Abstract
RANKL induces the formation of osteoclasts, which are responsible for bone resorption. Herein, we investigated the role of SWAP-70-like adapter of T cells (SLAT) in RANKL-induced osteoclastogenesis. Expression levels of SLAT were reduced during RANKL-induced osteoclastogenesis. Overexpression of SLAT in BMMs inhibited TRAP-positive multinuclear osteoclast formation and attenuated the expression of NFATc1, which is an important modulator in osteoclastogenesis. Furthermore, silencing of SLAT by RNA interference enhanced osteoclast formation as well as NFATc1 expression. In addition, SLAT was involved in RANKL-induced JNK activation in osteoclasts. Taken together, our data suggest that SLAT acts as a negative modulator of RANKL-induced osteoclastogenesis.
Keywords: bone, gene expression, osteoclast differentiation, RANKL, SLAT
INTRODUCTION
Bone is a highly dynamic tissue, and bone homeostasis is maintained by a delicate balance between bone formation and bone resorption under the regulation of systemic factors such as cytokines and hormones (Suda et al., 1999; Teitelbaum, 2000). Bone loss can be easily caused by an imbalance between osteoclasts, which resorb bone; and osteoblasts, which form new bone (Takayanagi, 2007; Walsh et al., 2006). Dysregulation of osteoclasts under various pathological conditions, such as inflammation and estrogen deficiency, leads to bone loss (Rodan and Martin, 2000). Importantly, osteoporotic fractures are a significant cause of mortality and morbidity in the elderly population and represent a substantial economic burden to society (Boonen and Singer, 2008; Harvey et al., 2010). Therefore, understanding the mechanism of osteoclast formation is important for the development of new therapeutic strategies for bone diseases.
Osteoclasts are derived from hematopoietic cells in the presence of macrophage colony stimulating factor (M-CSF) and RANKL. Binding of RANKL to its receptor RANK activates nuclear factor kappa B (NF-κB), c-Jun N-terminal kinase (JNK), p38, and extracellular signal-regulated kinase (ERK), which are important for osteoclast differentiation (Lee and Kim, 2003). RANKL regulates various transcription factors including NF-κB, c-Fos, and NFATc1, which act as positive modulators of osteoclast differentiation (Boyle et al., 2003; Teitelbaum, 2000). RANKL induces the expression of c-Fos and NFATc1 during osteoclastogenesis. NFATc1 induces the expression of target genes such as tartrate-resistant acid phosphatase (TRAP), cathepsin K, and osteoclast-associated receptor (OSCAR), which are important for osteoclast differentiation or function (Kim et al., 2005; So et al., 2003; Walsh et al., 2006).
A T cell receptor (TCR)-regulated protein called switching B-cell complex 70 kDa subunit (SWAP70)-like adapter of T cells (SLAT) was originally isolated based on its abundant expression in T helper 2 (Th2) cells and its homology with SWAP-70, a B cell-enriched guanine nucleotide exchange factor (GEF) (Borggrefe et al., 1998; Pearce et al., 2006; Shinohara et al., 2002; Tanaka et al., 2003). SLAT (also called Def-6 or IBP) is abundant in central and peripheral lymphoid tissues, with high amounts displayed in thymocytes and in peripheral T cells, and it translocates to the immunological synapse upon antigen stimulation (Becart et al., 2007; Gupta et al., 2003). SLAT-deficient mice revealed a role of SLAT in thymic DN1 cell expansion, T cell activation, and Th1 and Th2 cell inflammatory responses. The defect in Th1 and Th2 cell responses was traced to defective Ca2+-NFAT signaling (Becart et al., 2007). Although SLAT enhances TCR-mediated NFAT activation and Th1 and Th2 differentiation, the role of SLAT in RANKL-induced osteoclastogenesis has not yet been studied.
In this study, we investigated the role of SLAT in RANKL-induced osteoclastogenesis. Here we reported that SLAT is down-regulated by RANKL. In addition, we demonstrated that SLAT is involved in RANKL-induced JNK activation and NFATc1 induction. Therefore, our data suggested that SLAT is a modulator of RANKL-induced osteoclastogenesis.
