Summary
Human pluripotent stem cells (hPSCs) represent a promising source of patient-specific cells for disease modeling, drug screens, and cellular therapies. However, the inability to derive engraftable human hematopoietic stem and progenitor (HSPCs) has limited their characterization to in vitro assays. We report a strategy to re-specify lineage-restricted CD34+CD45+ myeloid precursors derived from hPSCs into multilineage progenitors that can be expanded in vitro and engraft in vivo. HOXA9, ERG, and RORA conferred self-renewal and multilineage potential in vitro and maintained primitive CD34+CD38− cells. Screening cells via transplantation revealed that two additional factors, SOX4 and MYB, were required for engraftment. Progenitors specified with all five factors gave rise to reproducible short-term engraftment with myeloid and erythroid lineages. Erythroid precursors underwent hemoglobin switching in vivo, silencing embryonic and activating adult globin expression. Our combinatorial screening approach establishes a strategy for obtaining transcription factor-mediated engraftment of blood progenitors from human pluripotent cells.
Introduction
Recent advances in reprogramming to induced pluripotent stem cells (IPSCs) has provided access to a wide array of patient-specific pluripotent lines that have the potential to give rise to any somatic cell type. A large number of pluripotent lines have been generated from patients with hematologic diseases, including Fanconi anemia (Muller et al., 2012), sickle cell anemia (Zou et al., 2011), Diamond Blackfan anemia (Garcon et al., 2013), Shwachman Diamond syndrome (Tulpule et al., 2013), chronic myelogenous leukemia (Kumano et al., 2012), JAK2V617F myelo-proliferative disorder (Ye et al., 2009), dyskeratosis congenita (Agarwal et al., 2010), Pearson Syndrome (Cherry et al., 2013), and others. These lines have the potential to become powerful models to gain insight into the molecular basis of disease and as platforms for drug screens (Cherry and Daley, 2013). To reveal the disease phenotype, IPSCs have to be differentiated into the target cell type of interest – hematopoietic stem and progenitor cells. Numerous protocols for hematopoietic differentiation of hPSCs into short-lived progenitors and mature cells have been established (Chadwick et al., 2003; Kennedy et al., 2012). However, no system presently exists to generate large numbers of transplantable cells from hPSCs, thereby precluding disease modeling in vivo and hampering the scope of experiments and screens that can be performed.
A major hurdle for generating engraftable HSPCs is the complex nature of hematopoietic ontogeny. It is now widely accepted that hematopoietic cells arise during mid-gestation in multiple temporal waves from hemogenic endothelial (HE) cells lining the major arteries (Bertrand et al., 2010; Boisset et al., 2010). Directed differentiation protocols attempt to recapitulate ontogeny by calibrated addition of morphogens such as BMP4, Activin A, and Notch ligands. These protocols can promote the emergence of HE and recapitulate the temporal waves of hematopoietic progenitors, but generate few if any transplantable cells (Choi et al., 2012; Kennedy et al., 2012). Prior reports of limited engraftment of hPSC-derived cells in immunodeficient mice have not been widely exploited owing to the heterogeneity among hPSC lines and variations among protocols (Ledran et al., 2008; Wang et al., 2005). More importantly, these protocols generate only small numbers of transplantable cells, and without the possibility of expanding them, it is difficult to move towards more practical models, such as in vivo engraftment of disease IPSCs.
One approach that has not been extensively explored in hematopoietic development is transcription factor-mediated specification and expansion of HSPCs. It was recently shown that a combination of Gata2, Gfi1b, Fos, and Etv6 promotes conversion of mouse fibroblasts into hematopoietic cells, suggesting that transcription factor reprogramming is a promising approach (Pereira et al., 2013). However, since fibroblasts are a distinct cell type, the precise conversion to HSPCs remains a challenge. We propose that conversions from closely related lineages, which minimize the “epigenetic distance” to a desired cell type, provide a more favorable context for precise alterations in cell fate. One possible approach is to promote specification of HE into transplantable HSPCs, which takes advantage of normal developmental cues. However, the process of endothelial to hematopoietic transition remains poorly understood, making it difficult to design rational interventions. An alternative approach is to start with committed hematopoietic progenitors and revert them to a more immature state. Such “re-specification” combines directed differentiation with transcription-based reprogramming to establish HSPC fate. A logical hypothesis is that the key regulatory factors that maintain HSCs can re-activate stem cell properties, such as self-renewal, in more mature progenitors. Molecular differences between primary human HSCs and progenitors have been well characterized by gene expression profiling, thereby enabling a rational approach to introduce candidate stem cell genes back into progenitors (Doulatov et al., 2010; Laurenti et al., 2013).
