Abstract
A total of 140,000 compounds were screened in a targetfree cell-based high throughput assay against HIV-1 infection, and a subset of 81 promising compounds was identified. Secondary screening of these 81 compounds revealed two putative human RNaseH2 inhibitors, RHI001 and RHI002, with IC50 value of 6.8 μM and 16 μM, respectively. RHI002 showed selective activity against human RNaseH2 while RHI001 inhibited HIV-RNaseH, E. coli RNaseH, and human RNaseH1 with IC50 value of 28.5 μM, 7.9 μM, and 31.7 μM, respectively. Kinetic analysis revealed that both inhibitors had non-competitive inhibitor-like properties. Because RNaseH2 is involved in the etiology of Aicardi-Goutier syndrome and has been suggested as an anticancer drug target, small molecule inhibitors modulating its activity would be useful for investigating the cellular function of this molecule.
Keywords: Aicardi-Goutier syndrome, cell-based screening, HIV inhibition, human RNaseH2
INTRODUCTION
Human RNaseH2 (hRNaseH2) is an enzyme complex consisting of three subunits, all of which are required for enzyme function. The catalytic subunit of eukaryotic RNaseH2, RNASEH2A, is well conserved and similar to the monomeric prokaryotic RNaseHII. In contrast, the RNASEH2B and RNASEH2C subunits from human and Saccharomyces cerevisiae share very little homology. Mutations in the subunits encoding hRNaseH2 cause Aicardi-Goutier syndrome (AGS), an autosomal recessive genetic disorder (Crow et al., 2006). AGS phenotypically mimics congenital viral infection, which elevates interferon alpha levels in cerebrospinal fluid (Aicardi and Goutieres, 1984; Goutieres, 2006; Goutieres et al., 1998). In addition, hRNaseH2 has been suggested as an anticancer drug target (Flanagan et al., 2009). hRNaseH2 is required for maintaining genome stability by removing ribonucleotides misincorporated by replicative polymerases (Hiller et al., 2012; Reijns et al., 2012). Furthermore, hRNaseH2 is essential for HIV replication (Genovesio et al., 2011). Fifty-six host genes including hRNaseH2 that affect HIV replication were previously identified using a genome-wide siRNA screen. In addition, depletion of human RNaseH2 (hRNaseH2) impairs HIV infection in Jurkat cells when siRNAs were transiently transfected. Therefore, small molecule inhibitors that modulate RNaseH2 activity may be useful tools for investigating the cellular function of this molecule.
We hypothesized that some anti-HIV compounds might also have inhibitory activity against hRNaseH2 and thus, against HIV, when the screening is performed in a target-free cell based assay which include the whole life cycle of HIV replication. Initially, we screened 140,000 compounds in our target-free cell-based screen for anti-HIV activity and identified 81 validated hit compounds. We then screened these 81 compounds using an enzymatic assay for RNaseH2 and identified two putative hRNaseH2 inhibitors, RHI001 and RHI002. In a selectivity test, RHI002 showed very good specificity, uniquely inhibiting hRNaseH2, while RHI001 inhibited all tested RNaseH species. Both compounds showed a non-competitive inhibitor-like pattern in a mode of inhibition test.
MATERIALS AND METHODS
Compound libraries
The compound library contained 140,000 synthetic compounds, which were purchased from ChemDiv (20,000) and Euroscreen (120,000).
Plasmids
Plasmid pET-hH2ABC, which bears three hRNaseH2 subunits (RNASEH2A, RNASEH2B, and RNASEH2C) with independent N-terminal His-tags, was provided by R. J. Crouch (Eunice Kennedy Shriver NICHD, USA) (Chon et al., 2009). The hRNaseH1 gene was amplified by reverse transcriptase-polymerase chain reaction (RT-PCR) using total RNA from HeLa cells as template. Two primers (5′-GGG CAT ATG TTC TAT GCC GTG AGG AGG GGC-3′ and 5′-GGG GGA TCC TCA GTC TTC CGA TTG TTT AGC-3′) were used for amplification. The DNA fragment was inserted into the NdeI/BamHI site of pET-28a (Novagene). The resulting plasmid, pET-hRNaseH1, with N-terminally His-tagged human RNaseH1, was generated. The amino acid sequence was identical to the reported human RNaseH1 gene (GenBank accession No. NM002936).