MATERIALS AND METHODS
Plasmid constructs and reagents
SLAT was generated by RT-PCR using RNA from bone marrow-derived macrophages (BMMs) and cloned into pMX-IRES-EGFP. The primers sequences were as follows: 5′-AAG ATC TAT GGC CCT GCG CAA GGA GCT GCT CAA G-3′ (5-SLAT-BglII) and 5′-CCT CGA GAT TCC CTG GTG CTG GAT CCA GTT TTT C-3′ (3-SLAT-XhoI). These amplified PCR fragments were digested with BamHI and XhoI, and were cloned into a pMX vector. SB203580, LY294002, PD98059, L-l-tosylamide-2-phenylethyl chloromethyl ketone (TPCK), BAY and SP600125 were purchased from Calbiochem (USA). All cell culture media and supplements were obtained from Hyclone.
Osteoclast formation and TRAP staining
Murine osteoclasts were prepared from bone marrow cells as previously described (Kim et al., 2009). In brief, femurs were aseptically removed from 6-8-week-old ICR mice, and bone marrow cells were flushed out with a sterile 21-gauge syringe. The cells were cultured in α-minimal essential medium (α-MEM; Hyclone, USA) containing 10% FBS (Hyclone) with MCSF (30 ng/ml) for 3 days. Floating cells were removed and adherent cells (BMMs) were used as osteoclast precursors. To generate osteoclasts, BMMs were cultured with M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 4 days. Cultured cells were fixed and stained for TRAP. TRAP-positive multinuclear cells (TRAP + MNCs), containing more than three nuclei were counted as osteoclasts.
Retroviral gene transduction
Retroviral infection was performed as previously described (Kim et al., 2008; 2012). To generate retroviral stock, generated recombinant plasmids and the parental pMX vector were transfected into the packaging cell line Plat E using FuGENE 6 (Roche Applied Sciences, USA). Plat E cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, and 2 × 105 cells were seeded in a 6-well plate. On the following day, the medium was changed to α-MEM containing 10% FBS. Viral supernatant was collected from cultured media 24–48 h after transfection. BMMs were incubated with viral supernatant for 8 h in the presence of 10 μg/ml polybrene (Sigma-Aldrich, USA). After removing the viral supernatant, BMMs were further cultured with M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 4 days.
RT-PCR and real-time PCR
Cells were washed with ice-cold PBS and lysed in Qiazol lysis Reagent (Qiagen). First-strand cDNA was transcribed from 2 μg RNA using Superscript II reverse transcriptase (Invitrogen). The reverse transcribed cDNA was amplified by PCR. For Real-time PCR, PCRs were performed using the QuantiTect SYBR Green PCR kit (Qiagen) in triplicates on Rotor-Gene 6000 (Corbett Research). The thermal cycling conditions were as follows: 15 min at 95°C, followed by 40 cycles of 95°C for 10 seconds, 58°C for 15 s, and 72°C for 20 s. All quantitations were normalized to an endogenous control GAPDH. The relative quantitation value for each target gene compared to the calibrator for that target gene is expressed as 2−(Ct-Cc) (Ct and Cc are the mean threshold cycle differences after normalizing to GAPDH). The relative expression levels of samples are presented by semilog plot. The primer sequences used for PCR amplification were as follows: NFATc1, forward 5′-CTC GAA AGA CAG CAC TGG AGC AT and reverse 5′-CGG CTG CCT TCC GTC TCA TAG-3′; OSCAR, forward 5′-TGC TGG TAA CGG ATC AGC TCC CCA GA-3′ and reverse 5′-CCA AGG AGC CAG AAC CTT CGA AAC T-3′; TRAP, forward 5′-CTG GAG TGC ACG ATG CCA GCG ACA-3′ and reverse 5′-TCC GTG CTC GGC GAT GGA CCA GA-3′; c-Fos, forward 5′-ATG GGC TCT CCT GTC AAC ACA CAG-3′ and reverse 5′-TGG CAA TCT CAG TCT GCA ACG CAG-3′; SLAT, forward 5′-TGC TCA AGT CTA TCT GGT ACG CC-3′ and reverse 5′-GAA GAG GCA CCA TAG ACG GAA AG-3′. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control.