Re-specification was likely first demonstrated in mouse ESCs using the homeobox transcription factor HoxB4. Expression of HoxB4 in lineage-restricted hematopoietic progenitors isolated from EB differentiation or from yolk sac endowed them with extended self-renewal and long-term multi-lineage engraftment capacity (Kyba et al., 2002). HOXB4 has not functioned similarly in human ESCs (Lee et al., 2008; Wang et al., 2005), nor HSCs (Amsellem et al., 2003), suggesting species-specific differences. These studies however prompted us to search for factors tailored for hPSCs. Here, we identify HOX- and ETS-family transcription factors HOXA9 and ERG as inducers of self-renewal and multi-lineage potential in hematopoietic progenitors differentiated from hPSCs. Addition of SOX4 and MYB modulates this network to enable myeloid and erythroid engraftment, paving the way for in vivo models of hematological diseases from human IPSCs.
Results
Selection of candidate transcription factors
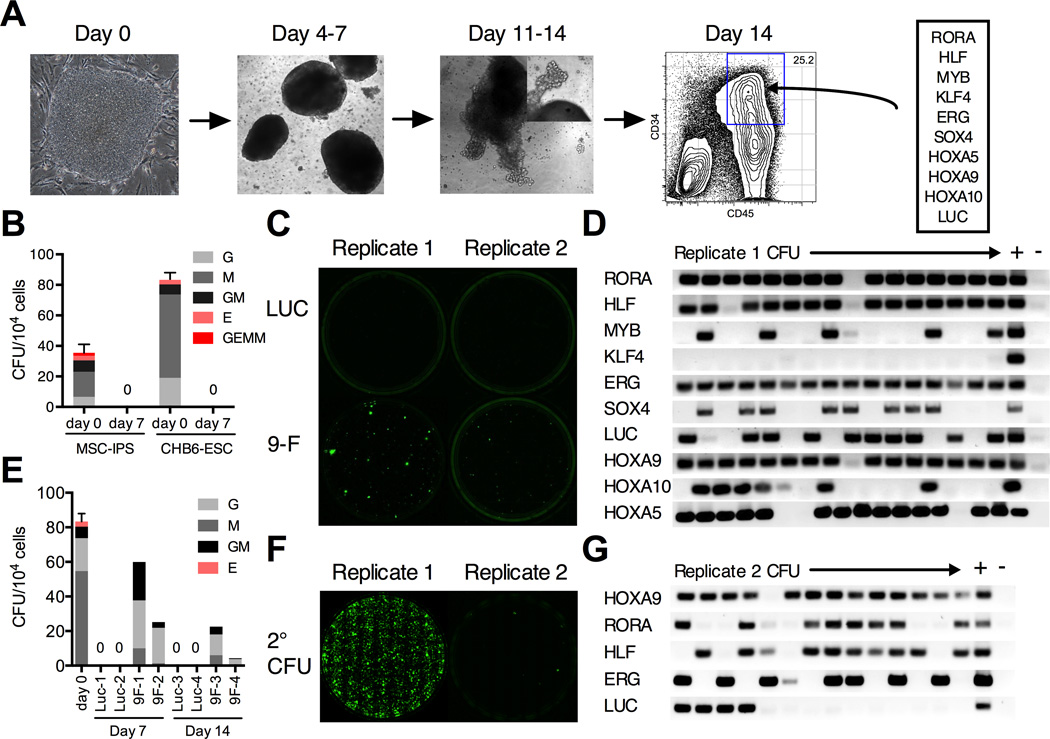
We hypothesized that introduction of stem cell transcription factors into hematopoietic progenitors derived from hPSCs would endow these cells with stem cell properties such as self-renewal potential. Gene expression of purified mouse and human HSCs has been extensively analyzed. Since functionally relevant factors are likely conserved, we used two independent mouse (Chambers et al., 2007; Ivanova et al., 2002) and two human datasets (Doulatov et al., 2010; Novershtern et al., 2011) to select transcriptional regulators more highly expressed in HSCs compared with progenitors or mature cells. Factors enriched in either of the two species-specific datasets were intersected to identify conserved regulators. These were then compared with gene sets differentially regulated during ontogeny to select factors upregulated in definitive HSCs (McKinney-Freeman et al., 2012). From this list of 32 HSC-specific transcription factors, 9 are significantly under-expressed in CD34+CD45+ EB progenitors compared with CD34+CD38− cord blood (CB) HSCs (Wang et al., 2005). These are ERG, KLF4, SOX4, MYB, RORA, HLF, HOXA5, HOXA9, and HOXA10. These open reading frames were cloned into a pSMAL-GFP lentiviral vector. Lentiviruses were produced as a mini-library of 9 factors (9F), plus luciferase (LUC) which serves as an internal control (Figure S1A).