HIV RNaseH consists of two subunits (p66 and p51); each subunit was cloned separately then combined in a plasmid. The genes were amplified by PCR using p96ZM651pol-opt (obtained through the NIH AIDS research and reference program, Division of AIDS, NIAID, NIH, G) as a template (Gao et al., 2003). For p66, two primers (5′-ATA TCC ATG GGA CCC ATC AGC CCC ATC GAG ACC-3′ and 5′-ATA TAA GCT TCA GCA CCT TGC GGA TGC CCT TGC-3′) were used and the PCR product was inserted into the NcoI/HindIII site of pET-28a. For p51, two primers (5′-ATA TCA TAT GCC CAT CAG CCC CAT CGA GAC C-3′ and 5′-ATA TCT CGA GCT AGA AGG TCT CGG CGC CCA CGA TGG-3′) were used, and the PCR product was inserted into the NdeI/XhoI site of pET-15b (Novagene). An EcoRV digested fragment of the p51 construct was then inserted into the EcoRV site of the p66 construct. The resulting plasmid, pET-HIVRNaseH, bearing two open reading frames (ORFs) with independent T7 promoters and N-terminal His-tags, was generated.
Expression and purification of proteins from E. coli
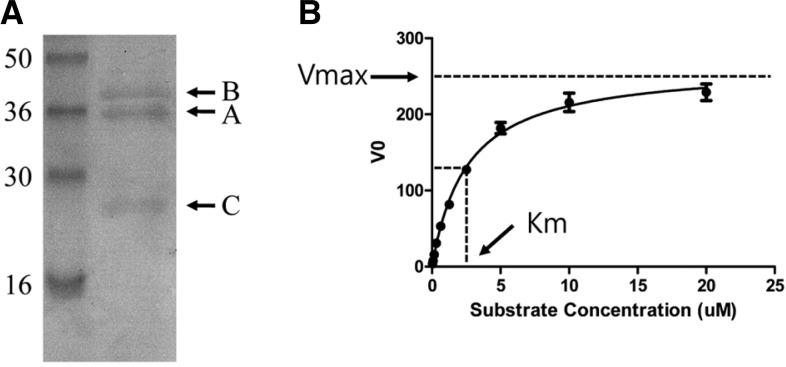
To express hRNaseH2, human RNaseH1, and HIV RNaseH, the plasmids pET-hH2ABC, pET-RNaseH1, and pET-HIVRNaseH were transformed into E. coli strains BL21 DE3 CodonPlus RIL (Stratagene), Rosetta (DE3) (Novagen), and BL21 (DE3) LysS (Promega), respectively. The induction conditions (IPTG concentration/duration/temperature) for each protein were as follows: 500 μM/5 h/20°C for hRNaseH2, 500 μM/15 h/25°C for hRNaseH1, and 100 μM/15 h/25°C for HIV RNaseH. The soluble fraction of E. coli lysate was subjected to histidine affinity chromatography (AKTA explorer, GE Healthcare) and the purified protein was analyzed by SDS-PAGE (see Fig. 2A for hRNaseH2; data not shown for hRNaseH1 and HIV RNaseH). Commercially available E. coli RNaseH (Takara) was used for the specificity study.
Fig. 2.
Overexpression and kinetic analysis of the hRNaseH2 enzyme. (A) Heterotrimeric hRNaseH2 was subjected to 12% SDS-PAGE after purification by histidine affinity chromatography. The deduced molecular weights from the amino acid sequences of subunits A, B, and C were 35.56, 37.31, and 20.01 kDa, respectively. Numbers represent the size of standard proteins in kDa. (B) Michaelis-Menten kinetic analysis of hRNaseH2. Vmax and Km were estimated to be 264.9 RFU/min and 2.556 μM, respectively.
Establishment of the RNaseH activity assay
An RNaseH activity assay was established applying the fluorescence resonance energy transfer (FRET) technique. The RNA/DNA heteroduplex substrate was generated by synthesizing modified oligonucleotides (5′-rGrArU rCrUrG rArGrC rCrUrG rGrGrA rGrCrU-FAM-3′ and 5′-DABCYL-dAdGdC dTdCdC dCdAdG dGdCdT dCdAdG dAdTdC-3′, TriLink Biotechnologies) and hybridizing them by boiling at 95°C for 10 min and then cooling to room temperature. Buffer conditions for each enzyme reaction are summarized in Table 1. Enzyme was pre-incubated with compound for 10 min at room temperature and the reaction was initiated by adding substrate. Reactions were performed in 384-well plates (Greiner Bio-one, Cat# 781091) in a total volume of 50 μl. After 30 min of reaction time, fluorescence was measured from the bottom of the plates using a SpectraMax M5 (Molecular Devices) at 490 nm/528 nm (excitation/emission) wavelength. The final DMSO concentration was adjusted to 1% throughout this study.