Western blot analysis
Cells from transduced BMMs were harvested after washing with ice-cold PBS and then lysed in extraction buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, and protease inhibitors). Cell lysates were subjected to SDS-PAGE and Western blotting. Primary antibodies used included IκB, phospho-p38, p38, phospho-JNK, JNK, phospho-ERK, ERK (Cell Signaling Technology, USA), actin (Sigma-Aldrich), Flag (Sigma-Aldrich). HRP conjugated secondary antibodies (Amersham Biosciences, USA) were probed and developed with ECL solution (Millipore). Signals were detected and analyzed by LAS3000 luminescent image analyzer (Fuji, Japan).
siRNA preparation and transfection
Predesigned mouse SLAT siRNAs (ID nos. J-045645-09, J-045645-10, J-045645-11 and J-045645-12) were purchased from Dharmacon (Thermo Fisher Scientific). Only the siRNA ID no. L-045645-01-0005 (5 nM) had a significant inhibitory effect on SLAT expression in BMMs; therefore, we used this siRNA for all studies. The siRNAs were transfected into BMMs using the DharmaFECT 1 transfection reagent (Thermo Fisher Scientific).
Statistical analysis
Results are presented as means ± SD. Statistical differences were determined by a paired Student’s t-test with significance level at p < 0.05.
RESULTS
RANKL down-regulates the expression of SLAT during osteoclastogenesis
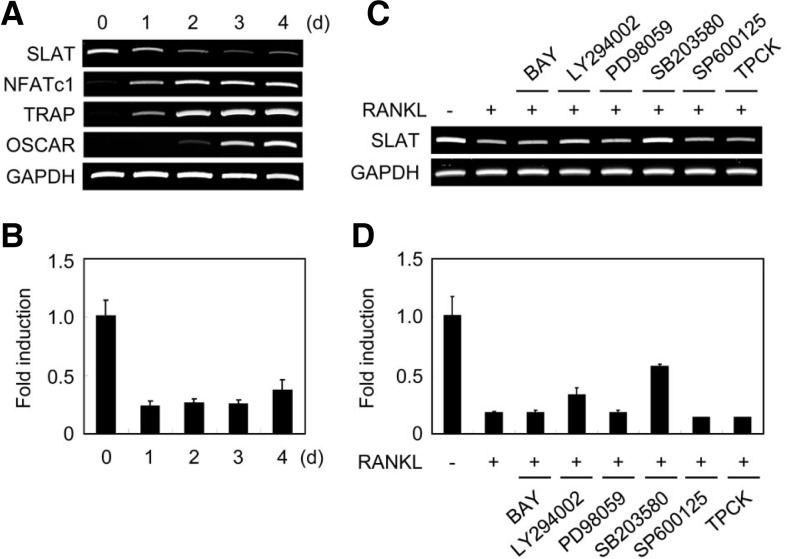
In order to examine whether the SLAT gene is expressed in osteoclast lineage cells, we observed mRNA expression of SLAT during the differentiation of osteoclasts. Osteoclast differentiation was induced by culturing BMM cells in the presence of M-CSF and RANKL. RANKL down-regulated SLAT expression during osteoclast differentiation, whereas osteoclast-associated genes such as NFATc1, TRAP, and OSCAR were strongly induced by RANKL (Figs. 1A and 1B). Since RANKL activates NF-κB, JNK, p38, ERK, and AKT during osteoclastogenesis, we tested various inhibitors to determine which signaling cascades are essential for RANKL-induced SLAT down-regulation. Among various inhibitors, p38 MAPK inhibitor (SB203580) strongly blocked RANKL-induced SLAT down-regulation (Figs. 1C and 1D). These results suggest that RANKL down-regulated expression of SLAT mostly through p38 MAPK pathways during osteoclastogenesis.
Fig. 1.
SLAT gene expression during osteoclastogenesis. (A, B) BMMs were cultured with M-CSF and RANKL for the indicated times. Total RNA was collected at each time point. (A) RT-PCR was performed to detect expression of the indicated genes. (B) Real-time PCR analysis was performed for the expression of SLAT and GAPDH. (C, D) BMMs were cultured with M-CSF and RANKL for 2 days in the presence of various inhibitors; Mock (DMSO), BAY (20 μM), LY294002 (10 μM), PD98059 (20 μM), SB203580 (20 μM), SP600125 (5 μM), TPCK (5 μM). RT-PCR (C) and Real-time PCR (D) were performed for the expression of SLAT and GAPDH.
