed in Supplementary Table 1. Briefly, pre-let-7 or mature let-7 RNA fractions isolated from 100 μg of total RNA were mixed with 32P-labeled primers in 1× RT buffer. The primer extension reaction, which was initiated by the addition of dNTPs and reverse transcriptase (Applied Biosystems), was carried out for 1 h at 42°C, and terminated by incubation at 85°C for 10 min. Products were resolved on 15% denaturing PAGE gels. 32P-labeled synthetic oligonucleotides with the let-7 sequence in single nucleotide increments (16 to 22 nt) were used as the size ladder. Gels were exposed to a phosphoimager screen for 2 h and analyzed using the Storm system.
Primary microRNA and mature miRNA-specific quantitative real-time RT-PCR
Total RNA was isolated with TRIzol® (Invitrogen) and treated with TURBO DNA-free kit (Ambion). For primary microRNA expression level analyses, random hexamer primers were used in the reverse transcription step. Pri-miRNA-specific primers (Supplementary Table 2) were designed for regions that did not include the mutation points and used for real-time PCR. Expression levels were normalized using GAPDH as an internal control. For mature miRNA, reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). Truncated 20 nt mature cel-let-7 and endogenous human let-7 have similar detection sensitivities in the TaqMan MicroRNA assay (Trujillo et al., 2010). The miRNA expression level was first normalized using U6 as an internal control and then subtracted from the endogenous human miR-NA signal. Measurements were normalized to U6 (ΔCt) and comparisons calculated as the inverse log of the ΔΔCT to give the relative fold change for all miRNA levels (Livak and Schmittgen, 2001). The PCR experiments were repeated three times, each using separate sets of samples.
Statistical analysis
All data reported represent at least three independent experiments. Data reported are means ± SD. Statistical differences were determined by a T test for comparison of two groups using Prism 5.0 software. P values < 0.05 were considered statistically significant.
RESULTS
A single loop nucleotide mutation rescues defective biogenesis of C. elegans let-7 in human cells
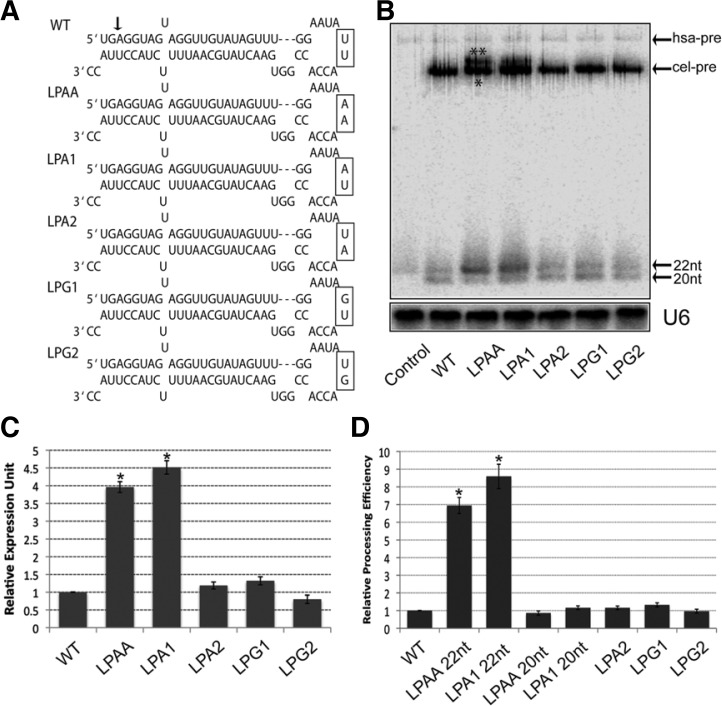
Our previous studies demonstrated that truncated pre and mature let-7 could be produced by ectopic expression of C. elegans let-7 gene (cel-let-7) in human cells (Trujillo et al., 2010) and one mutant (LPAA) (Yue et al., 2011) (Fig. 1A) with two AA nucleotides substituting for two UU nucleotides in the pri-let-7 terminal loop can partially restore defective cel-let-7 biogenesis. However, it remains unclear whether this rescue function is caused by changes in the terminal loop sequence or secondary structure, because LPAA has a different predicted loop structure from WT cel-let-7 (by RNA mfold prediction). To further dissect which nucleotides are critical for the rescue function, we mutated two pri-let-7 terminal loop nucleotides without affecting the loop secondary structure (by RNA mfold prediction) (Fig. 1A) and tested the effects of these mutations on the biogenesis of cel-let-7 (Figs. 1B and 1C). Both miRNA Northern blots and qPCR results confirmed our previous finding that pri-let-7 loop nucleotides have very different roles in the biogenesis of cel-let-7. Mutants LPA2, LPG1 and LPG2 had little or no effect on mature let-7 production. Strikingly, the LPA1 mutation resulted in a significant increase (> two-fold) in mature let-7 production and more pre-let-7 was detected in miRNA Northern blots, which phenocopies the LPAA rescue function (Fig. 1B). We also noticed that partially rescued pre-let-7 from LPAA and LPA1 mutants dramatically increased pre-let-7 processing efficiency (> seven-fold, 22 nt LPAA/LPA1 Fig. 1D). However, we did not observe this effect of truncated pre-let-7 made from the same LPAA and LPA1 mutants (20 nt LPAA/LPA1 Fig. 1D). Next, we tested whether LPA1 can rescue truncated pre and mature let-7 that lack the first two nucleotides at the 5′ end of mature let-7 produced from cel-let-7-WT. Primer extension analysis confirmed that part of the pre and mature let-7 produced by LPA1 mutant had the same 5′ end as the endogenous human let-7 (Supplementary Fig. 1). To exclude the possibility that our results came from different transfection and expression levels of mutant plasmids, we carried out specific qPCR assay to measure primary let-7 expression levels from loop mutants. We found that all the mutants express similar primary let-7 level as WT let-7 (Supplementary Fig. 2A). Although a previous study showed that the pri-miRNA loop is dispensable for miRNA processing in vitro (Han et al., 2006), our results strongly suggest that terminal loop nucleotides play a key role in Drosha/DGCR8 proces-sing of pri-miRNA to pre-miRNA in vivo.
Fig. 1.
Identification of a single loop mutation that partially rescues defective let-7 biogenesis. (A) Schematic diagram depicting the pre-let-7 miRNA sequences and structures of the wild-type cel-let-7 gene and loop mutants. An arrow shows the truncated site for wild-type pre-let-7 and mature let-7. (B) Expression levels of pre- and mature c-let-7 from wild-type and loop mutants, as determined by Northern blotting. Blots were also probed for U6 small nuclear RNA as a loading control. Truncated pre-let7 (*) and rescued pre-let-7 (**) (C) Expression level of mature let-7 from wild-type and loop mutants was measured by miRNA-specific QPCR assay. (D) Pre-let-7 processing efficiency was determined by Northern blotting. The steady status mature let-7 level, normalized to pre-let-7 level, was used to quantify the efficiency. Representative results of three independent transfections are shown.
Loop mutagenesis reveals different effects on miRNA processing efficiency
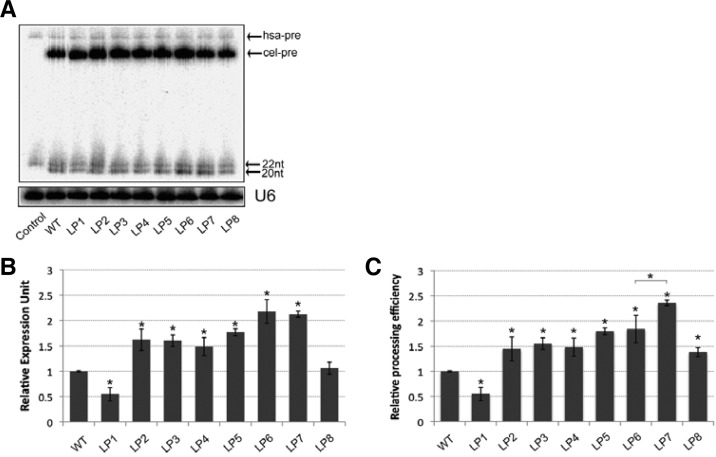
To test whether other nucleotides in the pri-let-7 loop region could also restore the defective biogenesis of the cel-let-7-WT, we further generated a series of single nucleotide mutations over the whole cel-let-7 loop, maintaining the same predicted secondary structure as WT cel-let-7 (Table 1), and measured their effects on the biogenesis of cel-let-7. All the loop mutants express similar primary let-7 level as WT let-7 (Supplementary Fig. 2B). We found that single nucleotide mutations in the pri-let-7 loop have both positive and negative effects on mature let-7 production. Some mutants (LP2, LP3, LP4, LP5 and LP8) had no effect, or only displayed a slight increase in mature let-7 production, whereas mutant LP1 reduced mature let-7 production by approximately 50%. Surprisingly, two mutants (LP6 and LP7) produced more than two-fold more mature let-7 (Figs. 2A and 2B). Although primer extension analyses of gel-purified small RNA fragments showed that none of these mutants rescued truncated mature let-7 production (data not shown), we cannot exclude the possibility that other nucleotide substitutions may rescue the defective biogenesis of cel-let-7. More importantly, these mutants displayed differential processing efficiency of pre-let-7 to mature let-7 (Fig. 2C). In fact, although the cel-let-7-LP7 mutant has a similar mature let-7 level as the cel-let-7-LP6 mutant, the pre-cel-let-7-LP7 mutant displayed nearly 40% higher processing efficiency than the pre-cel-let-7-LP6 mutant. Similarly, the pre-cel-let-7-LP8 mutant also displayed 40% higher processing efficiency than the WT, despite a similar mature let-7 expression level. Consistent with the finding by Schopman et al. (2010), showing that optimizing the loop sequence of shRNA can increase siRNA production and inhibition function, our results demonstrated that loop nucleotides modulate the efficiency of miRNA biogenesis.