Table 1.
Assay conditions for RNaseHs
| Enzyme | Vmax (RFU/min) | Km (μM) | Reaction time (min) | Temp (°C) | Buffer | |
|---|---|---|---|---|---|---|
| Human RNaseH2 | 25 ng | 265 | 2.56 | 30 | 37 | 50 mM Tric-C, pH 8.5, 50 mM NaCl, 5 mM MgCl2, 1 mM 2-mercaptoethanol, 50 μg/ml BSA, 1% glycerol |
| Human RNaseH1 | 30 ng | 136 | 0.32 | 30 | 30 | 50 mM Tris-Cl, pH 8.0, 60 mM KCI, 5 mM MgCl2 |
| HIV RNaseH | 250 ng | 629 | 0.48 | 30 | 37 | 50 mM Tris-Cl, pH 8.0 34 mM KCI, 6 mM MgCl2, 1 mM DTT, 1 mM EDTA |
| E. coli RNaseH | 0.25 unit | 2,646 | 1.24 | 30 | 37 | 40 mM Tris-Cl, pH 8.0, 4 mM MgCl2, 1 mM DTT, 4% glycerol, 30 μg/ml BSA |
A reaction progress curve was obtained to determine the initial velocity region of the enzymatic reaction and subsequent experiments were conducted in this linear range. Substrate concentration was varied to generate a saturation curve for the determination of Vmax (Fig. 2B). According to the Michaelis-Menten kinetic model, the substrate concentration at Vmax/2 is identified as the Km (Table 1).
To test substrate specificity, RNA/DNA or DNA/DNA duplexes with the same nucleotide sequence were used. Modified oligonucleotides, oligo1 (5′-FAM-rArCrG rUrArG rGrUrU rCrUrG rArGrG rGrUrG rGrCrG rGrUrA rCrUrA rArCrG rUrC-3′), oligo2 (5′-FAM-dAdCdG dTdAdG dGdTdT dCdTdG dAdGdG dGdTdG dGdCdG dGdTdA dCdTdA dAdCdG dTdC-3′), and oligo3 (5′-dTdAdG dTdAdC dCdGdC dCdAdC dCdCdT dCdAdG dAdAdC dCdTdA dCdGdT-DABCYL-3′) were synthesized. RNA/DNA and DNA/DNA duplexes were generated by hybridizing oligo1-oligo3 and oligo2-oligo3, respectively.
Human RNaseH2 inhibitor screening
The Z′ value of the hRNaseH2 activity assay was measured by comparing the difference in average fluorescence intensity by FRET of 16 repeated samples between 100% inhibition (reaction mixture without enzyme) and 0% inhibition (reaction mixture without any inhibitor). The equation for the Z′ value was 1 - (3× SD of 100% inhibition + 3× SD of 0% inhibition)/(mean of 0% inhibition-mean of 100% inhibition). Duplicates of each compound were analyzed at the final concentration of 50 μM.
Gel based hRNaseH2 assay
hRNaseH2 (75 ng) was pre-incubated with or without test compound (100 μM final) at room temperature for 10 min in a volume of 50 μl; the reaction was started by adding 1.6 μg RNA/DNA duplex substrate (using the same nucleotide sequence as in t he FRET b ased hRNaseH2 a c tivity a ssay without FAM/DABCYL modification). The reaction mixture was incubated at 37°C for 90 min and separated on a 20% native polyacrylamide gel in 1× TBE. After electrophoresis, the gel was stained using a SilverQuest silver staining kit (Invitrogen). Control 1 and control 2 were performed without or with excess (2 μg) enzyme, respectively, and run with the other samples.
Measurement of anti-HIV activity of the hRNaseH2 inhibitors
CG8 cells (Sommer et al., 2004), which bear a construct expressing green fluorescent protein (GFP) under the control of the HIV LTR, were seeded at a concentration of 2 × 104 cells/well in 96-well plates. Compounds, serially diluted in DMSO, were added and cells were infected with HIV-1 IIIB at a multiplicity of infection (MOI) of 0.1. After 3 days, GFP signal was read at 485 nm/535 nm (excitation/emission) using a Victor3V (Perkin Elmer).