Overexpression of SLAT inhibits osteoclast formation as well as NFATc1 expression
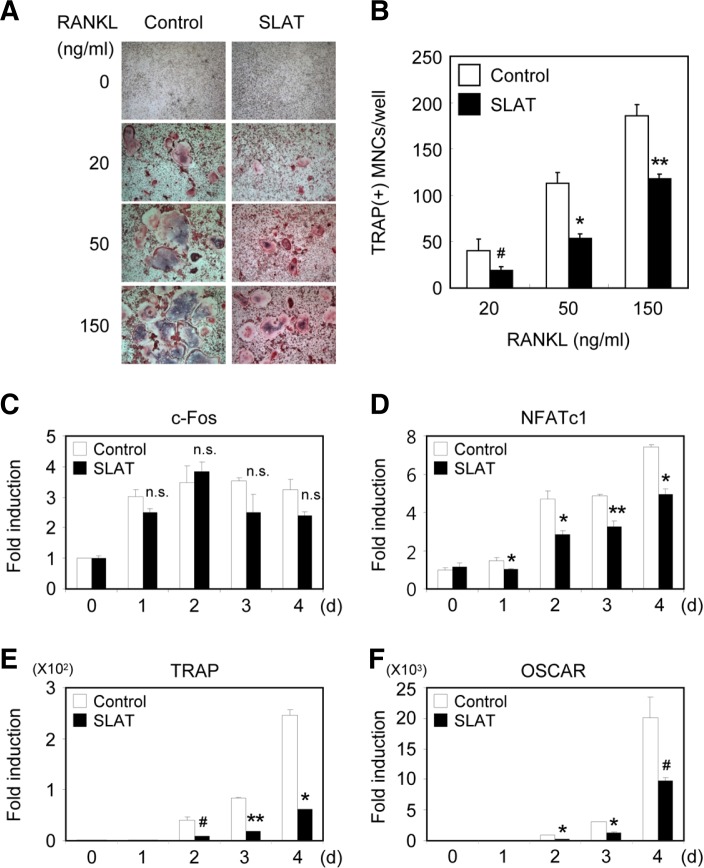
Since SLAT was expressed in osteoclast lineage cells, we tried to find out whether SLAT is required for efficient osteoclast differentiation and function. Transduced BMMs were cultured with M-CSF alone or M-CSF/RANKL, and were stained for TRAP (Fig. 2A). RANKL treatment of control vector-infected BMMs increased the number of TRAP+ MNCs in a dose-dependent manner. Compared with the control vector, the overexpression of SLAT in BMMs significantly decreased the formation of TRAP+ MNCs mediated by M-CSF and RANKL (Figs. 2A and 2B). These results suggest that SLAT may have a key role in RANKL-mediated osteoclast differentiation.
Fig. 2.
Overexpression of SLAT in BMMs inhibits osteoclast formation. (A, B) BMMs were transduced with pMX-IRES-EGFP (control) or SLAT retrovirus and cultured for 4 days with M-CSF alone or M-CSF and various concentrations of RANKL as indicated. (A) Cultured cells were fixed and stained for TRAP. (B) Numbers of TRAP+ multinucleated cells were counted (#P < 0.05, *P < 0.01, ** P < 0.005 vs Control. ns, not significant). (C-F) BMMs were transduced with pMX-IRES-EGFP (control) or SLAT retrovirus and cultured with MCSF and RANKL for the indicated times. Realtime PCR analysis was performed to detect the indicated genes in control or SLAT-over-expressing samples.
Based on our observation that overexpression of SLAT inhibits RANKL-mediated osteoclast differentiation, we examined the expression levels of various genes, which are known to be important for osteoclastogenesis. Overexpression of SLAT attenuated RANKL-mediated induction of NFATc1, TRAP, and OSCAR. However, the expression of c-Fos was not affected by SLAT overexpression during osteoclastogenesis (Figs. 3C–3F). Taken together, our data indicate that overexpression of SLAT attenuates RANKL-induced osteoclastogenesis as well as NFATc1 gene expression.
Fig. 3.