Table 1.
Loop mutations in pre-c-let-7 and effects on miRNA processing efficiency
| c-let-7 | Loop sequence | Mature level | Pre-miRNA processing |
|---|---|---|---|
| WT | GGAAUAUUACCACC | 1 | 1 |
| LP1 | GGC⃞AUAUUACCACC | 0.55 ± 0.13 | 0.55 ± 0.11 |
| LP2 | GGAU⃞UAUUACCACC | 1.62 ± 0.21 | 1.44 ± 0.24 |
| LP3 | GGAAA⃞AUUACCACC | 1.60 ± 0.11 | 1.55 ± 0.12 |
| LP4 | GGAAUU⃞UUACCACC | 1.49 ± 0.18 | 1.48 ± 0.18 |
| LP5 | GGAAUAUUG⃞CCACC | 1.77 ± 0.07 | 1.80 ± 0.07 |
| LP6 | GGAAUAUUAG⃞CACC | 2.18 ± 0.23 | 1.84 ± 0.27 |
| LP7 | GGAAUAUUACG⃞ACC | 2.12 ± 0.07 | 2.36 ± 0.06 |
| LP8 | GGAAUAUUACCG⃞CC | 1.06 ± 0.12 | 1.38 ± 0.10 |
Fig. 2.
Loop mutations that modulate miRNA biogenesis efficiency. (A) Northern blot showing the expression levels of pre- and mature let-7 made from wild-type and loop mutants. (B) miRNA-specific qPCR assay measuring the expression level of mature let-7 from wild-type and loop mutants. (C) Pre-let-7 processing efficiency was determined by Northern blotting. The steady status mature let-7 level, normalized to pre-let-7 level, was used to quantify the efficiency. Representative results of three independent transfections are shown (*p < 0.05).
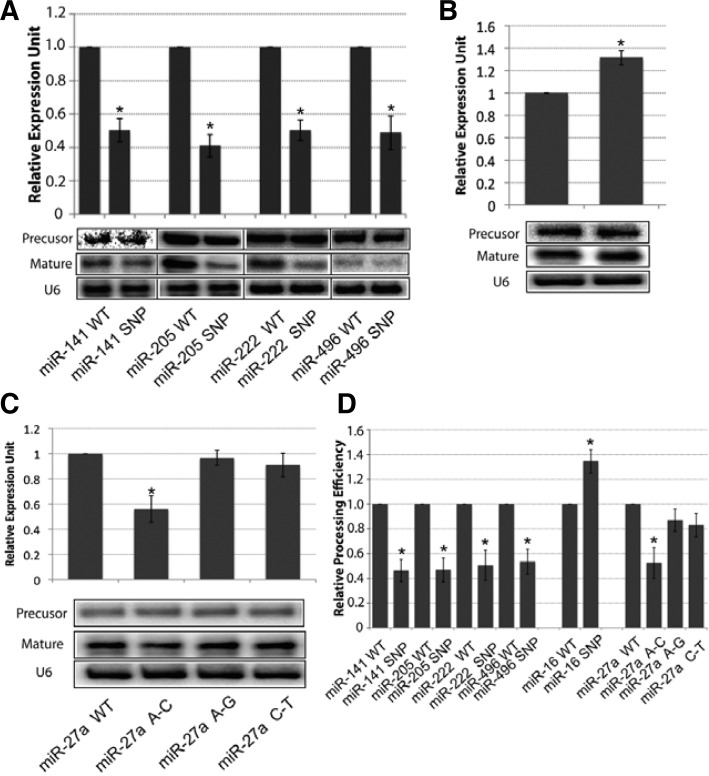
Loop SNPs in human miRNA affect mature miRNA production
Several groups reported that SNPs in miRNA flanking and stem regions could affect the biogenesis of miRNAs, both in vivo and in vitro (Duan et al., 2007; Sun et al., 2009). Thus, we attempted to investigate whether and how SNPs in the loop region of human miRNA would impact on miRNA biogenesis. To address this question, we mapped the human SNPs (NCBI dbSNP 135 for human) onto human pre-miRNA genes and identified 32 SNPs in 21 miRNA loop regions (Table 2 and Supplementary Table 3). We observed that several miRNAs had more than one SNP in the loop region, such as miR-27a and miR-559. We chose 12 SNPs in diseases-related microRNAs (Table 2) and tested how these loop SNPs affected mature miRNA production. WT and SNP mutant plasmids were transfected into BOSC 23 cells, and mature microRNA levels were measured by TaqMan MicroRNA specific qPCR assay. Consistent with our findings for the loop’s effects on cel-let-7 processing, we found that these SNPs have different effects on mature miRNA production. Some SNPs caused a significant decrease (> 50%) in mature miRNA expression (Fig. 3A), whereas one SNP, in the miR-16 loop region, resulted in more than 30% increased mature miR-16 expression (Fig. 3B). Interestingly, although miR-27a