Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) mediates neuronal cell death in a variety of pathological conditions involving severe DNA damage. Poly(ADP-ribose) (PAR) polymer is a product synthesized by PARP-1. Previous studies suggest that PAR polymer heralds mitochondrial apoptosis-inducing factor (AIF) release and thereby, signals neuronal cell death. However, the details of the effects of PAR polymer on mitochondria remain to be elucidated. Here we report the effects of PAR polymer on mitochondria in cells in situ and isolated brain mitochondria in vitro. We found that PAR polymer causes depolarization of mitochondrial membrane potential and opening of the mitochondrial permeability transition pore early after injury. Furthermore, PAR polymer specifically induces AIF release, but not cytochrome c from isolated brain mitochondria. These data suggest PAR polymer as an endogenous mitochondrial toxin and will further our understanding of the PARP-1-dependent neuronal cell death paradigm.
Keywords: apoptosis-inducing factor, mitochondria, neuronal cell death, poly(ADP-ribose) polymer
INTRODUCTION
Poly(ADP-ribose) polymerase-1 (PARP-1) is a nuclear protein that serves as a sensor of DNA breaks (Virag and Szabo, 2002). PARP-1 is activated by binding to DNA strand breaks and facilitates damage repair through poly(ADP-ribosyl)ation of target proteins. Numerous proteins can be poly(ADP-ribosyl)ated including histones, transcription factors and PARP-1 itself (Diefenbach and Burkle, 2005). Poly(ADP-ribosyl)ation confers a highly negative charge on acceptor proteins and causes electrostatic repulsion between DNA and poly(ADP-ribosyl)ated proteins, thereby allowing chromatin remodeling, DNA repair and signaling (Virag and Szabo, 2002). However, over-activated PARP-1 also induces cell death under conditions causing severe DNA damage, such as glutamate excitotoxicity, ischemia-reperfusion injury and inflammation (Wang et al., 2004; Yu et al., 2002). Previously PARP-1-induced cell death was considered necrotic because over-activation of PARP-1 results in exhaustion of cellular ATP (Herceg and Wang, 1999). PARP-1 cleaves NAD+ into nicotinamide and ADP-ribose, and uses the latter for generation of poly(ADP-ribose) (PAR) polymer. Once PARP-1 consumes NAD+, it subsequently depletes ATP to convert it to NAD+. However, recent studies have demonstrated that PARP-1-induced cell death exhibits both apoptotic and necrotic features with apoptosis-inducing factor (AIF) as an essential mediator of cell death (Yu et al., 2002; 2003; 2006). To distinguish this special biochemical pathway of programmed cell death from apoptosis or necrosis, a new term “parthanatos” was dubbed (Yu et al., 2006).
PARP-1 and AIF-dependent cell death has gained significant interest in recent years, because of its potential implication as a therapeutic target in a variety of pathological conditions. The PARP-1 product PAR polymer, has been identified as a novel cell death signal, triggering AIF release from mitochondria and subsequent cell death (Yu et al., 2006). It seems likely that there is a bi-directional communication for coordination of cell death between the nucleus and mitochondrion. PAR polymer generated in the nucleus, directly interacts with mitochondria, especially with the small pool of AIF located on the cytosolic side of the outer mitochondrial membrane (Yu et al., 2009), thus causing AIF release. Once released, AIF translocates to the nucleus and initiates caspase-independent cell death leading to execution of parthanatos.
It is now well recognized that mitochondria play an important role in the regulation of survival and death of cells. Many cell death signals converge on mitochondria and cause mitochondrial dysfunction through the dissipation of mitochondrial membrane potential or opening of the mitochondrial permeability transition (MPT) pore. These events can lead to liberation of many cell death proteins from mitochondria (Tait and Green, 2010). Though one aspect of PAR toxicity on mitochondria, i.e. AIF release, has begun to be known, there still remain many intriguing questions regarding the effects of PAR on mitochondria dysfunction and its molecular mechanism of action. In particular, it is not known whether PAR polymer can affect mitochondrial physiology and if so, to what extent PAR can induce changes in mitochondrial parameters. Therefore, thorough investigation of the effects PAR exert on mitochondria is imperative to further our understanding of the molecular mechanisms of PAR polymer-induced parthanatos cell death pathway. In addition, its therapeutic potential for tissue protection has not been explored fully. In this study, we investigated the effects of PAR on mitochondria in cells in situ and isolated mitochondria in vitro. We found that PAR causes depolarization of mitochondria membrane potential and opening of the MPT pore. Furthermore, PAR specifically induces AIF release, but not cytochrome c from isolated mitochondria. These data reveal the cytotoxic effects of PAR and further contribute to our understanding of the PARP-1 and AIF-dependent cell death paradigm.
MATERIALS AND METHODS
Materials and reagents
The concentration of synthetic PAR polymer (Trevigen, USA) was expressed as a molar concentration of monomeric ADP-ribose units in polymers (Yu et al., 2006). Fluorometric analysis of mitochondrial membrane potential was achieved by 5,5′,6,6′- tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1, Cayman Chemical Company, USA) and tetramethylrhodamine ethyl ester perchlorate (TMRE, Sigma, USA). The Image-iT® LIVE Mitochondrial Transition Pore Assay Kit (Invitrogen, USA) was used to monitor the opening of the MPT pore in cells. All other reagents used for the isolation of mitochondria, measurement of mitochondrial respiration, and immunoblot analysis were purchased from Sigma.
Cell culture and delivery of PAR polymer to cells
HeLa cells (human cervical adenocarcinoma cells) were grown in Dulbecco’s modified Eagle’s medium (Invitrogen) media supplemented with 10% fetal bovine serum (FBS, Invitrogen) and antibiotics (50 units/ml of penicillin and 50 μg/ml of streptomycin, Invitrogen). Cells were maintained in a humidified incubator containing 5% (v/v) CO2 at 37°C up until cells were 90% confluent. Cells were seeded in culture plates (Corning Life Science, USA) or imaging chamber with a glass bottom (NuncLab-Tek® II Chamber Slide System, Thermo Scientific, USA) at a density of 3 × 105 cells/cm2. The next day, cells were subjected to treatment and then used for assays at the indicated time points. BioPORTER® (Genlantis, USA), a lipid-based delivery system of macromolecules, was used for delivery of PAR polymer into cells. In FBS-free media, a mixture of PAR polymer and BioPORTER reagent was added to cells for 3 h at 37°C. Following replacement of mixture with growing media, cells were used for assays.