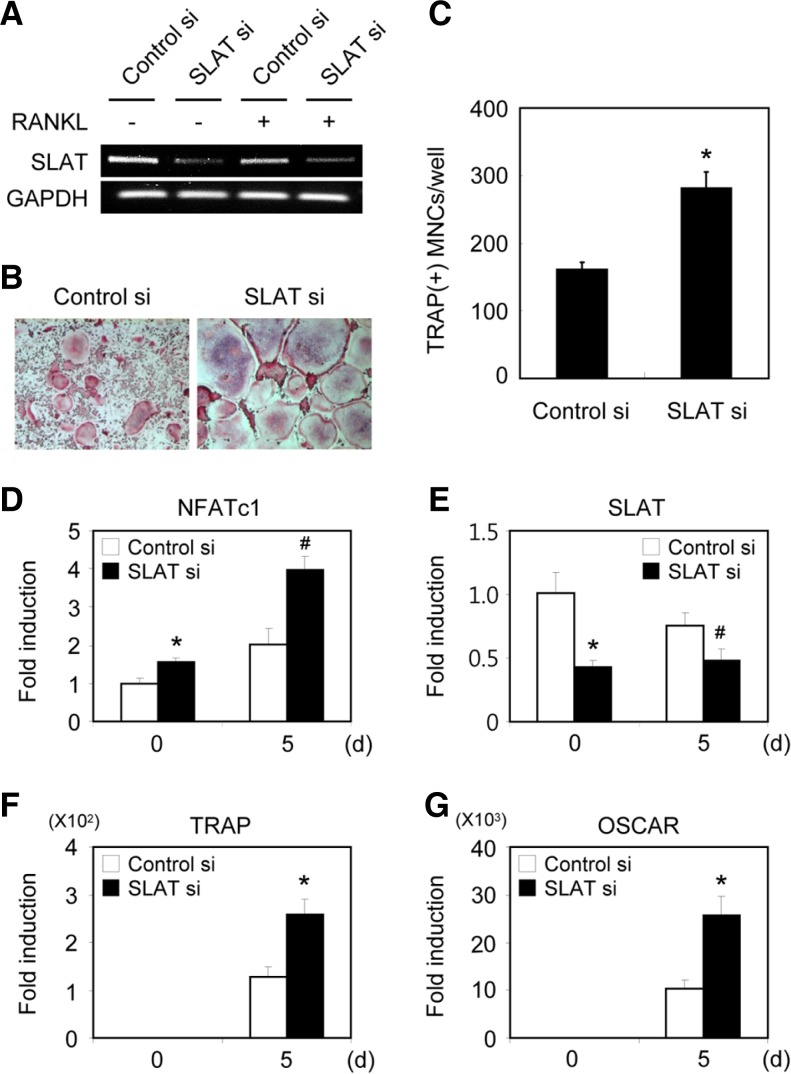
Silencing of SLAT enhances osteoclast formation and induction of NFATc1. (A) BMMs transfected with GFP or SLAT siRNA were cultured for 2 days in the absence or presence of RANKL. RTPCR was performed. (B) BMMs transfected with GFP or SLAT siRNA were cultured for 5 days in the presence of M-CSF and RANKL. Cultured cells were fixed and stained for TRAP. (C) TRAP(+) MNCs were counted as osteoclasts. (D-G) BMMs transfected with GFP or SLAT siRNA were cultured with M-CSF and RANKL for the indicated times. Real-time PCR analysis was performed with the primers specific for SLAT, NFATc1, OSCAR, TRAP, and GAPDH (control). #P < 0.05, *P < 0.01 vs control.
Down-regulation of SLAT enhances osteoclast formation as well as NFATc1 expression
To investigate down-regulation of SLAT gene in BMMs by specific-siRNAs, BMMs were transfected with control siRNA, or SLAT siRNA. After 48 h of transfection, the expression of SLAT was observed by real-time PCR and RT-PCR. The expression of SLAT was reduced by SLAT siRNA, not by control GFP-specific siRNA (Figs. 3A and 3E).
To examine the physiological role of SLAT in osteoclastogenesis, BMMs transfected with control siRNA or SLAT siRNA were cultured for 5 days with M-CSF and RANKL. Compared with the control siRNA, the silencing of SLAT in BMMs resulted in a significant increase in the formation of TRAP+ MNCs mediated by RANKL (Figs. 3B and 3C). Furthermore, silencing of SLAT also enhanced the expression of NFATc1 as well as of OSCAR and TRAP during RANKL-mediated osteoclastogenesis (Figs. 3D–3G). These results suggest that SLAT may have a key role in RANKL-mediated osteoclast differentiation.