has three SNPs in different loop regions, only the A–C mutation caused a 40% decrease of mature miR-27a expression; the other two SNPs did not have such an effect (Fig. 3C). We also observed that all loop SNPs tested had little or no effects on pri and pre-miRNAs production (For pri-miRNAs, Supplementary Fig. 2C; for pre-miRNAs, Northern blot, Figs. 3A–3C) and only affected pre-miRNAs’ processing efficiency (Fig. 3D). In addition, several SNPs displayed minimal effects on mature miRNA production (Supplementary Fig. 3). Together with the findings by Zhang and Zeng (2010), which showed that the terminal loop region controls miRNA processing by Drosha and Dicer in vitro, our results represent the first evidence that demonstrates directly that the terminal loop plays a critical role in human miRNA processing in vivo.
Table 2.
Experimentally validated single nucleotide polymorphism(SNP) in pre-miRNA
| miRNA | Loop sequence (SNP) | SNPID | Effects |
|---|---|---|---|
| hsa-mir-16-1 | CGTTAAGATTCTAAAATTATCT(C) | rs72631826 | Gain |
| hsa-mir-27a | GGGTCCACA(C/G/T)CCAAGTCG | rs895819 | Loss/No |
| hsa-mir-27a | GGGTC(T)CACACCAAGTCG | rs11671784 | No |
| hsa-mir-30e | TGTGAGCAATAG(A)TAAGGAA | rs112439044 | No |
| hsa-mir-34a | TGTAAGGTGTTC(T)AGAGGA | rs72631823 | No |
| hsa-mir-130b | ATAGGCCG(A)CTGGG | rs72631822 | No |
| hsa-mir-135a2 | AGTAATAAAGT(C)C | rs113322127 | No |
| hsa-mir-141 | GGT(C)CTAATTGTGAAGCT | rs111718468 | Loss |
| hsa-mir-184 | TTTG(T)TGACTGTAAGT | rs41280052 | Loss |
| hsa-mir-205 | TCTCATACCCAACC(T) | rs113859371 | Loss |
| hsa-mir-222 | GTCTTTCG(A)TAATC | rs72631825 | Loss |
| hsa-mir-496 | TTTA(C)TTTATG | rs79307187 | Loss |
Fig. 3.
Loop SNPs in human miRNAs affect mature miRNA production. (A) Precursor and mature miRNAs made from mir-141, miR-205, miR-222, mir-496 and their SNP miRNAs were measure by miRNA qPCR (top) and Northern blotting (bottom). (B) miRNA-specific qPCR (top) and Northern blotting (bottom) showing the expression level of precursor and mature miR-16 from the wild-type and SNP mutant. (C) Precursor and mature miR-27a level from three loop SNPs in the miR-27a loop, measured by miRNA qPCR (top) and Northern blotting (bottom), showing that only the A to C mutation affected miR-27a processing. (D) Northern blot showing pre-miRNA processing efficiency. Efficiency was quantified by the steady status mature miRNA level normalized to the pre-miRNA level. Representative results of two or three independent transfections are shown (*p < 0.05).