Cytotoxicity
HeLa (ATCC CCL-2), HEK293 (ATCC CRL1573), HepG2 (ATCC HB-8065), and SK-N-SH (ATCC HTB-11) cells were seeded at the concentration of 1.5 × 104 cells/well in 96 well plates. The next day, serially diluted compounds in DMSO (final concentration was 1%) were added and further incubated for 3 days. Culture supernatant was replaced by fresh media supplemented with 10 μg/ml resazurin (Sigma, Cat# R7017) and further incubated for 2 h. Fluorescence was measured using a SpectraMax M5 (Molecular Devices) at 530 nm/590 nm (excitation/emission).
Data analysis
The Vmax, Km, and IC50 were analyzed using GraphPad Prism 5 (GraphPad Software, Inc.).
RESULTS
Cell-based high throughput screening
A commercial small molecule library comprising 140,000 diverse small molecules was screened in a cell-based high throughput assay adapted from a novel phenotypic assay [manuscript in preparation]. In this assay, the reporter cell line includes a GFP ORF under the control of the HIV LTR whose expression is driven by the HIV tat protein so that the level of replication is proportional to the measured GFP signal. When the cells are infected with HIV at low MOI (0.1) and allowed a few more replication cycles, the assay includes the full life cycle of HIV.
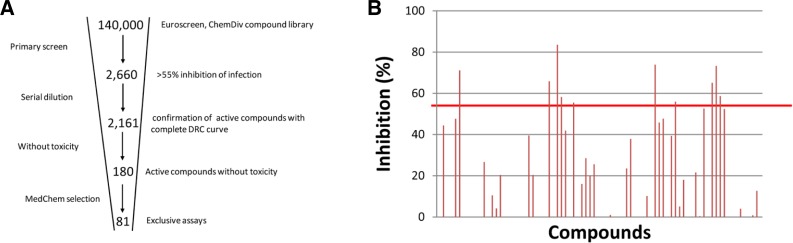
From the primary screen, 2,660 compounds were identified that showed a statistically significant inhibition of HIV-1 infection. These compounds were then assessed for their potency (IC50) and potential cytotoxicity, and 2,161 of the compounds were reconfirmed to have activity. Of these, 180 exhibited limited or no cytotoxicity. These 180 compounds were then assessed for their novelty and amenability to chemical modification, and 81 were selected as hits and repurchased (Fig. 1A). These 81 compounds were again tested in the primary screening assay, as well as in a resazurin-based cytotoxicity assay.
Fig. 1.
Screening of anti-HIV compounds and hRNaseH2 inhibitors. (A) Selection of 81 active hits from the 140,000 compounds. (B) Duplicates of each compound were analyzed at a final concentration of 50 μM, and data were expressed as percent inhibition. The cutoff line for hits representing 3× SD (55.4%) is indicated.
Recently, we identified RNaseH2 as a host factor essential for HIV-1 replication by siRNA screening (Genovesio et al., 2011). We hypothesized that some anti-HIV compounds might have inhibitory activity against hRNaseH2, and thus against HIV, because the 81 selected hits were identified in a target-free screen which included the full life cycle of HIV. To confirm the hypothesis, we established an in vitro hRNaseH2 enzyme activity assay to determine the effect of these hit compounds on this enzyme.
Establishment of the hRNaseH2 assay system
Plasmid pET-hH2ABC was transformed into the E. coli BL21 DE3 CodonPlus RIL strain. Expression of each RNaseH2 subunit with an N-terminal His-tag was driven by independent T7 promoters. The soluble fraction of E. coli lysate was subjected to histidine affinity chromatography and the purified protein was analyzed by SDS-PAGE. The purity was greater than 95% and the subunits were present in roughly a 1:1:1 ratio (Fig. 2A). The kinetic constants (Vmax and Km) of hRNaseH2 were measured using a FRET based assay with RNA/DNA heteroduplex as substrate, as described in the Materials and Methods. From Michaelis-Menten kinetic analysis, Vmax and Km were estimated to be 264.9 RFU/min and 2.556 μM, respectively (Fig. 2B, Table 1). The substrate specificity of hRNaseH2 was determined by comparing the generation of fluorescence using RNA/DNA or DNA/DNA duplex with the same nucleotide sequence as the substrate. As expected, hRNaseH2 could degrade the RNA/DNA duplex regardless of its nucleotide sequence, while the DNA/DNA duplex remained intact (data not shown).