SLAT is involved in RANKL-induced JNK signaling pathway
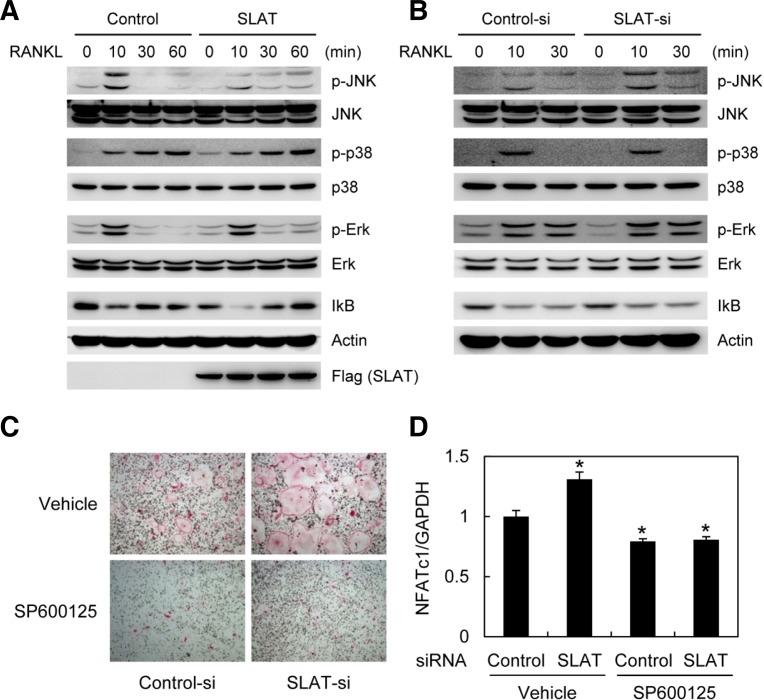
RANKL activates various signaling pathways such as NF-κB, JNK, p38 MAPK, and ERK. To investigate the role of SLAT in RANKL-induced signaling cascades, BMMs were infected with control or SLAT retrovirus. Overexpression of SLAT inhibited RANKL-induced JNK activation, whereas other signaling pathways such as p38, ERK, and NF-κB were not affected (Fig. 4A). Furthermore, silencing of SLAT by RNA interference enhanced only RANKL-induced JNK activation (Fig. 4B). These data indicate that SLAT is involved in RANKL-induced JNK activation in osteoclasts.
Fig. 4.
SLAT is involved in RANKL-induced JNK activation. (A) BMMs were transduced with control (pMX-IRES-EGFP) or SLAT retrovirus. (B) BMMs transfected with GFP or SLAT siRNA were cultured for 2 days in the presence of M-CSF. (A, B) Cells were stimulated with RANKL (500 ng/ml) for the indicated times. Whole cell extracts were subjected to Western blot analysis with specific antibodies as indicated. (C, D) BMMs transfected with control or SLAT siRNA were cultured with M-CSF and RANKL in the absence or presence of SP600125 (5 μM). (C) Cultured cells were fixed and stained for TRAP. (D) Real-time PCR analysis was performed with the primers specific for NF ATc1 and G APDH. * P < 0.05 vs control.
Next, we investigated whether JNK signaling cascade is involved in SLAT-mediated NFATc1 regulation. Treatment with JNK inhibitor (SP600125) strongly inhibited RANKL-induced osteoclast formation even in the SLAT knockdown samples (Fig. 4C). Furthermore, RANKL-mediated induction of NFATc1 was strongly blocked by treatment with JNK inhibitor both in control and SLAT knockdown samples (Fig. 4D). These data suggest that SLAT can regulate NFATc1 via JNK signaling pathways.
DISCUSSION
SLAT is abundant in central and peripheral lymphoid tissues, with high amounts found in thymocytes and peripheral T cells. Def6-/- mice are resistant to Th1- and Th2-mediated inflammation in a model of lung inflammation. This resistance reflects the essential role of SLAT/Def6 in promoting TCR-induced Ca2+ release from endoplasmic reticulum stores, and hence, subsequent steps in the Ca2+ signaling pathway, including activation of the transcription factor NFAT (Becart et al., 2007).