An in vitro screen for self renewal capacity
To generate hematopoietic progenitors, hPSCs were differentiated as EBs in the presence of BMP4 and cytokines, as previously described (Chadwick et al., 2003). An ESC line, CHB6, and an IPSC line, MSC-IPS1, were selected based on their robust hemogenic potential (Figure S1B). In these conditions, CD34+CD45+ progenitors appear at day 10 and expand until day 14 (Figure 1A). Since EBs in our protocol are cultured with serum, the day 14 progenitors likely represent a mixture of primitive and definitive cell types (Figure S1C). Progenitors isolated by flow sorting at day 14 possess robust myeloid colony-forming activity, but produce few erythroid or mixed colonies (Figure 1B). When cultured in serum-free media with cytokines, they rapidly differentiate and lose this potential, and after 7 days in culture no colonies are ever observed, indicating a complete lack of proliferative and self-renewing potential (Figure 1B). We reasoned that if a combination of factors allowed progenitors to proliferate and undergo self-renewal divisions - a basic property of HSCs, these would appear as rare colonies after extended culture. To test the candidate transcription factors in this simple screen, CD34+CD45+ progenitors were infected with the 9F library, cultured for 7 days, and plated in colony assays. Both CHB6 and MSC-IPS1 progenitors infected with 9F, but not LUC alone, consistently gave rise to colonies in independent experiments (Figure 1C). Single colonies were isolated and analyzed for the presence of integrated viral transgenes (Figure 1D). The frequency of LUC integrations controlled for infection efficiency, with more frequent integrations being under positive selection. Each colony harbored 4 – 8 integrations, with HLF, ERG, HOXA9, and RORA present in almost every colony. In an independent experiment with a lower transduction efficiency, HOXA9 and RORA were also under selection (Figure S1D). The colony-forming capacity could be sustained for at least 3 weeks in culture (Figure 1E). To further investigate this renewal potential, we carried out serial replating. Some, but not all, 9F–infected replicates had robust replating potential (Figure 1F). Almost all colonies from the replicates with high replating potential had HLF, ERG, HOXA9, and RORA integrations. By contrast, colonies from the poorly replating replicates harbored fewer integrations (Figure 1G) suggesting that higher-order combinations were important.
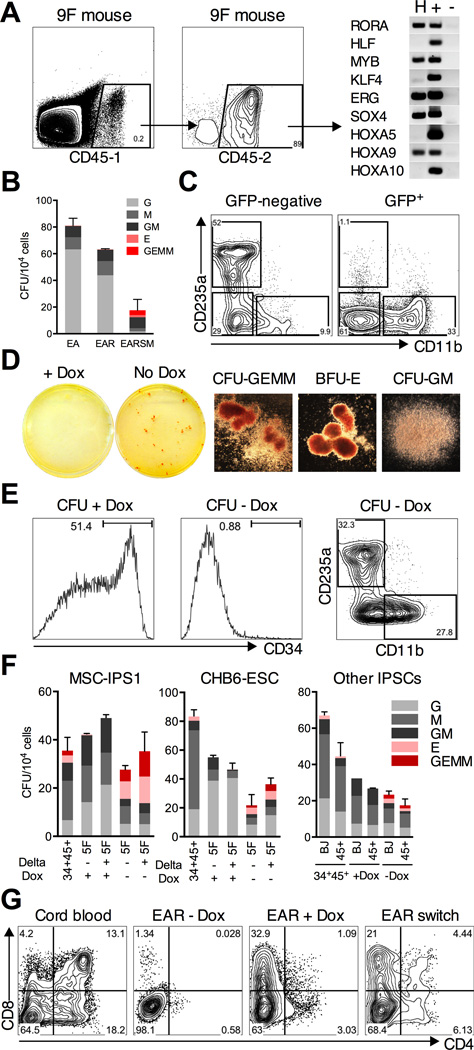
Figure 1. In vitro screen for progenitor re-specification.
(A) The scheme for EB differentiation of human hPSCs into hematopoietic progenitors. EBs cultured in serum, BMP4, and cytokines are dissociated after 14 days. CD34+CD45+ progenitors are isolated by flow sorting and transduced with a 9F lentiviral library. (B) Colony-forming capacity of CD34+CD45+ EB progenitors from MSC-IPS1 and CHB6-ESC after sorting (day 0) is lost after 7 days of culture in serum-free media (day 7). (C) Representative plates of CHB6 EB progenitors transduced with the 9F library showing emergence of de novo hematopoietic colonies after 7 days of culture. (D) Retrospective analysis of transgene insertions in individual colonies isolated from replicate #1 in (C). LUC is an internal control for transduction efficiency. Columns ‘+’ and ‘–’ are positive and negative controls for PCR. (E) Colony counts and types observed after 7 or 14 days of culture of luciferase (Luc) or 9F–transduced progenitors. (F) Serial replating potential of 9F replicates. (G) Transgene insertions for a non-replating sample (replicate #2 in C) show that most colonies do not contain higher-order combinations of the common integrations. Data are shown as mean ± SEM of 3 independent samples. See also Figure S1.