Cell viability and cell death assay
Cell viability was measured by a colorimetric method using the water-soluble tetrazoliumcompound [3-(4,5-dimethylthiazol-2- yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt, MTS] following the manufacturer’s protocol (Thermo Scientific). The optical density was measured at 490 nm using a micro-plate reader. Cell viability was expressed as a percentage of the control group (vehicle only). Cell death was assessed fluorometrically using propidium iodide (PI) in the absence or presence of digitonin (Kim et al., 2003). Cells in a 96-black well plate were treated with 30 μM of PI. Immediately after incubation at 37°C for 20 min, fluorescence intensity (Fldead) corresponding to the dead cell population was measured using a fluorescence micro-plate reader. Following membrane permeabilization with 200 μM of digitonin and further incubation for 20 min at 37°C, fluorescence intensity (Fltotal) corresponding to the whole cell population was measured. The cell death in each well was expressed as a percentage value from the ratio between Fldead and Fltotal.
Isolation of mouse brain mitochondria
Mice (C57Bl/6J, male, 8–10 weeks-old) were maintained in the animal facility (12 h light/dark cycle, 22 ± 1°C), and provided with food and tap water ad libitum. All experiments using live animals were conducted following the protocol approved by the Daegu Gyeongbuk Institute of Science and Technology Institutional Animal Care & Use Committee. Nonsynaptic mitochondria from mouse brain were isolated following the Percoll gradient method as described previously (Sims and Anderson, 2008). Following decapitation, the mouse brain was quickly removed and placed in ice-cold mitochondria isolation buffer (MIB, 300 mM sucrose, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), and 1 mM ethylene glycol tetraacetic acid (EGTA), pH 7.4 at 4°C). The tissue was homogenized with a Dounce homogenizer, and unbroken tissue was removed by centrifugation (1,300× g, 3 min, 4°C). The supernatant was further centrifuged (21,000× g, 10 min, 4°C) to collect the crude mitochondrial fraction. The resulting pellet was resuspended in 15% (v/v) Percoll (GE Healthcare, USA) solution and loaded on the top of a 24% (v/v) and 40% (v/v) discontinuous Percoll layer. Following centrifugation (30,700× g, 10 min, 4°C), the non-synaptosomal mitochondria (NSM) band located in the interface between the 24% and 40% Percoll layer was recovered. Brain mitochondria were finally recovered by successive centrifugation in MIB without EGTA. The protein concentration was determined using the Bradford protein assay (Thermo Scientific).
Measurement of mitochondrial respiration
The respiratory activity of isolated mitochondria was measured using an oxygen monitor (Oxygraph, Hansatech Instruments, UK) equipped with a Clark-type oxygen electrode following the method of Brown et al. (2006). Protein (100 μg) isolated from brain mitochondria was suspended in a sealed, constantly stirred, and thermostatically controlled chamber at 37°C in 2 ml of KCl respiration buffer [125 mM KCl, 0.1% (w/v) bovine serum albumin, 20 mM HEPES, 2 mM MgCl2, and 2.5 mM KH2PO4, pH 7.2]. The real-time oxygen concentration in the reaction mixture was measured in the presence of oxidative substrates and reagents. The rate of oxygen consumption was calculated based on the rate of the response of isolated mitochondria to the successive administration of oxidative substrates (5 mM pyruvate and 2.5 mM malate): 150 μM ADP added twice in 1 min intervals; 1 μM oligomycin; 1 μM carbonyl cyanide 3-chlorophenylhydrazone (CCCP) and finally 1 mM succinate. The respiratory control ratio (RCR) was determined by dividing the rate of oxygen consumption/min for state III (in the presence of ADP, second addition) by state IV (in the absence of ADP and presence of oligomycin A).
Assessment of mitochondrial membrane potential in cells
Imaging or fluorometric measurement of mitochondrial membrane potential in cells was achieved using membrane potential- dependent fluorescence dyes including JC-1 and TMRE. Cells were stained with JC-1 (10 μg/ml in media) or TMRE (100 nM) for 15 min at 37°C, followed by washing with buffer provided in the kit or normal buffer (130 mM NaCl, 5.5 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, 25 mM glucose, 20 mM HEPES, pH 7.4), respectively. After washing with each buffer, stained cells were visualized by confocal microscopy (FluoView 1000 LSM, Olympus) or fluorescence microscope (TE2000-U Inverted Microscope, Nikon). Separately, the relative ratio of red/green fluorescence from JC-1 stained cells was measured using a fluorescence micro-plate reader.
Assessment of MPT pore opening in cells
Determination of mitochondrial membrane permeabilization in cells was assessed using the mitochondrial transition pore assay kit (Invitrogen) following the manufacturer’s protocol. Labeling solution containing 1.0 μM calcein-AM, 200 nM MitoTracker Red CMXRos, 1.0 μM Hoechst 33342 dye, and 1.0 mM CoCl2 in Hanks’ Balanced Salt Solution (with sodium bicarbonate, calcium, and magnesium) was supplemented with 10 mM HEPES and 2 mM L-glutamine.succinate (100 μM) was freshly prepared and added before use. At the end of incubation, cells in the imaging chamber were incubated with warm labeling solution for 15 min at 37°C. A separate group of cells were treated with ionomycin for 10 min before labeling and served as a positive control for Ca2+-mediated pore opening. Imaging of stained live cells was visualized by confocal microscopy (FluoView 1000 LSM, Olympus).