In osteoclasts, ectopic expression of NFATc1 causes osteoclast precursors to undergo efficient differentiation in the absence of RANKL (Kim et al., 2005; Takayanagi et al., 2002). In addition, NFATc1-deficient embryonic stem cells fail to differentiate into osteoclasts in response to RANKL (Takayanagi et al., 2002), thus suggesting that NFATc1 acts as a master regulator of osteoclastogenesis. Since SLAT could enhance TCR-mediated NFAT activation in immune cells, we supposed that SLAT might positively regulate RANKL-induced osteoclastogenesis via NFATc1 activation. However, overexpression of SLAT in osteoclast precursors attenuates osteoclast differentiation via down-regulation of JNK/NFATc1 cascades, while knockdown of SLAT in BMMs enhances RANKL-induced osteoclastogenesis as well as JNK activation and NFATc1 induction. These data indicate that SLAT has different roles in different cells, immune cells, and osteoclasts. It is interesting that SLAT acts as a positive modulator for TCR-mediated NFAT activation via Ca2+ release in immune cells, and acts as a negative regulator for RANKL-induced NFATc1 induction via blocking JNK activation in osteoclasts. One of the possible reasons for the dual role of SLAT could be that different molecules might be involved in different cell signaling pathways.
RANKL is a potent activator of MAP kinase superfamily member, JNK. Activation of JNK by RANKL results in phosphorylation of c-jun, an activator protein 1 (AP-1) component, in osteoclasts. JNK is also known to be activated by cell stressors such as UV radiation and pro-inflammatory cytokines including TNF-α and IL-1. JNK stimulates AP-1 transcription factor activity and up-regulates CaMKII and CaMKIV in committed preosteoclasts. The elevated CaMK level in pre-osteoclasts leads to a sustained increase in NFATc1 level, which is required to maintain the expression of osteoclast-associated genes in osteoclasts (Chang et al., 2008). Our data revealed that SLAT is involved in RANKL-induced JNK activation as well as in NFATc1 induction in osteoclasts; however, the underlying mechanism of SLAT-mediated RANKL-induced JNK activation is unknown. A further study will be needed for understanding how SLAT plays a role in differentiation of osteoclasts via JNK signaling cascades.
In conclusion, this study demonstrates that SLAT acts as a negative regulator of RANKL-induced osteoclastogenesis by regulating JNK and NFATc1. Therefore, SLAT might be a potential target for treating bone diseases such as osteoporosis.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant (MRC for Gene Regulation, 2011-0030132) funded by the Korea government [Ministry of Science, Ict & Future Planning (MSIP)] and a Grant HI11C0674 of the Korean Health Technology R&D Project, Ministry of Health & Welfare (to K.K.).
REFERENCES
- Becart S, Charvet C, Canonigo Balancio AJ, De Trez C, Tanaka Y, Duan W, Ware C, Croft M, Altman A. SLAT regulates Th1 and Th2 inflammatory responses by controlling Ca2+/NFAT signaling. J Clin Invest. 2007;117:2164–2175. doi: 10.1172/JCI31640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonen S, Singer AJ. Osteoporosis management: impact of fracture type on cost and quality of life in patients at risk for fracture I. Curr Med Res Opin. 2008;24:1781–1788. doi: 10.1185/03007990802115796. [DOI] [PubMed] [Google Scholar]
- Borggrefe T, Wabl M, Akhmedov AT, Jessberger R. A B-cell-specific DNA recombination complex. J Biol Chem. 1998;273:17025–17035. doi: 10.1074/jbc.273.27.17025. [DOI] [PubMed] [Google Scholar]
- Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- Chang EJ, Ha J, Huang H, Kim HJ, Woo JH, Lee Y, Lee ZH, Kim JH, Kim HH. The JNK-dependent CaMK pathway restrains the reversion of committed cells during osteoclast differentiation. J Cell Sci. 2008;121:2555–2564. doi: 10.1242/jcs.028217. [DOI] [PubMed] [Google Scholar]