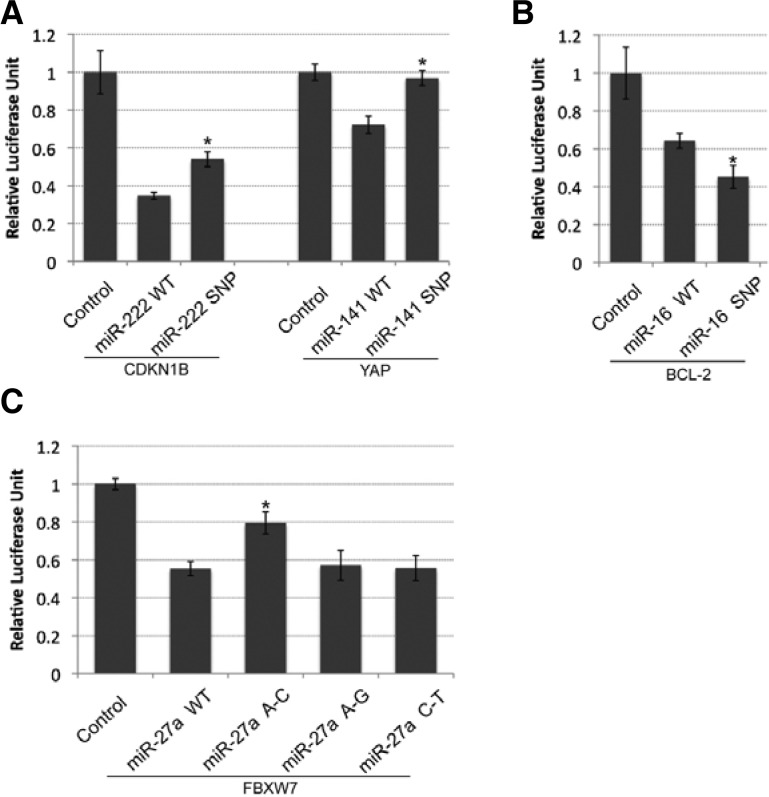
Loop SNPs in human miRNAs affect miRNA post-transcriptional regulatory functions
To further investigate the role of the loop SNPs in miRNA target regulatory function, we selected four cancer-related miRNA targets to test the SNPs’ effects on miRNA post-transcriptional function, using a luciferase reporter assay. MiR-222 was previously reported to repress the expression of CDKN1B (p27), a key regulator of cell cycle in chronic lymphocytic leukemia (CLL) (Frenquelli et al., 2010). As shown in Fig. 4A, a G-A SNP in the pri-miR-222 loop resulted in a remarkable decrease in target repression of CDKN1B from ∼65% repression by WT miR-222 to ∼45% repression by the mutant. More strikingly, a G-A SNP in pri-miR-141 completely abolished miR-141 repression of its target gene, YAP (Fig. 4A), which was reported to act as a tumor suppressor in breast cancer (Yuan et al., 2008). We also observed that a G-A SNP in the pri-miR-16 loop slightly enhanced miR-16 repression of anti-apoptosis gene BCL-2, from 40% repression by WT mir-16 to 55% by the mutant (Fig. 4B). Consistent with the mature miR-27a expression pattern, only the A–C SNP mutation in the pri-miR-27 loop induced a 50% reduction in repression activity of the target gene, FBXW7, compared with WT mir-27a. By contrast, the other two SNPs had a similar repression activity as WT mir-27a (Fig. 4C). Overall, these results suggest that loop SNPs have different effects on miRNA regulatory function by affecting mature miRNA production.
Fig. 4.
Loop SNPs in human miRNAs affect miRNA repression activity on the 3′ UTRs of target genes. (A) Dual-luciferase assay showing the repression activity of miR-222 and SNP miR-222 on the 3′ UTR of CDKN1B (left) and the repression activity of miR-141 and SNPmiR-141 on the 3′ UTR of the YAP gene (right). (B) Dual-luciferase assay performed with miR-16 and SNP miR16 on the 3′ UTR of the BCL-2 gene. (C) A–C mutation in the mir-27a loop region attenuated miR-27a repression activity on the 3′ UTR of the FBXW7 gene. Representative results of at least six independent trials (± S.D.) (*p < 0.05).