Screening of hRNaseH2 inhibitors
To determine if there were any RNaseH2 inhibitors among the anti-HIV hits that we identified, we screened the compounds using an established hRNaseH2 assay system. Statistical analysis of the assay showed that the mean RFU of 0% inhibition (DMSO treated) and 100% inhibition (reaction mixture without enzyme) were 1,932 ± 246 and 599 ± 15, respectively. The Z′ value of the assay was calculated as 0.58 (n = 16). Duplicates of each compound were analyzed at a final concentration of 50 μM and data were expressed as percent inhibition. Ten compounds showing inhibition greater than 3 × SD (55.4%) were selected as hits (Fig. 1B).
Confirmation of anti-hRNaseH2 activity
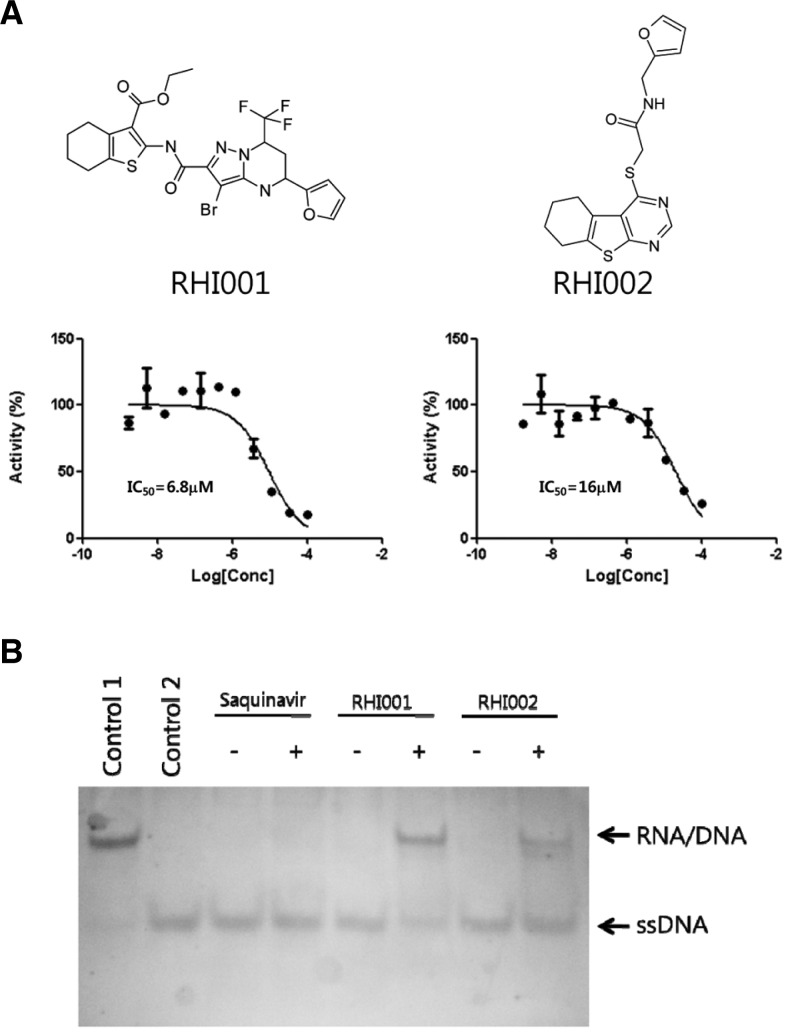
Further validation of the 10 selected compounds was performed by a dose response curve (DRC) analysis. As a result, two inhibitors, RHI001 and RHI002, were isolated (Fig. 3A). The IC50s of RHI001 and RHI002 were 6.8 μM and 16 μM, respectively. The auto-fluorescence and quenching properties of the two inhibitors were determined by adding the compounds before and after the hRNaseH2 degraded its substrate. Neither of the compounds was shown to interfere with the fluorescence of the substrate (data not shown).
Fig. 3.
Confirmation of anti-hRNaseH2 activity. (A) Human RNaseH2 was pre-incubated with serially diluted compound for 10 min at room temperature, and then substrate was added to a final concentration of 2 μM. After a 30 min reaction, the fluorescence was measured. The values are expressed as percent activity of DMSO control. Each concentration was done in duplicate. (B) The hRNaseH2 reaction was performed in the presence or absence of inhibitors. Control 1, reaction mixture without enzyme; Control 2, reaction mixture with excess enzyme (2 μg per reaction).