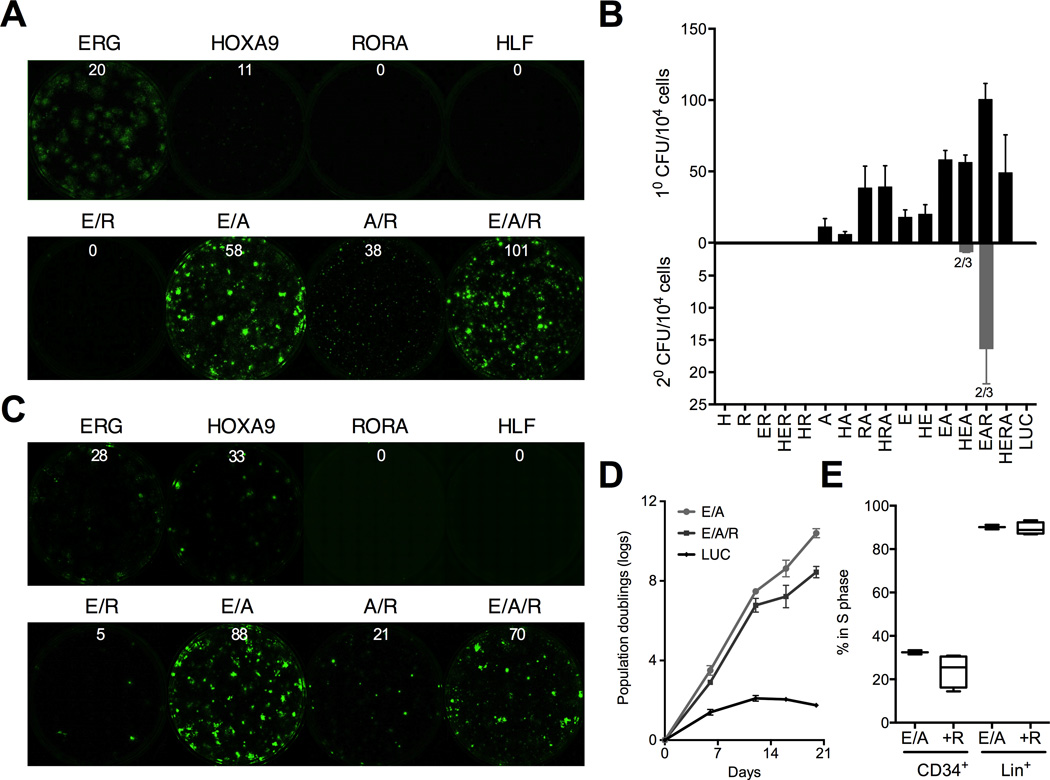
Retrospective analysis has identified HLF, ERG, HOXA9, and RORA as candidate factors that endow embryonic hematopoietic progenitors with self-renewal capacity. To test them prospectively, we infected CD34+CD45+ cells with all 16 combinations of the four factors, and assayed each by plating colonies at day 7 of culture. Rare colonies appeared in single ERG- or HOXA9-transduced samples, but both ERG and HOXA9 (abbreviated ‘EA’) were required for robust colony-forming activity in both CHB6 and MSC-IPS1 progenitors (Figure 2A–C and Figure S2A–B). The addition of RORA (abbreviated ‘EAR’) enhanced colony formation, while addition of HLF did not alter the number of colonies for any combination of factors (Figure 2B). EA colonies were larger in size, whereas EAR specified more colonies. Consistent with this, EA cultures proliferated more rapidly than EAR, which contained more progenitors with slower cell cycle kinetics (Figure 2D and 2E). No colonies were observed in any single-infected samples after 14 days of culture. ERG or HOXA9 single-transduced samples also never gave rise to secondary colonies implying that multiple factors are required for re-specification. By contrast, most of the EAR-transduced replicates could be serially replated (Figure 2B and S2A–B). Based on these data we included RORA in subsequent experiments.
Figure 2. Self-renewal is driven by HOXA9, ERG, and RORA.
Prospective analysis of HOXA9 (A), ERG (E), HLF (H), and RORA (R) combinations in CHB-ESC (A and B) and MSC-IPS (C) (see also Figure S2A) progenitors using the in vitro colony assay. Independent pluripotent lines are shown to demonstrate reproducibility of the factor combinations. Representative plates for combinations of HOXA9, ERG, and RORA are shown in (A) and (C), and the full quantitation of primary and secondary colony-forming efficiency in (B). Numbers above the plates indicate an average number of colonies per 104 cells plated. Numbers below secondary bars indicate the fraction of replicates that gave rise to secondary colonies. (D) Proliferation of ERG, HOXA9 and RORA (EAR)- transduced progenitors compared with ERG and HOXA9 without RORA (EA), and LUC control. (E) Cell cycle analysis of EA- and EAR-induced CD34+ cells and the more mature lineage-positive (Lin+; defined as CD14+, CD15+, or CD11b+) cells with or without RORA after 14 days of culture. All data are shown as mean ± SEM of at least 3 independent replicates. See also Figure S2.
Our data suggest that activation of self-renewal in hPSC-derived progenitors requires at least two transcription factors – HOXA9 and ERG. Notably, EA also promoted renewal of CB CD34+ progenitors in vitro, and the possibility of re-specifying adult progenitors to HSPCs warrants further exploration (Figure S2C). HOXA9 is the key homeotic gene that defines HSPC identity (Di-Poi et al., 2010; Lawrence et al., 2005). ERG is a component of HSC renewal during embryogenesis and stress hematopoiesis (Loughran et al., 2008). ERG is also a transcriptional target of HOXA9 suggesting a basis for their functional cooperation in our assay (Huang et al., 2012).