DISCUSSION
Our work reveals that nucleotides in the pri-RNA loop region affect miRNA processing fidelity (Fig. 1). We used a combination of Northern blotting and primer extension assays to demonstrate that loop nucleotides modulate the accuracy of the Drosa/DGCR8 Complex processing of the 5′ seed region of pre-miRNA or mature miRNA would indirectly diversify the miRNA target repertoire selection, because the 5′ seed region is essential for miRNA target recognition and regulation. Intriguingly, there are controversial roles for pri-miRNA loops in miRNA processing and mature miRNA biogenesis. Previously, a mir-16-1 in vitro processing assay suggested that the loop is dispensable for pri-mir-16-1 processing and mature miR-16 production (Han et al., 2006). However, Zhang and Zeng (2010) reported that a flexible terminal loop region is critical for miRNA processing, using a similar in vitro processing assay for different miRNAs. Our findings strongly support the latter model, and suggest that further analyses are required to precisely dissect the role of the pri-miRNA loop in miRNA processing in vivo. Although several regulatory proteins have been demonstrated to bind to pri/pre-miRNA loops and affect the production of mature miRNA (Heo et al., 2008; 2009; Michlewski and Caceres, 2010; Newman et al., 2008; Trabucchi et al., 2009; Viswanathan et al., 2008) and conserved loop sequences are required for the interaction between these regulatory factors (Heo et al., 2009; Newman et al., 2008), none of these proteins have been shown to be involved in the regulation of the processing accuracy of the mature miRNA 5′ seed region. Additionally, 5′ end polymorphisms have been detected in miRNA profiles of CD8 T cells (Wu et al., 2007), but there is little evidence to explain the heterogeneity of the 5′ end of mature miRNAs. A recent study reported that the loop position is critical for the accuracy of dicer processing (Gu et al., 2012), but it should be noted that dicer processing inaccuracy is mainly involved in the generation of the 3′ end polymorphisms. Therefore, our finding presents a novel mechanism for 5′ end polymorphism generation and suggests that a class of molecules may exist that recognize pri-miRNA loops and affect the recognition fidelity of Drosha catalytic domain, which controls the accuracy of miRNA 5′ seed generation. This finding is consistent with a recent report by Auyeung et al. (2013) showing that the primary-sequence determinant GUG motif in the loop region distinguishes pri-miRNAs from other hairpin-containing transcripts for efficient processing. Recently, 5′-end recognition by Dicer has been shown to be critical for precise and effective biogenesis of miRNAs (Park et al., 2011). In our study, we also observed rescued pre-let-7 (with a canonical 5′ end) from LPAA and LPA1 mutants dramatically increased mature cel-let-7 production (> four-fold) and Pre-let-7 processing efficiency (> sevenfold). It is quite plausible that the restoration of the canonical 5′ end cel-pre-let-7 structure may enhance Dicer enzyme processing efficiency. In addition, our results also suggest that the terminal loop can function independently of 5′ end recognition to affect Dicer processing efficiency (Figs. 2C and 3D).
Furthermore, our findings that mutagenesis of loop nucleotides can modulate miRNA processing efficiency and mature miRNA production (Fig. 2) provides new insights into optimal shRNA design for gene silencing in vivo. RNA interference (RNAi) is a key tool to investigate gene function in vivo and target gene knockdown efficiency is an important factor in RNAi-based studies (Paddison and Hannon, 2002). Most studies used the original shRNA design of the pSuper system (Brummelkamp et al., 2002); however, several studies indicated that shRNA molecules do not always exhibit the same silencing effects as the corresponding synthetic siRNA for the same target sequence (Hinton et al., 2008; Li et al., 2007b; Miyagishi et al., 2004). Therefore, the efficiency during processing from hairpin shRNA to small interfering RNA (siRNA) is a key issue. Similar to our findings for the loop’s role in miRNA processing efficiency, Schopman et al. (2010) reported that optimization of the loop sequence of the hairpin shRNA can significantly improve the inhibitory function of the shRNA in different cell types by increasing small interfering RNA production. In our study, we observed that LPA1 produced four-fold more mature let-7 when substituted with an A nucleotide in the pri-let-7 loop (Figs. 1B and 1C). However, a G nucleotide substitution (LPG1, Figs. 1B and 1C) at the same site in the loop region had almost no effect on mature let-7 production, indicating that the loop sequence specificity is important for pri/pre-miRNA processing. Recently, a human miR-30-based shRNA design showed promising efficiency in gene knockdown in several cell culture and mammal systems (Silva et al., 2005; Xia et al., 2006). Our observations strongly suggested that the efficiency and specificity of miRNA-based shRNAs could be further improved by optimizing the pri-miR-30 loop sequence.