The inhibitory effects of RHI001 and RHI002 on hRNaseH2 activity were further confirmed using a gel-based assay. The assay conditions were re-optimized for the purpose of comparing band intensities; the FRET based assay had been performed under initial velocity conditions (initial linear portion of the enzyme reaction when less than 10% of substrate has been depleted). With RHI001 or RHI002 present in the hRNaseH2 reaction, degradation of the RNA/DNA duplex was inhibited (Fig. 3B). By comparison, the negative control compound, Saquinavir, a known HIV protease inhibitor, showed no effect on hRNaseH2 activity.
Anti-HIV activity of hRNaseH2 inhibitors in cells
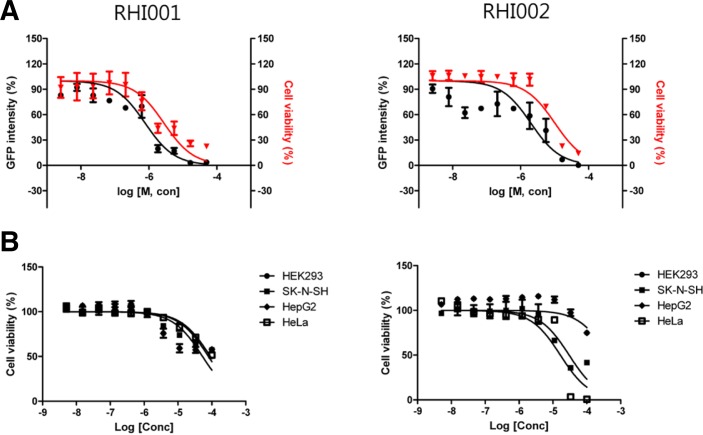
Antiviral activity of the two putative hRNaseH2 inhibitors was re-confirmed in a cell-based assay in a 96-well format. CG8 cells were treated with the compounds and infected with HIV-1 IIIB at a MOI of 0.1. After 3 days, GFP signal was read and data were expressed as percent virus titer. As shown in Fig. 4A, the EC50s of RHI001 and RHI002 against HIV were 0.8 μM and 2.0 μM, respectively. A resazurin assay revealed that the EC50s of RHI001 and RHI002 against CG8 cells were 2.8 μM and 10.0 μM, respectively.
Fig. 4.
Anti-HIV activity and cytotoxicity of hRNaseH2 inhibitors. (A) CG8 cells were treated with serially diluted compounds and infected with HIV-1 IIIB at a MOI of 0.1. After 3 days, GFP signal was read. For cytotoxicity, a resazurin assay was performed. Right and left Y-axis represents percentage of residual GFP intensity and viable cells, respectively. Each concentration was done in duplicate. (B) Cytotoxicity of inhibitors on HEK293, SK-N-SH, HepG2, and HeLa cells. Cells were treated with compounds for 3 days and a resazurin assay was performed. Each concentration was done in duplicate.
Cytotoxicity of compounds in four additional cell lines with different origins, HepG2 (hepatocellular carcinoma), HEK293 (embryonic kidney), HeLa (cervical cancer), and SK-N-SH (neuroblastoma) were tested (Fig. 4B). The CC50 of RHI001 in HEK293, SK-N-SH, HepG2, and HeLa were calculated as 52.98 μM, 79.31 μM, 52.97 μM, and 90.43 μM, respectively. The CC50 of RHI002 were >100 μM, 30.07 μM, > 100 μM, and 16.55 μM, respectively. With the exception of CG8 cells where HIV-1 is replicating, the other cells tested showed CC50 values high enough to be used for functional studies.
Selectivity of the inhibitors
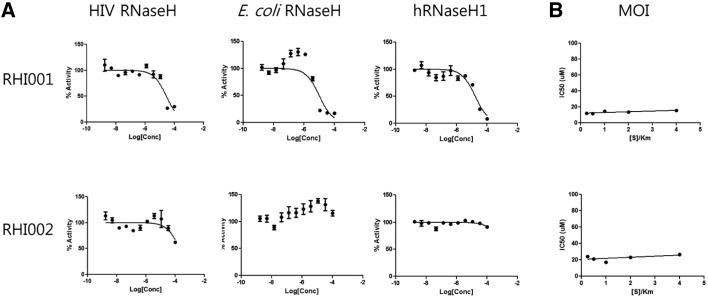
To further characterize the properties of the inhibitors, crossreactivity with HIV RNaseH, E. coli RNaseH, and human RNaseH1 was determined. Assay conditions for each enzyme were optimized using a kinetic approach and summarized in Table 1. RHI001 inhibited activity of HIV RNaseH, E. coli RNaseH, and human RNaseH1 with an IC50 of 28.5, 7.9, and 31.7 μM, respectively. By comparison, RHI002 did not show clear inhibition of the three RNaseHs (Fig. 5A). These results indicate that RHI001 may be a general inhibitor of most RNaseHs, whereas RHI002 may be a specific inhibitor of hRNaseH2. The two inhibitors were tested in two additional HIV-specific enzymatic assays, HIV reverse transcriptase (RT) and HIV integrase, and found to have no effect (data not shown).