Assessment of mitochondrial membrane potential in isolated brain mitochondria
For fluorometric measurement of mitochondrial membrane potential with isolated brain mitochondria in vitro, brain mitochondria (4.0 mg/ml of mitochondria in MIB without EGTA) were prestained with JC-1 (10 μg/ml in MIB without EGTA) at 37°C for 15 min. After addition of two volumes of MIB without EGTA, the remaining dye was washed by centrifugation (10,000 × g, 3 min, 4°C). The resulting pellet was resuspended with assay buffer (125 mM KCl, 2 mM MgCl2, 20 mM HEPES, 2 mM KH2PO4, 5 mM pyruvate, and 2.5 mM malate, pH 7.4, 4.0 mg/ml of mitochondria) and kept on ice until ready for use. Prestained mitochondria (0.8 mg/ml mitochondria diluted with assay buffer) was mixed with 50 μl of PAR polymer solution in a 96-black well plate and red/green fluorescence of JC-1 from the reaction mixture was measured with a fluorescence microplate reader for 40 min (2 min interval, 37°C). For experiment with TMRE, brain mitochondria were incubated with PAR polymer for 10 min at 37°C (0.5 mg/ml mitochondria in 100 μl of reaction mixture in assay buffer). Reaction mixture was further incubated for 5 min in the presence of 100 nM of TMRE. At the end of incubation, residual TMRE in the supernatant was recovered by centrifugation (10,000× g, 3 min, 4°C) and transferred to a fresh 96-black well plate. Fluorescence intensity in the supernatant was measured using a fluorescence microplate reader.
AIF and cytochrome c release assay
The AIF and cytochrome c release assay was performed following the method of Polster et al. (2005). Release assay buffer (125 mM KCl, 2 mM K2HPO4, 20 mM HEPES, pH 7.0) supplemented with 5 mM succinate, 2 μM rotenone, 4 mM MgCl2, 3 mM ATP, and 1 mM ADP was freshly prepared before the assay. Brain mitochondria were incubated with PAR polymer for 30 min at 37°C in release assay buffer (1.0 mg/ml mitochondria in 100 μl of reaction mixture). Following recovery of the supernatant by centrifugation (10,000× g, 5 min, 4°C), AIF and cytochrome c released from brain mitochondria were resolved by polyacrylamide gel electrophoresis and analyzed by immunoblot analysis.
Immunoblot analysis
Resolved proteins in polyacrylamide gel (Bio-Rad, USA) were transferred onto polyvinylidene fluoride membranes (Millipore, USA). Immunoblot analysis was performed using primary antibodies against PSD-95 (postsynaptic density protein 95, Cell Signaling, USA), β-tubulin (Sigma), AIF (Epitomics, USA), VDAC-1 (voltage-dependent anion channel 1, Affinity BioReagents, USA), COX-IV (cytochrome c oxidase or Complex IV, Cell Signaling), and cytochrome c (BD Biosciences, USA). After incubation with secondary antibodies conjugated with horse radish peroxidase, blots were exposed to chemiluminescence reagent and imaged. Densitometric analysis was performed using ImageJ software (National Institute of Health, USA).
Statistical analysis
All data were generated from at least three independent experiments and values are the means ± the standard error of means (SEM). Significant differences to the control were determined by the t-test or one-way analysis of variance (ANOVA, Duncan’s multiple range tests) at the indicated p value.
RESULTS
PAR polymer depolarizes mitochondrial membrane potential in HeLa cells
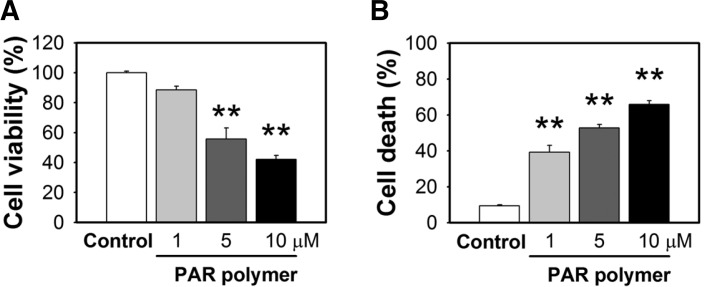
A substantial amount of PAR polymer is produced in response to a variety of toxic conditions in which PARP-1 plays an important role (Luo and Kraus, 2012). Previous studies revealed that PAR polymer can serve as an AIF-releasing factor and death signal (Yu et al., 2006). However, other effects of PAR on mitochondria are not well understood. For a detailed study of the effects of PAR on mitochondria, we delivered synthesized PAR polymer into HeLa cells and assessed mitochondrial functional parameters. This approach was used to exclude the consequences of PARP-1 activation on other cellular signaling events and focus on the direct cytotoxic outcome of PAR polymer on mitochondria. Because PAR polymer cannot cross the plasma membrane owing to its large size and negative charge, commercially available lipid-based formulation (BioPORTER) was used for PAR polymer delivery into cells. This reagent has been used for delivery of macromolecules including proteins, peptides, antibodies, and DNA through direct fusion with membrane or endocytosis and has been successfully utilized for PAR delivery in the previous study (Yu et al., 2006). BioPORTER-mediated transport of PAR polymer induced a large reduction in cell viability in a dose-dependent manner (Fig. 1A), which was proportional to an increase in cell death (Fig. 1B). PAR polymer-treated cells underwent morphological changes distinct from control cells, which included a decrease in cytosolic area, and an increase in vacuoles and cell debris-like particles that are supposed to represent dying cells (data not shown). These results are in accordance with the previous study that reported the nature of the PAR polymer as a novel death signal (Andrabi et al., 2006). We used half the dose of the BioPORTER reagent recommended by the manufacturer, which did not show cytotoxicity (data not shown).
Fig. 1.
PAR polymer induces cell death. (A) PAR polymer-induced loss of cell viability. HeLa cells were treated with PAR polymer for 3 h at 37°C. After replacement with fresh media, cells were further incubated for 21 h and cell viability was measured using the MTS assay. (B) PAR polymer-induced cell death. At the same set of treatment conditions, cell death was measured by the cell death assay using PI staining without/with digitonin. Bar represents % value to control (mean ± SEM). Significant difference to control was determined by one-way ANOVA (Duncan’s multiple range test, **P < 0.01).