Recently, SNPs in miRNA seed regions have been shown to be associated with human diseases, including hereditary diseases and cancers (Chin et al., 2008; Jazdzewski et al., 2009; Mencia et al., 2009; Sun et al., 2009; 2010). Gong et al. (2012) reported more than 700 SNPs in human miRNA genes and identified 50 SNPs in human miRNA seed regions, and predicted the target gain and loss effects for these SNPs. Although SNPs in miRNA seed regions would directly influence the miR-NA target binding and selection, one caveat for SNP study in miRNA seed regions is that these SNPs may completely change the wild-type miRNA target repertoire and create a new target regulatory network in vivo, because SNPs may form totally new seeds in the mutant miRNAs. In this study, we specifically identified 32 SNPs in human miRNA loop regions and further examined their roles in miRNA processing and target regulatory function. We observed that half of our tested SNPs (6/12) had effects on miRNA processing, which provided a new molecular mechanism for SNPs in human diseases by modulating miRNA production. For example, the minor allele C (A → C) of SNP rs895819 in the miR-27a gene is associated with reduced risk of gastric cancer (Zhou et al., 2012). We found that only the A–C SNP significantly reduced the mature miR-27a expression level (50%) (Fig. 3C). The SNP attenuated miR-27a’s inhibition of its target protein, FBW7 (Fig. 4C), a tumor suppressor that functions as a substrate component for a SCF ubiquitin ligase (Lerner et al., 2011). Thus, it is plausible that SNP rs895819 (A → C) leads to upregulation of the FBW7 protein level by reducing miR-27a processing. Upregulated FBW7 protein will cause downregulation of multiple oncoproteins and reduce the chance of tumorigenesis. MiR-16 was the only miRNA whose expression level was upregulated by loop SNPs in our study. MiR-16 plays a critical role as a tumor repressor in the development of chronic lymphocytic leukemia (CLL) (Cimmino et al., 2005). CLL is characterized by overexpression of the antiapoptotic B cell lymphoma 2 (Bcl2) protein, and miR-16 expression was reported to be inversely correlated with Bcl2 expression. We used a reporter system to confirm that Bcl-2 is a bona fide target for miR-16. More importantly, SNP rs7263 1826 induced a 30% increase in mature miR-16 production and repression of oncogene BCL-2, which may be used as a prognostic indicator for subpopulations of CLL.
In summary, we identified regulatory information in the terminal loop region of miRNAs that may play a critical role in miRNA processing fidelity and efficiency. The results have broad implications for understanding the mechanisms underlying miRNA biogenesis, miRNA-related diseases and shRNA design.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no. 81271274). The authors would like to thank Dr. Song S. Wang for helpful discussion and comments on the manuscript. The authors have no conflicting financial interests.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Arnold CP, Tan R, Zhou B, Yue SB, Schaffert S, Biggs JR, Doyonnas R, Lo MC, Perry JM, Renault VM, et al. MicroRNA programs in normal and aberrant stem and progenitor cells. Genome Res. 2011;21:798–810. doi: 10.1101/gr.111385.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Chaulk SG, Thede GL, Kent OA, Xu Z, Gesner EM, Veldhoen RA, Khanna SK, Goping IS, MacMillan AM, Mendell JT, et al. Role of pri-miRNA tertiary structure in miR-17∼92 miRNA biogenesis. RNA Biol. 2011;8:1105–1114. doi: 10.4161/rna.8.6.17410. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- Frenquelli M, Muzio M, Scielzo C, Fazi C, Scarfo L, Rossi C, Ferrari G, Ghia P, Caligaris-Cappio F. MicroRNA and proliferation control in chronic lymphocytic leukemia: functional relationship between miR-221/222 cluster and p27. Blood. 2010;115:3949–3959. doi: 10.1182/blood-2009-11-254656. [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Tong Y, Zhang HM, Wang K, Hu T, Shan G, Sun J, Guo AY. Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum Mutat. 2012;33:254–263. doi: 10.1002/humu.21641. [DOI] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis PN, Kay MA. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Hinton TM, Wise TG, Cottee PA, Doran TJ. Native microRNA loop sequences can improve short hairpin RNA processing for virus gene silencing in animal cells. J RNAi Gene Silencing. 2008;4:295–301. [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jazdzewski K, Liyanarachchi S, Swierniak M, Pachucki J, Ringel MD, Jarzab B, de la Chapelle A. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc. Natl. Acad. Sci USA. 2009;106:1502–1505. doi: 10.1073/pnas.0812591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lerner M, Lundgren J, Akhoondi S, Jahn A, Ng HF, Akbari Moqadam F, Oude Vrielink JA, Agami R, Den Boer ML, Grander D, et al. MiRNA-27a controls FBW7/hCDC4-dependent cyclin E degradation and cell cycle progression. Cell Cycle. 2011;10:2172–2183. doi: 10.4161/cc.10.13.16248. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Li L, Lin X, Khvorova A, Fesik SW, Shen Y. Defining the optimal parameters for hairpin-based knockdown constructs. RNA. 2007a;13:1765–1774. doi: 10.1261/rna.599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007b;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Liu G, Min H, Yue S, Chen CZ. Pre-miRNA loop nucleotides control the distinct activities of mir-181a-1 and mir-181c in early T cell development. PLoS One. 2008;3:e3592. doi: 10.1371/journal.pone.0003592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7 a biogenesis. Nat Struct Mol Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishi M, Sumimoto H, Miyoshi H, Kawakami Y, Taira K. Optimization of an siRNA-expression system with an improved hairpin and its significant suppressive effects in mammalian cells. J Gene Med. 2004;6:715–723. doi: 10.1002/jgm.556. [DOI] [PubMed] [Google Scholar]
- Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Ouyang YB, Lu Y, Yue S, Xu LJ, Xiong XX, White RE, Sun X, Giffard RG. miR-181 regulates GRP78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol Dis. 2012a;45:555–563. doi: 10.1016/j.nbd.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang YB, Lu Y, Yue S, Giffard RG. miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion. 2012b;12:213–219. doi: 10.1016/j.mito.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, Hannon GJ. RNA interference: the new somatic cell genetics? Cancer Cell. 2002;2:17–23. doi: 10.1016/s1535-6108(02)00092-2. [DOI] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopman NC, Liu YP, Konstantinova P, ter Brake O, Berkhout B. Optimization of shRNA inhibitors by variation of the terminal loop sequence. Antiviral Res. 2010;86:204–211. doi: 10.1016/j.antiviral.2010.02.320. [DOI] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Sun G, Yan J, Noltner K, Feng J, Li H, Sarkis DA, Sommer SS, Rossi JJ. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009;15:1640–1651. doi: 10.1261/rna.1560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Gu H, Zeng Y, Xia Y, Wang Y, Jing Y, Yang L, Wang B. Hsa-mir-27a genetic variant contributes to gastric cancer susceptibility through affecting miR-27a and target gene expression. Cancer Sci. 2010;101:2241–2247. doi: 10.1111/j.1349-7006.2010.01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo RD, Yue SB, Tang Y, O’Gorman WE, Chen CZ. The potential functions of primary microRNAs in target recognition and repression. EMBO J. 2010;29:3272–3285. doi: 10.1038/emboj.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naive, effector and memory CD8 T cells. PLoS One. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XG, Zhou H, Samper E, Melov S, Xu Z. Pol II-expressed shRNA knocks down Sod2 gene expression and causes phenotypes of the gene knockout in mice. PLoS Genet. 2006;2:e10. doi: 10.1371/journal.pgen.0020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Tomlinson V, Lara R, Holliday D, Chelala C, Harada T, Gangeswaran R, Manson-Bishop C, Smith P, Danovi SA, et al. Yes-associated protein (YAP) functions as a tumor suppressor in breast. Cell Death Differ. 2008;15:1752–1759. doi: 10.1038/cdd.2008.108. [DOI] [PubMed] [Google Scholar]
- Yue SB, Trujillo RD, Tang Y, O’Gorman WE, Chen CZ. Loop nucleotides control primary and mature miRNA function in target recognition and repression. RNA Biol. 2011;8:1115–1123. doi: 10.4161/rna.8.6.17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 2010;38:7689–7697. doi: 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wu GQ, Zhang Y, Feng ZY, Zhu SM. Propofol induces apoptosis of hepatocellular carcinoma cells by upregulation of microRNA-199a expression. Cell Biol Int. 2013;37:227–232. doi: 10.1002/cbin.10034. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Du WD, Chen G, Ruan J, Xu S, Zhou FS, Zuo XB, Lv ZJ, Zhang XJ. Association analysis of genetic variants in microRNA networks and gastric cancer risk in a Chinese Han population. J Cancer Res Clin Oncol. 2012;138:939–945. doi: 10.1007/s00432-012-1164-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.