HOXA9, ERG, and RORA establish a hematopoietic hierarchy
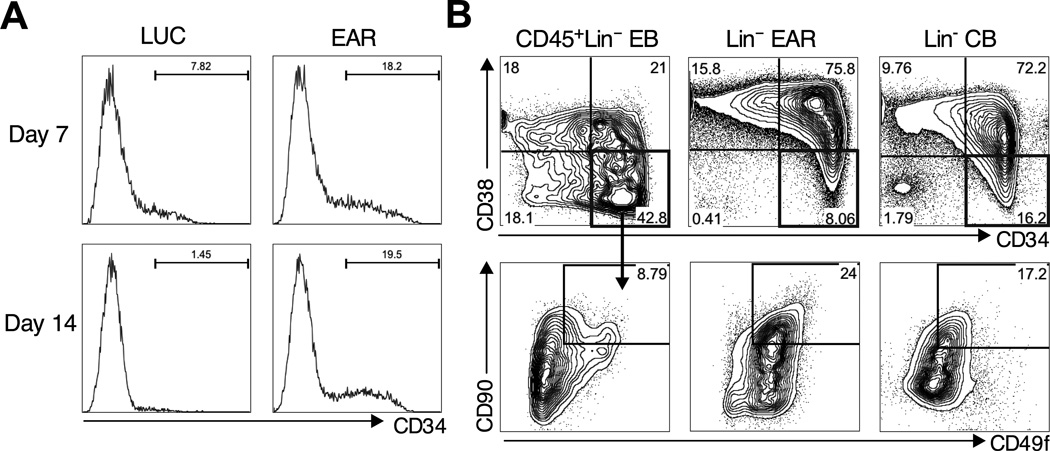
Individual lineages in the hematopoietic hierarchy can be accurately distinguished by a combination of cell surface antigens. We next examined if EAR-induced hematopoiesis generated progenitor subsets that resemble those found in CB or adult marrow. Cultured EB CD34+CD45+ progenitors completely differentiate into CD34− cells after 7 days of culture consistent with loss of clonogenic capacity (Figure 3A). By contrast, a CD34+ population was maintained in EAR cultures, expanding between 7 – 14 days, and persisting after that (Figure 3A). CD34+ cells were also found in dissociated primary colonies, accounting for serial replating. CD34 expression segregated with the lineage-negative population that did not express myeloid markers CD14, CD15 or CD11b. Human HSCs are found exclusively in the CD34+CD38− compartment, and the loss of stem cell potential is correlated with the gradual acquisition of CD38 and loss of CD34 expression (Mazurier et al., 2003). The starting EBs lack a clearly defined CD34+CD38−population (Figure 3B). By contrast, EAR established a pattern of CD34 and CD38 closely resembling that of lineage-depleted CB (Figure 3B). CD90+CD49f+ cells could be further identified within the CD34+CD38− EAR compartment, which are highly enriched for HSCs in CB (Figure 3B) (Notta et al., 2011). Thus, the three factors establish a rudimentary HSPC hierarchy, even in the presence of constitutive transgene expression.
Figure 3. Factor-based establishment of hematopoietic hierarchy.
(A) Progressive loss of CD34+ progenitors in control serum-free cultures (left panels, LUC), while EAR maintains and expands CD34+ cells (right panels). (B) Detailed analysis of the phenotypic HSC compartment in EBs, EAR cultures (from dissociated colonies), and primary CB. Lineage-positive cells are gated out (using a combination of CD14/CD15/CD11b for EBs and EAR). CD34 and CD38 delineates CD34+CD38− HSCs and CD34+CD38+ progenitors (top). CD34+CD38 fraction is further marked by CD90 and CD49f (bottom) with double positive cells being the most primitive in CB.