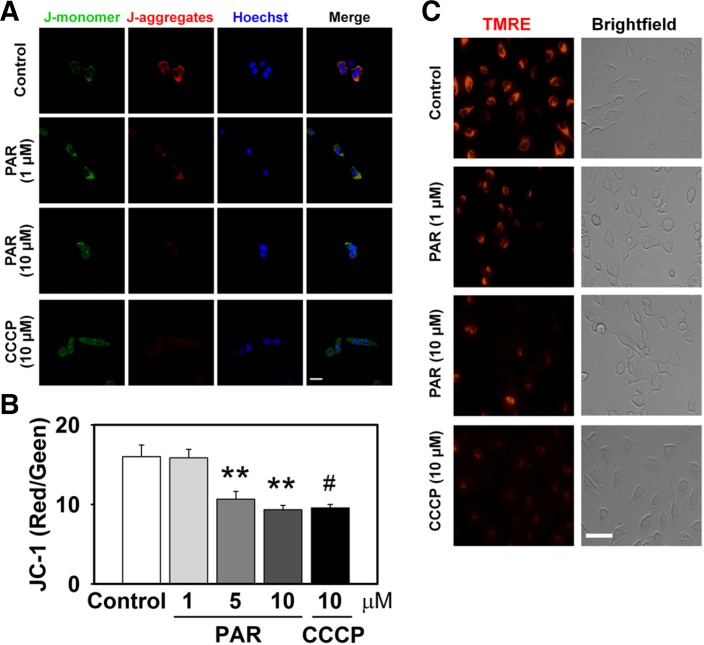
To further examine the effects of PAR on mitochondria function, we observed mitochondrial membrane potential in PAR polymer-delivered HeLa cells. Mitochondrial transmembrane potential was measured using potential dependent fluorescent probes. JC-1 is a dual-color probe and selectively accumulates in live mitochondria in a membrane potential-dependent manner (Green and Reed, 1998; Reers et al., 1991). Upon entering the cells, JC-1 accumulates inside healthy mitochondria maintaining high mitochondria membrane potential and forms complexes (J-aggregates) with intense red fluorescence. On the other hand, in dying or unhealthy cells with low mitochondrial membrane potential, JC-1 does not accumulate in mitochondria and remains in a monomeric form with green fluorescence. In vehicle-treated control cells, red fluorescence of J-aggregates was predominant over the green signal of J-monomers, suggesting functional mitochondria with a high membrane potential. However, 3 h treatment with PAR polymer reduced red fluorescence in a dose-dependent manner, suggesting PAR polymer-induced mitochondrial depolarization. Representative images are shown in Fig. 2A. Quantitative measurements of the dose-dependent effect of PAR polymer on depolarization were assessed using a fluorescence microplate reader and displayed a substantial decrease in the ratio of red to green fluorescence (Fig. 2B). CCCP, an uncoupler of oxidative phosphorylation, was used as a positive control for depolarization of mitochondrial potential and cells were treated for 30 min before staining. TMRE is a potential sensitive, cell-permeant fluorescent dye that is readily sequestered by active mitochondria (Ehrenberg et al., 1988; Farkas et al., 1989). Because it is rapidly and reversibly taken up by intact mitochondria in a potential-dependent manner, TMRE has been described as one of the best fluorescent dyes for dynamic and in situ quantitative measurements of mitochondrial potential by means of imaging or flow cytometric analysis (Umegaki et al., 2008). Similar to the results with JC-1 staining, cells treated with PAR polymer or CCCP lost TMRE fluorescence signal, indicating loss of mitochondrial membrane potential (Fig. 2C). Taken together, these data suggest that PAR polymer induces depolarization of mitochondrial transmembrane potential.
Fig. 2.
PAR polymer depolarizes mitochondrial membrane potential in cells. HeLa cells were treated with PAR polymer for 3 h at 37°C. After replacement with fresh media, cells were labeled for imaging. CCCP was applied 30 min before labeling. (A) Confocal microscope images of PAR polymer-treated HeLa cells after staining with JC-1. Cells were labeled with JC-1 and visualized by confocal microscopy after counter staining of nuclei with Hoechst 33342 (scale bar represents 20 μm). JC1 (mono), JC-1 in monomeric form; JC1 (aggre), JC-1 in aggregate form. (B) Fluorometric analysis of PAR polymer-treated HeLa cells after staining with JC-1. Fluorescent intensity of J-aggregates (red) and Jmonomers (green) in cells stained with JC-1 were measured using a fluorescence microplate reader. Bar represents ratio of red to green intensity (mean ± SEM). Significant difference to control was determined by one-way ANOVA (Duncan’s multiple range test, **P < 0.01) or t-test (#P < 0.05). (C) Fluorescence microscope images of PAR polymer-treated HeLa cells after staining with TMRE. Cells were labeled with TMRE and visualized by fluorescence microscopy (scale bar represents 100 μm).