Fig. 5.
Characterization of inhibitors. (A) Specificity of inhibitors. Optimization of reaction conditions for each enzyme was done independently and summarized in Table 1. RHI001 inhibited activity of HIV RNaseH, E. coli RNaseH, and human RNaseH1 with IC50s of 28.5, 7.9, and 31.7 μM, respectively. In comparison, RHI002 showed no clear inhibitory effect on the RNaseHs. Each concentration was done in duplicate. (B) Modality of inhibitors. IC50s in various substrate concentrations were measured and plotted (see text for details).
Modality of inhibitors
The mechanism of interaction between inhibitor and enzyme was studied by monitoring the effect of the [S]/Km ratio on the IC50 value (Copeland, 2003). IC50s in various substrate concentrations (0.625, 1.25, 2.5, 5, and 10 μM representing 0.25, 0.5, 1, 2, and 4 [S]/Km ratios, respectively) were measured and plotted (Fig. 5B). The IC50 of RHI001 was 12.1, 11.4, 14.5, 13.6, and 15.5 μM when the [S]/Km ratio was 0.25, 0.5, 1, 2, and 4, respectively. The IC50 of RHI002 was 24.3, 21.3, 17.1, 23.1, and 26.6 μM when the [S]/Km ratio was 0.25, 0.5, 1, 2, and 4, respectively. These results indicate that neither inhibitor’s activity was sensitive to substrate concentration. This type of inhibitor is referred to as a noncompetitive inhibitor, which may bind to both the free enzyme and ES complex.
DISCUSSION
Previous reports indicated that hRNaseH2 is involved in at least three biological functions. First, mutations in the subunits encoding hRNaseH2 cause Aicardi-Goutier syndrome (AGS), an autosomal recessive genetic disorder (Crow et al., 2006). Second, hRNaseH2 is required for maintaining genome stability by removing ribonucleotides misincorporated by replicative polymerases (Hiller et al., 2012; Reijns et al., 2012). And third, hRNaseH2 has essential roles in the HIV replication process (Genovesio et al., 2011). Here, we described two hRNaseH2 inhibitors that demonstrated anti-HIV activity. Although antiviral activity was observed, they were not regarded as drug candidates due to the narrow selectivity index (SI) in HIV permissive cells. However, much less cytotoxicity was observed in other cell lines such as HepG2 (hepatocellular carcinoma), HEK293 (embryonic kidney), HeLa (cervical cancer), and SK-N-SH (neuroblastoma) (Fig. 4B). Therefore, the two inhibitors may provide a good tool for the investigation of the cellular function of human RNaseH2.
AGS caused by hRNaseH2 mutation phenotypically mimics congenital viral infection, with elevation of interferon alpha in cerebrospinal fluid. Therefore, it is hypothesized that inhibition of hRNaseH2 results in the accumulation of short RNA-DNA hybrids generated during the normal cellular life cycle (such as the initiating molecule of Okazaki fragments and replication of transposons), subsequently triggering the same innate immune response as viral infection. However, the narrow SI hampers the ability to study the nature of the antiviral activity of the inhibitors. Currently, we are working on a structure-activity-relationship (SAR) study to generate derivatives with a wider SI. At the same time, monitoring of genome wide changes induced by the inhibitors, such as microarray and PCR array, is ongoing.
Genetic studies of AGS patients have shown that biallelic mutations in RNASEH2A, RNASEH2B, and RNASEH2C were observed in 3, 47, and 18 cases, respectively (Rice et al., 2007). Moreover, of five mutations analyzed in human RNASE2B and RNASE2C linked to AGS, only one, R69W in the RNASEH2C protein, exhibited a significant reduction in specific activity (Chon et al., 2009). The role of RNASEH2B and RNASEH2C in the overall activity of RNaseH2 is not yet clear. Previously, it was reported that purification of the three-subunit complex from stable HeLa cells transfected with tagged-RNASEH2A in a 1:1:1 ratio, with little or no excess RNASEH2A suggesting that any protein not interacting with the RNASEH2B and RNASEH2C proteins did not survive as a separate protein (Chon et al., 2009). We hypothesized that mutations in RNASEH2B and RNASEH2C might fail to form complex with the catalytic subunit, and inappropriately configured proteins would be degraded by the proteasome-mediated pathway. In this case, overall hRNaseH2 activity would then be decreased.