HOXA9, ERG, and RORA re-activate expression of HSC genes
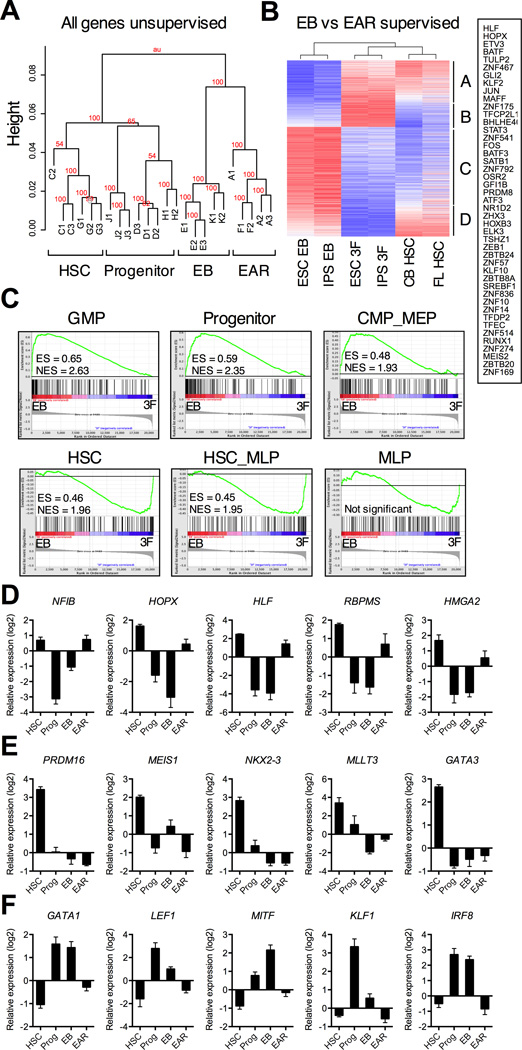
To clarify the molecular basis for self-renewal of re-specified progenitors, we performed microarray profiling of CD34+CD38− cells isolated by flow sorting from EBs and EAR cultures, as well as primary CD34+CD38− HSCs and CD34+CD38+ progenitors from CB and fetal liver (FL). Unsupervised clustering showed that these populations segregated to distinct branches indicating broad differences in gene expression (Figure 4A). To define the molecular program initiated by the factors, we identified genes induced or repressed greater than 2-fold by EAR in EB progenitors (ttest, FDR <0.05; Table S1). The majority of these 2070 genes were concordantly expressed in HSCs (Figure 4B), including genes on in HSCs activated by EAR (group A) and genes off in HSCs repressed by EAR (group C). The transcription factors induced by EAR include RUNX1, HLF, AP-1 family (FOS, JUN, MAFF), and GFI1B, all critical elements of a normal HSC program (Figure 4B). To analyze this using a different metric, we employed refined signatures for the human HSPC hierarchy (Laurenti et al., 2013). Six signatures capture the major patterns of early transcription: HSC, MLP (early lymphoid), HSC_MLP (shared stem and lymphoid), GMP (myeloid), CMP_MEP (erythroid), and Progenitor (shared between all progenitors). HSC and HSC_MLP signatures were significantly enriched in EAR compared to EBs (Figure 4C, bottom row). By contrast, progenitor signatures were enriched in EBs (Figure 4C, top row). These data show that while factor-transduced progenitors retain similarity to EBs, EAR initiates a gene expression program that is shared with normal HSCs.
Figure 4. Gene expression analysis of re-specified progenitors.
CD34+CD38− cells re-specified with EAR (fractions A and F) were compared to their respective starting CHB6-ESC and MSC-IPS EBs (fractions E and K), primary CB and FL HSCs (fractions C and G) and CD34+CD38+ progenitors (fractions D and H) by Affymetrix microarray profiling. Each sample was isolated by flow sorting with 2 or 3 biological replicates (#1–). (A) Unsupervised global clustering of all probes. Bootstrap values indicate confidence levels. (B) Supervised clustering of 2070 genes differentially expressed between CD34+CD38−EBs and EAR progenitors re-specified from them (>2-fold; ttest, FDR <0.05). Clusters of differential genes are marked A-D. Transcription factors significantly upregulated by EAR (from clusters A and B) are boxed on the right. See Table S1 for a full list of genes. (C) Gene set enrichment of human HSC and progenitor signatures in EBs compared with EAR progenitors. ES = enrichment score; NES = normalized enrichment score; all comparisons were significant with FDR <10−4, except MLP. (D-F) Expression levels of top differentially expressed transcription factors (ranked by fold change) between CB and FL HSCs and progenitors (>2-fold; ttest, FDR <0.05; see Table S2 for a full gene list). Expression values are median transformed relative to all probes. (D) HSC transcription factors significantly upregulated by EAR. (E) HSC transcription factors not induced EAR. (F) Progenitor transcription factors significantly repressed by EAR. Data are shown as mean ± SEM of 6 total biological replicates for HSCs, or 5 replicates for progenitors, EBs, and EAR. See also Figure S3, Table S3 and S4.
To further analyze transcriptional changes induced by the three factors, we performed gene-set enrichment analysis for annotated gene sets in the MSigDB database. HSC gene sets and HOXA9 targets were enriched in EAR, while cell cycle, serum response, and oncogene-induced gene sets were enriched in EBs (Figure S3A). To test if the factors turn on some HSC-specific genes and repress progenitor-specific genes, we compiled a list of genes differentially expressed between CB/FL HSCs and progenitors (ttest, FDR <0.05; Table S2). Genes on in HSCs and off in progenitors were largely off in EBs indicating a loss of stem cell gene expression. EAR activated some of these genes, although the trend did not reach significance (Figure S3B). This was mirrored by HSC-specific transcription factors, most of which were under-expressed in EB progenitors (Figure 4D and 4E). We discerned two patterns of expression in response to EAR: factors re-activated by EAR to levels similar in HSCs, including HLF, NFIB, HOPX, HMGA2, and RBPMS (Figure 4D), and transcription factors not affected by EAR, such as GATA3, PRDM16, EVI1, MLLT3, and NKX2–3 (Figure 4E). HOXA9 co-factors MEIS1 and PBX1 also belonged to this group. Genes off in HSCs and on in progenitors, including myeloid and erythroid transcription factors, such as GATA1, KLF1, LEF1, CEBPA, and MITF, were highly expressed in EBs, and significantly downregulated by EAR (Figure 4F and Figure S3B). These data suggest that EB progenitors lack the functional properties of HSCs not only because they fail to upregulate HSC-specific genes, but also because they express negative regulators of stem cell function that define differentiated cells (see Table S3 for EB vs HSC gene list). While re-specified progenitors remain different from HSCs (see Table S4 for EAR vs HSC gene list), EAR upregulated a subset of HSC-specific genes and repressed progenitor genes, which likely accounts for their capacity to induce self-renewal in progenitors.