PAR polymer induces opening of the MPT pore in HeLa cells
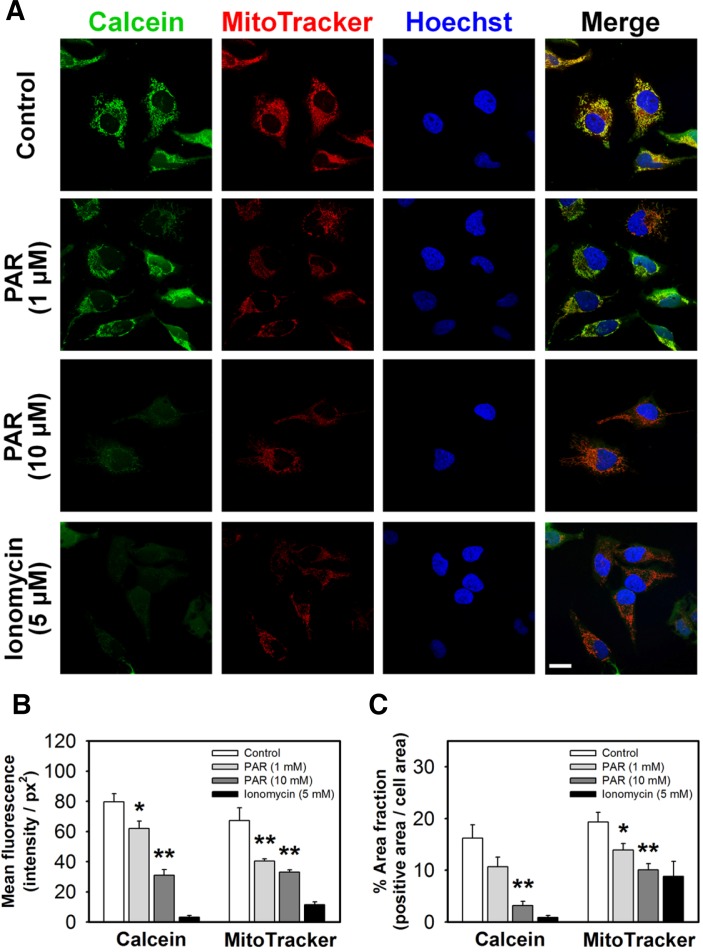
The MPT pore seems to be involved in regulation of mitochondrial function and release of pro-apoptotic molecules during cell death (Tait and Green, 2010). To study the effects of PAR polymer on the MPT pore in HeLa cells, we monitored opening of the MPT pore using a combination of fluorescent probes (calcein dye) and a quencher (Co2+) (Petronilli et al., 1998). The acetoxymethyl ester modification of calcein (calcein AM) facilitates diffusion of the dye across the plasma membrane and accumulation in the cytosolic compartment including mitochondria. After cleavage of acetoxymethy esters by intracellular esterases inside cells, the very polar fluorescent dye calcein is retained within mitochondria. Whereas the fluorescence from cytosolic calcein can be quenched by the addition of CoCl2, the mitochondria remain brightly fluorescent until mitochondrial pore opening allows entry of Co2+ and permits quenching of the fluorescence. Location of mitochondria was confirmed by co-staining with a potential sensitive probe (MitoTracker Red CMXRos). While control cells exhibited intense green signal from calcein overlapping with mitochondrial staining by MitoTracker Red, PAR polymer induced a significant decrease in green fluorescence intensity (Fig. 3A). A separate group of cells was treated with ionomycin, an ionophore that causes influx of excessive amount of Ca2+, as a positive control for MPT pore opening. Incubation of HeLa cells with ionomycin for 10 min resulted in loss of green signal, indicating opening of the MPT pore (Fig. 3A). Because MitoTracker Red is also dependent on mitochondrial membrane potential, a decrease in red fluorescent signal accompanied the loss of green fluorescence from calcein (Fig. 3A).
Fig. 3.
PAR polymer induces mitochondrial membrane permeabilization in cells. (A) Confocal microscope images of PAR polymertreated HeLa cells after staining with calcein and MitoTracker red. HeLa cells were treated with PAR polymer for 3 h at 37°C. Ionomycin was applied 10 min before labeling. At the end of the incubation, cells were stained with labeling solution containing calcein, Co2+, and MitoTracker Red for 15 min. After washing, labeled cells were visualized by confocal microscopy after counterstaining of nuclei with Hoechst 33342 (scale bar represents 20 μm). (B) Mean fluorescence and (C) area fraction of calcein (green) or MitoTracker Red (red) in cells treated with PAR polymer. Cell area (pixel2), fluorescence intensity (arbitrary unit), and green or red positive area (pixel2) were calculated within the defined area enclosing the boundary of a cell. Mean fluorescence (sum intensity of green or red/cell area) and area fraction (green or red positive area/cell area) in a cell were calculated. At least 10 cells in a group were randomly chosen for analysis. Bar represents mean fluorescence and % area fraction in a cell (mean ± SEM). Significant difference to control was determined by one-way ANOVA (Duncan’s multiple range test, *P < 0.05 and **P < 0.01).
To evaluate the quantitative effect of PAR polymer on depolarization and MPT pore opening, each cell from confocal images was analyzed and the mean fluorescence intensity of either calcein or MitoTracker signal was quantified. Cell area (pixel2), fluorescence intensity (arbitrary unit), and green- or red-positive area (pixel2) were calculated within the defined area enclosing the boundary of a cell. Mean fluorescence intensity of green or red in a cell (sum intensity of green or red/cell area) was supposed to be a marker for mean mitochondrial function in a cell, with green representing the intact MPT pore and red representing polarized mitochondria. The area fraction in a cell (green or red positive area/cell) represented the population of functional mitochondria in a cell. Because of its threadlike shape in live cells, area-based analysis was more practical approach than counting mitochondria. More than 10 cells in a group were randomly chosen and subjected to quantitative analysis. As expected, PAR polymer decreased cellular mean fluorescent intensity as well as the positive area fraction of calcein or MitoTracker in a dose-dependent manner, suggesting that PAR polymer impaired mitochondrial potential and reduced the functional population of mitochondria having intact membrane permeability as well (Figs. 3B and 3C). These data reveal the nature of PAR polymer as a possible endogenous mitochondrial toxin in HeLa cells.
Functionality and purity of isolated brain mitochondria
To demonstrate the direct interaction between PAR polymer and mitochondria without interference of cytosolic components, we isolated mitochondria from mouse brain and evaluated the effect of PAR polymer on dissipation of mitochondrial membrane potential. A Percoll density gradient was used for preparation of mitochondria. Following Percoll discontinuous density gradient centrifugation, nonsynaptic brain mitochondria was enriched at the interface of 24% (v/v) and 40% (v/v) Percoll solution. Synaptosomes, which are the pinched off structure of brain cells during mechanical homogenization and contain synaptosomal mitochondria, were found at the interface of 15% (v/v) and 24% (v/v) Percoll solution. Because nonsynaptic mitochondria are the main population of mitochondria compared with the small population of synaptosomal mitochondria in the brain, we collected only nonsynaptic mitochondria from the third layer and used them for in vitro experiments.