- Gupta S, Fanzo JC, Hu C, Cox D, Jang SY, Lee AE, Greenberg S, Pernis AB. T cell receptor engagement leads to the recruitment of IBP, a novel guanine nucleotide exchange factor, to the immunological synapse. J Biol Chem. 2003;278:43541–43549. doi: 10.1074/jbc.M308960200. [DOI] [PubMed] [Google Scholar]
- Harvey N, Dennison E, Cooper C. Osteoporosis: impact on health and economics. Nat Rev Rheumatol. 2010;6:99–105. doi: 10.1038/nrrheum.2009.260. [DOI] [PubMed] [Google Scholar]
- Kim K, Kim JH, Lee J, Jin HM, Lee SH, Fisher DE, Kook H, Kim KK, Choi Y, Kim N. Nuclear factor of activated T cells c1 induces osteoclast-associated receptor gene expression during tumor necrosis factor-related activation-induced cytokine-mediated osteoclastogenesis. J Biol Chem. 2005;280:35209–35216. doi: 10.1074/jbc.M505815200. [DOI] [PubMed] [Google Scholar]
- Kim K, Lee SH, Ha Kim J, Choi Y, Kim N. NFATc1 induces osteoclast fusion via up-regulation of Atp6v0d2 and the dendritic cell-specific transmembrane protein (DCSTAMP) Mol Endocrinol. 2008;22:176–185. doi: 10.1210/me.2007-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim K, Jin HM, Song I, Youn BU, Lee J, Kim N. Silibinin inhibits osteoclast differentiation mediated by TNF family members. Mol. Cells. 2009;28:201–207. doi: 10.1007/s10059-009-0123-y. [DOI] [PubMed] [Google Scholar]
- Kim K, Kim JH, Moon JB, Lee J, Kwak HB, Park YW, Kim N. The transmembrane adaptor protein, linker for activation of T cells (LAT), regulates RANKL-induced osteoclast differentiation. Mol. Cells. 2012;33:401–406. doi: 10.1007/s10059-012-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ZH, Kim HH. Signal transduction by receptor activator of nuclear factor kappa B in osteoclasts. Biochem Biophys Res Commun. 2003;305:211–214. doi: 10.1016/s0006-291x(03)00695-8. [DOI] [PubMed] [Google Scholar]
- Pearce G, Angeli V, Randolph GJ, Junt T, von Andrian U, Schnittler HJ, Jessberger R. Signaling protein SWAP-70 is required for efficient B cell homing to lymphoid organs. Nat Immunol. 2006;7:827–834. doi: 10.1038/ni1365. [DOI] [PubMed] [Google Scholar]
- Rodan GA, Martin TJ. Therapeutic approaches to bone diseases. Science. 2000;289:1508–1514. doi: 10.1126/science.289.5484.1508. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Terada Y, Iwamatsu A, Shinohara A, Mochizuki N, Higuchi M, Gotoh Y, Ihara S, Nagata S, Itoh H, et al. SWAP-70 is a guanine-nucleotide-exchange factor that mediates signalling of membrane ruffling. Nature. 2002;416:759–763. doi: 10.1038/416759a. [DOI] [PubMed] [Google Scholar]
- So H, Rho J, Jeong D, Park R, Fisher DE, Ostrowski MC, Choi Y, Kim N. Microphthalmia transcription factor and PU.1 synergistically induce the leukocyte receptor osteoclast-associated receptor gene expression. J Biol Chem. 2003;278:24209–24216. doi: 10.1074/jbc.M302940200. [DOI] [PubMed] [Google Scholar]
- Suda T, Takahashi N, Udagawa N, Jimi E, Gillespie MT, Martin TJ. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocrine Rev. 1999;20:345–357. doi: 10.1210/edrv.20.3.0367. [DOI] [PubMed] [Google Scholar]
- Takayanagi H. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat Rev. 2007;7:292–304. doi: 10.1038/nri2062. [DOI] [PubMed] [Google Scholar]
- Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Bi K, Kitamura R, Hong S, Altman Y, Matsumoto A, Tabata H, Lebedeva S, Bushway PJ, Altman A. SWAP-70-like adapter of T cells, an adapter protein that regulates early TCR-initiated signaling in Th2 lineage cells. Immunity. 2003;18:403–414. doi: 10.1016/s1074-7613(03)00054-2. [DOI] [PubMed] [Google Scholar]
- Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000;289:1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- Walsh MC, Kim N, Kadono Y, Rho J, Lee SY, Lorenzo J, Choi Y. Osteoimmunology: interplay between the immune system and bone metabolism. Ann Rev Immunol. 2006;24:33–63. doi: 10.1146/annurev.immunol.24.021605.090646. [DOI] [PubMed] [Google Scholar]