In vivo engraftment requires SOX4 and MYB
We next tested if transcription factor re-specified progenitors can successfully engraft in irradiated mice. 1 × 106 EAR cells were transplanted after 2 weeks of primary culture into NOD/SCID-IL2Rγnull (NSG) mice. We found no detectable human engraftment 4 weeks after transplant using two independent CD45 antibodies to accurately detect human chimerism (n = 10). This indicated that either EAR was insufficient for even short-term engraftment, or that changes induced during culture impaired their capacity for homing and retention in the bone marrow. At the same time, we also transplanted 9F library-transduced progenitors. One mouse transplanted into the right femur had a 0.2% human engraftment after 4 weeks (Figure 5A). The integration pattern in CD45+ cells isolated by flow sorting showed the presence of SOX4 and MYB transgenes in addition to EAR (Figure 5A). This finding suggested that different transgenes were selected during in vivo reconstitution, and led us to believe that the addition of SOX4 and MYB to our cocktail of transcription factors would enable engraftment of EB-derived progenitors.
Figure 5. Re-specification of myeloid-restricted precursors into multi-lineage progenitors.
(A) Detailed analysis of human engraftment in a mouse transplanted with 9F cells after 14 days of culture. Two independent CD45 antibodies (CD45–1 and −2) were used to label human cells. (Right) Transgene insertions in CD45+ cells (column H) isolated from this mouse by flow sorting show SOX4 and MYB integrations. (B) Colony-forming efficiency of EB progenitors transduced with EA, EAR (+R), and EARSM (+SM) after 14 days of culture. (C) Flow analysis of dissociated EARSM colonies using CD235a (erythroid) and CD11b (myeloid) lineage markers. At least ten GEMM colonies were pooled, and plots are gated on GFP+ or -negative populations. (D) Colonies plated with or without Dox in the inducible transgene system. Erythroid and mixed colonies appear only upon Dox withdrawal. (Right) Representative images of CFU-GEMM, BFU-E, and CFU-GM colonies on plates without Dox. (E) Self-renewing CD34+ progenitors differentiate into myeloid and erythroid cells upon Dox removal. Flow analysis of dissociated EARSM colonies using the CD34 progenitor marker (left panels) and lineage markers CD11b and CD235a (right panel). (F) Re-specification of EB myeloid precursors into mixed lineage CFU-GEMM progenitors from MSC-IPS1 (left), CHB6-ESC (middle), and two other myeloid-restricted IPSC lines (right). EB progenitors were infected with EARSM and cultured with or without immobilized Delta1 ligand. Each group was plated with or without Dox. (G) T cell potential of re-specified cells in an OP9-DL1 stromal co-culture. EAR-induced cells were cultured on OP9-DL1 without Dox (EAR - Dox), with Dox (EAR + Dox), or with Dox for the first 2 weeks followed by Dox removal (EAR switch). Development of CD4+CD8+ T cells was assessed following 35 days of culture, compared with CB CD34+ cells. Plots are gated on CD45+ cells. Data in (B) and (F) are shown as mean ± SEM of at 3 independent replicates. See also Figure S4 and S5.
Transcription factors re-specify myeloid-restricted precursors into multi-lineage progenitors
To investigate the functional outcome of combining SOX4 and MYB with HOXA9, ERG, and RORA (EARSM), we infected CD34+CD45+ progenitors as before, and carried out colony assays after 14 days of culture. While EAR gave rise to only myeloid colonies, the addition of SOX4 and MYB prompted the appearance of a large number of blast erythroid (BFU-E) and mixed myelo-erythroid (CFU-GEMM) colonies (Figure 5B). The appearance of GEMM colonies was notable, since these are typically not detected, or very rare, among CD34+CD45+ EB progenitors. Mixed colonies contained both CD11b+ myeloid and CD235a+ erythroid cells. Myelo-erythroid cells were only found in the GFP-negative fraction, whereas GFP+ cells were CD11b+ or CD34+ (Figure 5C), suggesting that erythroid potential was only realized upon transgene silencing.