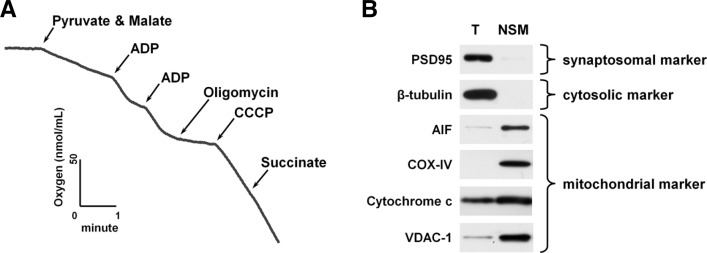
Respiration rate of isolated mitochondria was measured using a Clark-type oxygen electrode, because bioenergetics through oxidative phosphorylation is fundamental to the function of mitochondria (Fig. 4A) (Brownet al., 2006). The rate of oxygen consumption increased drastically after addition of ADP in the presence of oxidative substrates (pyruvate and malate), reflecting increased oxygen demand for the conversion of ADP to ATP by ATP synthase in mitochondria (State III). Following inhibition of ATP synthase by oligomycinA, the rate of oxygen consumption was minimized (State IV). RCR is defined as the ratio of the oxygen consumption rate for state III over state IV and has been widely used as an indicator of functional and structural integrity of isolated mitochondria (Brown et al., 2006). In general, mitochondria preparations with a RCR value higher than 5 are considered suitable for in vitro analysis. Our mitochondrial preparations typically yielded RCR values of 9.7 ± 1.2, which are far above the standard, demonstrating functional integrity of isolated mitochondria. Purity of isolated mitochondria was tested by immunoblot analysis using antibodies against synaptosomal (PSD95), cytosolic (β-tubulin), and mitochondrial markers (AIF, COX-IV, cytochrome c, and VDAC-1) (Fig. 4B). PSD95 and β-tubulin were only found in total brain homogenates, but not in isolated mitochondria, suggesting the absence of cytosolic or synaptosomal contamination in our mitochondrial preparations. Mitochondrial markers were enriched in isolated mitochondria when compared with total brain homogenates. Taken together, our isolated mitochondria were intact and free from contamination, and were therefore appropriate for further in vitro experiments.
Fig. 4.
Respiration rate measurement and purity analysis of isolated mouse brain mitochondria. (A) Representative traces of oxygen consumption with isolated mouse brain mitochondria. Non-synaptosomal mitochondria (NSM) were isolated from the whole brain of mouse and used for experiments. Mitochondrial oxygen consumption rate was measured using a Clark-type oxygen electrode in the presence of oxidative substrates (pyruvate and malate), ADP, oligomycin, CCCP, and succinate. The respiratory control ratio was determined by dividing the rate of oxygen consumption/min for state III (in the presence of ADP, second addition) by state IV (in the absence of ADP and presence of oligomycin). (B) Immunoblot analysis of isolated mouse brain mitochondria. Protein (total homogenate and isolated brain mitochondria; 20 μg) was subjected to immunoblot analysis using the indicated antibodies. T, total homogenate; NSM, non-synaptosomal mitochondria.
PAR polymer depolarizes mitochondrial membrane potential in isolated mitochondria
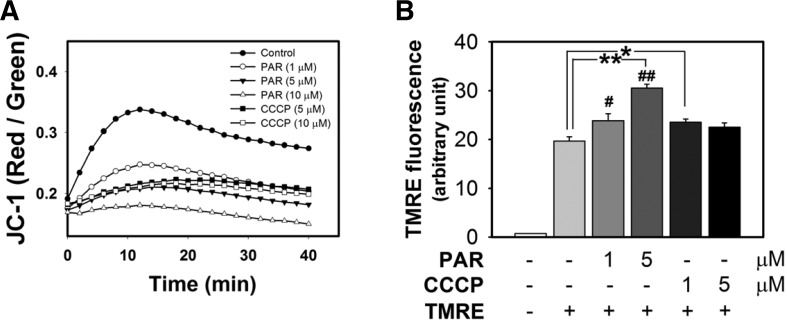
The methods were designed to assess the effects of PAR polymer on isolated mitochondria. One was monitoring the change of the dual-color probe (JC-1) which was deposited in mitochondria prior to PAR polymer treatment. The other method involved measuring the residual amount of TMRE in the supernatant of the mitochondria reaction mixture, which reflected failure of accumulation. In this experimental setting, TMRE was added post PAR polymer treatment. First, isolated mitochondria were prestained with JC-1 and red and green fluorescence was monitored upon addition of PAR polymer or CCCP using a microplate reader in a thermostatic chamber set at 37°C. The ratio of red to green fluorescence, a real-time indicator of in situ mitochondrial membrane potential, was plotted for 40 min (Fig. 5A). Compared with the high ratio of red to green signal in the control, PAR polymer significantly depolarized membrane potential of isolated mitochondria in a dose-dependent manner. Interestingly, PAR polymer was a highly potent depolarizer when compared with CCCP. In the second set of experiments, isolated mitochondria were incubated with PAR polymer or CCCP for 10 min at 37°C, followed by TMRE staining for an additional 5 min. Residual TMRE in the supernatant was recovered by centrifugation and measured by fluorometric analysis (Fig. 5B). PAR polymer increased TMRE levels in the supernatant dose-dependently, suggesting inhibition of TMRE accumulation in mitochondria. Taken together, these results demonstrate the direct effects of PAR polymer on depolarization of mitochondria.
Fig. 5.
PAR polymer depolarizes mitochondrial membrane potential in isolated mouse brain mitochondria. (A) Measurement of PAR polymer-induced depolarization of mitochondria with JC-1. Isolated mouse brain mitochondria were preincubated with JC-1. After addition of PAR polymer or CCCP to mitochondria containing JC-1, real-time fluorescence intensity of JC-1 aggregates (red) and monomer (green) in the reaction mixture was measured using a fluorescence microplate reader. Dot and trace represented the mean of red- to green-fluorescence ratio. (B) Measurement of PAR polymer-induced depolarization of mitochondria with TMRE. Isolated mouse brain mitochondria were treated with PAR or CCCP for 10 min at 37°C prior to incubation with TMRE for an additional 10 min. After centrifugation, TMRE fluorescence in the supernatant of the reaction mixture was measured using a fluorescence microplate reader. Bar represents the TMRE fluorescent level in the supernatant (mean ± SEM). Significant difference to control was determined by one-way ANOVA (Duncan’s multiple range test, *P < 0.05 and **P < 0.01) or t-test (#P < 0.05).