Pathogen-associated molecular patterns (PAMPs) are recognized by pattern recognition receptors (PRRs). The PRRs can be categorized into three major classes designated retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), Toll-like receptors (TLRs), and nucleotide oligomerization domain (NOD)-like receptors (NLRs). As reviewed by Takeuchi and Akira (2009), the PRRs recognize viral components, such as genomic DNA, double stranded RNA (dsRNA), single-stranded RNA (ssRNA), RNA with 5′-triphosphate ends, and viral proteins. However, PRR recognition of RNA-DNA hybrids has never been discovered. We hypothesize that accumulation of RNA-DNA hybrids following hRNaseH2 inhibition might trigger the innate immune response. More detailed research, including genome- and proteome- wide studies of host changes induced by the inhibitors and the response of other viruses to the inhibitors, may help identify novel PRR(s) against RNA-DNA hybrids.
Acknowledgments
This research was supported by the Pioneer Research Center Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2011-0001679). We would like to thank Dr. Michel Liuzzi for his helpful comments in preparing this manuscript.
REFERENCES
- Aicardi J, Goutieres F. A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis. Ann Neurol. 1984;15:49–54. doi: 10.1002/ana.410150109. [DOI] [PubMed] [Google Scholar]
- Chon H, Vassilev A, DePamphilis ML, Zhao Y, Zhang J, Burgers PM, Crouch RJ, Cerritelli SM. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNaseH2 complex. Nucleic Acids Res. 2009;37:96–110. doi: 10.1093/nar/gkn913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland RA. Mechanistic considerations in high-through-put screening. Anal Biochem. 2003;320:1–12. doi: 10.1016/s0003-2697(03)00346-4. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- Flanagan JM, Funes FM, Henderson S, Wild L, Carey N, Boshoff C. Genomics screen in transformed stem cells reveals RNASEH2A, PPAP2C, and ADARB1 as putative anticancer drug targets. Mol Cancer Ther. 2009;8:249–260. doi: 10.1158/1535-7163.MCT-08-0636. [DOI] [PubMed] [Google Scholar]
- Gao F, Li Y, Decker JM, Peyerl FW, Bibollet-Ruche F, Rodenburg CM, Chen Y, Shaw DR, Allen S, Musonda R, et al. Codon usage optimization of HIV type 1 subtype C gag, pol, env, and nef genes: in vitro expression and immune responses in DNA-vaccinated mice. AIDS Res. Hum. Retroviruses. 2003;19:817–823. doi: 10.1089/088922203769232610. [DOI] [PubMed] [Google Scholar]
- Genovesio A, Kwon YJ, Windisch MP, Kim NY, Choi SY, Kim HC, Jung S, Mammano F, Perrin V, Boese AS, et al. Automated genome wide profiling of cellular proteins involved in HIV infection. J. Biomol. Screen. 2011;16:945–958. doi: 10.1177/1087057111415521. [DOI] [PubMed] [Google Scholar]
- Goutieres F. Aicardi-Goutieres syndrome. Brain Dev. 2005;27:201–206. doi: 10.1016/j.braindev.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Goutieres F, Aicardi J, Barth PG, Lebon P. Aicardi-Goutieres syndrome: an update and results of interferon-alpha studies. Ann Neurol. 1998;44:900–907. doi: 10.1002/ana.410440608. [DOI] [PubMed] [Google Scholar]
- Hiller B, Achleitner M, Glage S, Naumann R, Behrendt R, Roers A. Mammalian RNaseH2 removes ribonucleotides from DNA to maintain genome integrity. J Exp Med. 2012;209:1419–1426. doi: 10.1084/jem.20120876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, Artuch R, Montalto SA, Bacino CA, Barroso B, et al. Clinical and molecular phenotype of Aicardi-Goutieres Syndrome. Am J Hum Genet. 2007;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer P, Vartanian JP, Wachsmuth M, Henry M, Guetard D, Wain-Hobson S. Anti-termination by SIV Tat requires flexibility of the nascent TAR structure. J Mol Biol. 2004;344:11–28. doi: 10.1016/j.jmb.2004.09.042. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]