PAR polymer induces AIF release, but not cytochrome c
To assess key effectors triggering cell death following PAR polymer-induced mitochondrial membrane permeabilization, we tested the effect of PAR polymer on the release of apoptogenic proteins from isolated mitochondria. The apoptogenic protein release assay was performed in KCl-based assay medium including ATP, ADP, succinate (oxidative substrate of complex II), and rotenone (complex I inhibitor). Succinate was used as a complex II substrate in the presence of rotenone to maintain pyridine nucleotides in a reduced state (Polster et al., 2005). AIF and cytochrome c were chosen as target apoptogenic proteins responsible for the executioner of caspase-independent and -dependent cell death, respectively. The isolated mitochondria without or with PAR in the assay medium were incubated at 37°C for 30 min. Supernatant was recovered from the reaction mixture by centrifugation. The amount of released AIF and cytochrome c in the supernatant was determined by immunoblot analysis. We compared the response of isolated mitochondria to Ca2+ overload as a positive control that facilitates AIF and cytochrome c release through mitochondrial membrane swelling and permeabilization (Fig. 6A). While only basal levels of AIF and cytochrome c were found to be released in control mitochondria, Ca2+ (3 mM) induced significant release of both AIF (4-fold) and cytochrome c (12-fold) from mitochondria, suggesting that our mitochondrial preparation exhibited typical response to Ca2+ and preserved the capability to release apoptogenic proteins (Azarashvili et al., 2007; Polster et al., 2005) (Fig. 6B). PAR polymer induced significant release of AIF (4-fold) from mitochondria, but not cytochrome c (Fig. 6B). Cytochrome c is a predominant apoptogenic protein released by Ca2+ that controls the cell death pathway in a caspase- dependent manner. However, PAR polymer only acted on AIF release, an executioner of caspase-independent cell death through parthanatos. Overall, our findings indicate that PAR polymer induces lethal dysfunction of mitochondria in cells and isolated mitochondria including depolarization and MTP pore opening, thereby triggering AIF release and cell death. PAR polymer, a product of PARP-1 activation, seems likely to be an endogenous mitochondrial toxin and potent AIF-releasing factor that leads to caspase-independent cell death.
Fig. 6.

PAR polymer induces AIF release from isolated mouse brain mitochondria. (A) Representative immunoblot images of AIF and cytochrome c release. Isolated mouse brain mitochondria were incubated with Ca2+ (3 mM) o r PAR polymer (5 μM) for 30 min at 37°C in the KCl-based assay buffer containing ATP (3 mM), ADP (0.8 mM), succinate (5 mM) and rotenone (2 μM). Released levels of AIF or cytochrome c in the supernatant of the reaction mixture were analyzed by immunoblotting. Mitochondrial pellets were probed with anti-VDAC-1 antibody to confirm that equal amounts of mitochondria were used in the experiments. (B) Quantitative analysis of immunoblot images by densitometric analysis. Immunoblot results were analyzed by ImageJ software. Bar represents fold-increase of released AIF or cytochrome c to control (n = 3, mean ± SEM). Significant difference to control was determined by paired t-test (**P < 0.01).
DISCUSSION
The major finding of our study is that PAR polymer induces mitochondrial dysfunction, i.e., depolarization of mitochondrial membrane potential and opening of the MPT pore, both in cells and isolated mitochondria. Furthermore, PAR polymer induced only AIF release, but not cytochrome c from mitochondria, in concordance with the caspase-independent nature of the parthanatos cell death program. Our report provides further links between PAR polymer and mitochondria and unveils the detailed effects that PAR exerts on the mitochondria. Current data nicely corroborates the previous works and substantially advance our understanding of the mechanistic action of PAR polymer in parthanatos.
Mitochondrial outer membrane permeabilization (MOMP) is an irreversible, ‘point-of-no-return’ event that gears up cell death machinery (Kroemer et al., 2009; Tait and Green, 2010). MOMP facilitates the release of apoptogenic proteins including AIF and cytochrome c. The MPT pore, a dynamic regulator of MOMP, is a putative protein complex constituted by VDAC, the adenine nucleotide translocase, cyclophilin D, hexokinase, and peripheral benzodiazepine receptor (Fulda et al., 2010; Tait and Green, 2010). Because PAR polymer induced only AIF release in isolated mitochondria, it is likely that PAR polymer regulates MOMP by specific interaction with the PAR polymer-binding proteins in a manner different from Ca2+-induced MOMP. However, it is still unclear how this highly negatively charged polymer can regulate mitochondrial membrane permeability and induce selective release of AIF. One candidate mechanism is through interaction with AIF itself. Small population of AIF are located on the cytosolic side of the mitochondrial outer membrane (Yu et al., 2009) and AIF has a putative PAR binding sequence and binding of PAR to AIF seems critical for PARP- 1-dependent cell death (Wang et al., 2011). Recently, Iduna was reported to protect the brain from cell death by interfering with PAR polymer-induced cell death (Andrabi et al., 2011). Therefore, future studies to identify mitochondrial targets of PAR polymer and their application to therapeutic design for tissue protection could be a promising approach.
PARP-1-dependent, AIF-mediated cell death is preceded by early generation of PAR polymer. Pathological conditions causing DNA damage, such as glutamate excitotoxicity, oxidative stress or inflammation elicit robust activation of PARP-1 and generation of PAR polymer. On the other hand, pharmacological inhibition or genetic ablation of PARP-1 activity yields significant neuroprotection against a variety of neurological insults. The intracellular level of PAR polymer in dying cells can be regulated by pharmacological or genetic manipulation of PARP- 1 activity. However, PAR polymer anabolism and catabolism is a dynamic process and can be affected by various cellular signaling as well as PARP-1 activity. Therefore, it would be desirable to come up with experimental settings where PAR polymer can be directly administered to cells without affecting other signaling pathways. To that end, we used synthetic PAR polymer throughout this study with a specific interest on the effects of PAR polymer on mitochondria. A lipid-based BioPORTER system was used for intracellular delivery of PAR polymer, as reported previously (Andrabi et al., 2006; Yu et al., 2006). We also adopted a cell-free in vitro system to assess the outcomes of direct, physical interaction of PAR polymer with isolated brain mitochondria. Functional and structural integrity of mitochondrial preparations were thoroughly characterized and ascertained above the standard (Fig. 4). In some cases, fluorescent probes intended to monitor mitochondrial functional parameters exhibit nonspecific action depending on doses and durations, or stain non-target cellular organelles other than mitochondria. These artifacts may lead to false interpretation (Ehrenberg et al., 1988; Farkas et al., 1989). To avoid these issues, we used various fluorescent probes that are supposed to have different actions including JC-1, TMRE, MitoTracker Red and calcein in combination with appropriate controls (ionomycin or CCCP). Our data consistently suggest that PAR polymer causes mitochondrial dysfunction, as evidenced by depolarization of mitochondrial membrane potential, opening of the MPT pore and AIF release.
In summary, this study aimed to evaluate the effects of PAR polymer on the mitochondrial cell death pathway with diverse biochemical in vitro approaches from cells to isolated mitochondria and assorted relevant assays. Identification and characterization of PAR polymer-binding proteins and understanding PAR polymer signaling mechanisms may provide a novel opportunity for brain protection against various neurological diseases.
Acknowledgments
This work was supported by the DGIST Convergence Science Center Program (13-BD-04) and MIREBrain program, and by the Korea Research Foundation Grant (KRF-2007-357-E00003).
REFERENCES
- Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi SA, Kang HC, Haince JF, Lee YI, Zhang J, Chi Z, West AB, Koehler RC, Poirier GG, Dawson TM, et al. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat Med. 2011;17:692–699. doi: 10.1038/nm.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarashvili T, Grachev D, Krestinina O, Evtodienko Y, Yurkov I, Papadopoulos V, Reiser G. The peripheral-type benzodiazepine receptor is involved in control of Ca2+-induced permeability transition pore opening in rat brain mitochondria. Cell Calcium. 2007;42:27–39. doi: 10.1016/j.ceca.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Brown MR, Sullivan PG, Geddes JW. Synaptic mitochondria are more susceptible to Ca2+ overload than nonsynaptic mitochondria. J Biol Chem. 2006;281:11658–11668. doi: 10.1074/jbc.M510303200. [DOI] [PubMed] [Google Scholar]
- Diefenbach J, Burkle A. Introduction to poly(ADP-ribose) metabolism. Cell Mol Life Sci. 2005;62:721–730. doi: 10.1007/s00018-004-4503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg B, Montana V, Wei MD, Wuskell JP, Loew LM. Membrane potential can be determined in individual cells from the nernstian distribution of cationic dyes. Biophys J. 1988;53:785–794. doi: 10.1016/S0006-3495(88)83158-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas DL, Wei MD, Febbroriello P, Carson JH, Loew LM. Simultaneous imaging of cell and mitochondrial membrane potentials. Biophys J. 1989;56:1053–1069. doi: 10.1016/S0006-3495(89)82754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov. 2010;9:447–464. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Herceg Z, Wang ZQ. Failure of poly(ADP-ribose) polymerase cleavage by caspases leads to induction of necrosis and enhanced apoptosis. Mol Cell Biol. 1999;19:5124–5133. doi: 10.1128/mcb.19.7.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Qian T, Lemasters JJ. Mitochondrial permeability transition in the switch from necrotic to apoptotic cell death in ischemic rat hepatocytes. Gastroenterology. 2003;124:494–503. doi: 10.1053/gast.2003.50059. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronilli V, Miotto G, Canton M, Colonna R, Bernardi P, Di Lisa F. Imaging the mitochondrial permeability transition pore in intact cells. BioFactors. 1998;8:263–272. doi: 10.1002/biof.5520080314. [DOI] [PubMed] [Google Scholar]
- Polster BM, Basanez G, Etxebarria A, Hardwick JM, Nicholls DG. Calpain I induces cleavage and release of apoptosis-inducing factor from isolated mitochondria. J Biol Chem. 2005;280:6447–6454. doi: 10.1074/jbc.M413269200. [DOI] [PubMed] [Google Scholar]
- Reers M, Smith TW, Chen LB. J-aggregate formation of a carbocyanine as a quantitative fluorescent indicator of membrane potential. Biochemistry. 1991;30:4480–4486. doi: 10.1021/bi00232a015. [DOI] [PubMed] [Google Scholar]
- Sims NR, Anderson MF. Isolation of mitochondria from rat brain using Percoll density gradient centrifugation. Nat Protoc. 2008;3:1228–1239. doi: 10.1038/nprot.2008.105. [DOI] [PubMed] [Google Scholar]
- Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- Umegaki T, Okimura Y, Fujita H, Yano H, Akiyama J, Inoue M, Utsumi K, Sasaki J. Flow cytometric analysis of ca-induced membrane permeability transition of isolated rat liver mitochondria. J Clin Biochem Nutr. 2008;42:35–44. doi: 10.3164/jcbn.2008006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag L, Szabo C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- Wang H, Yu SW, Koh DW, Lew J, Coombs C, Bowers W, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor substitutes for caspase executioners in NMDA-triggered excitotoxic neuronal death. J Neurosci. 2004;24:10963–10973. doi: 10.1523/JNEUROSCI.3461-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kim NS, Haince JF, Kang HC, David KK, Andrabi SA, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci Signal. 2011;4:ra201–13. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- Yu SW, Wang H, Dawson TM, Dawson VL. Poly(ADP-ribose) polymerase-1 and apoptosis inducing factor in neurotoxicity. Neurobiol Dis. 2003;14:303–317. doi: 10.1016/j.nbd.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc. Natl. Acad. Sci. USA. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SW, Wang Y, Frydenlund DS, Ottersen OP, Dawson VL, Dawson TM. Outer mitochondrial membrane localization of apoptosis-inducing factor: mechanistic implications for release. ASN Neuro. 2009;1:275–281. doi: 10.1042/AN20090046. [DOI] [PMC free article] [PubMed] [Google Scholar]