To explore this possibility, we switched to an inducible lentiviral system in which rtTA is expressed from the same vector as the transgene (Meerbrey et al., 2011). Progenitors were infected with five inducible lentiviruses and cultured in doxycycline (Dox) before plating colonies with or without Dox. As predicted, GEMM colonies formed only upon Dox withdrawal (Figure 5D). Colonies plated with Dox contained a mixture of CD11b+ cells and CD34+ self renewing progenitors; rare erythroid cells were always derived from transgene silencing (Figure 5E). Upon Dox withdrawal, CD34+ progenitors differentiated completely into CD235a+CD71+ erythroid or CD11b+ cells (Figure 5E). Rapid cessation of self-renewal upon Dox withdrawal is explained by the lack of endogenous HOXA9 and ERG transcription, making the system sensitive to induced transgene expression (Figure S4). This presents an attractive system with a high degree of control over self-renewal versus differentiation to model normal and disease hematopoiesis.
Two hypotheses can be put forward to account for the appearance of mixed colonies after factor transduction. One possibility is expansion of the rare GEMM progenitors present in the day 14 EBs. Due to heterogeneity among hPSC lines, rare GEMM colonies were in fact detected in MSC-IPS1, but not CHB6 EB progenitors (Figure 5F). In stark contrast, the emergence of GEMM colonies after EARSM transduction was efficient (up to 25% of all colonies) and highly reproducible with both CHB6 and MSC-IPS1 lines (Figure 5F). We tested the effect of culture on immobilized Delta1, which has been shown to support the expansion of HSPCs by providing a low level Notch signal (Delaney et al., 2005). Delta1-cultured progenitors trended towards higher numbers of GEMM colonies, but the trend did not reach significance. To examine this further, we screened a panel of IPSC lines and identified two lines - BJ-IPS1 and CD45-IPS1, derived from normal foreskin fibroblasts and CD45+ peripheral blood cells respectively, that did not give rise to BFU-E or GEMM colonies (Figure 5F, “Other IPSCs”). Both lines reproducibly gave rise to erythroid and mixed colonies after induction with EARSM (Figure 5F). These data argue against the possibility that the factors simply expand progenitors present in the starting EBs, and instead suggest that they actively re-specify myeloid-restricted precursors to a myelo-erythroid fate.
Lymphoid potential is critical evidence for re-specification, since EB progenitors do not generate B or T cells (Martin et al., 2008). To test lymphoid potential, we seeded EAR-transduced cells on OP9-DL1 stroma that promotes T cell differentiation. CB CD34+ cells progress through well-defined stages of differentiation starting with the acquisition of early lymphoid markers CD7 and CD5, and culminating with CD4+CD8+ lymphocytes. We noted that EAR cultures from which Dox was removed failed to proliferate; however continued exposure to Dox reduced maturation past the CD7+ stage. Cultures in which Dox was added for the first 2 weeks only showed improved maturation, with more than 4% of lymphocytes reaching the CD4+CD8+ stage after 5 weeks (Figure 5G). Similar frequencies of CD1a+ and CD3+ cells were observed (Figure S5). By comparison, >13% of CB CD34+ cells were CD4+CD8+ with robust co-expression of CD1a and CD3 (Figure 5G and S5). These data show that factor-induced progenitors harbor lymphoid potential indicative of re-specification to a definitive cell type. However their lymphoid potential is considerably less than for adult progenitors, and further optimization of transcription factor combinations is needed.
Re-specified progenitors mediate short-term myeloid and erythroid engraftment
The establishment of a robust and reproducible transplantation model from hPSCs is an important goal. To test the engraftment potential of re-specified progenitors, we used the inducible transgene system to expand EB-derived progenitors for 14 days with Dox, and transplanted 0.8 – 1.2 million cells (expanded from <5,000 EB CD34+CD45+ cells) into the femur of adult NSG mice. Human engraftment was evaluated after 4 or 8 weeks using two human CD45 antibodies, erythroid (CD235a), myeloid (CD33) and lymphoid (CD19, CD5) lineage markers.
Mice transplanted with MSC-IPS1 EARSM progenitors were engrafted after 4 weeks in the injected femur, and at lower levels, in the rest of bone marrow (Figure 6A and Figure S6A). We observed only minimal engraftment in mice when Dox was removed (Figure S6B). When Dox was administered, almost all injected mice were engrafted, with the average engraftment across several independent experiments and culture conditions of 2.1% (9 of 12 mice engrafted >0.1%). Similar engraftment levels were observed with CHB6-ESC progenitors (average 0.63%, 3 of 4 mice engrafted >0.1%) (Figure 6B). In most recipients, both myeloid and erythroid cells were present, however in some mice only myeloid, or only erythroid engraftment was found (Figure 6C). By contrast with CB-transplanted mice, we did not detect CD19+ B cells. To test the reproducibility of this system using independent IPSC lines, we transplanted EARSM-transduced progenitors derived from BJ-IPS and CD45-IPS lines. We observed a similar myelo-erythroid pattern of engraftment in these mice (Figure 6D). Taken together, 16 of 21 mice transplanted from four hPSC lines were engrafted, with the average engraftment of 2.7% (Figure 6E).
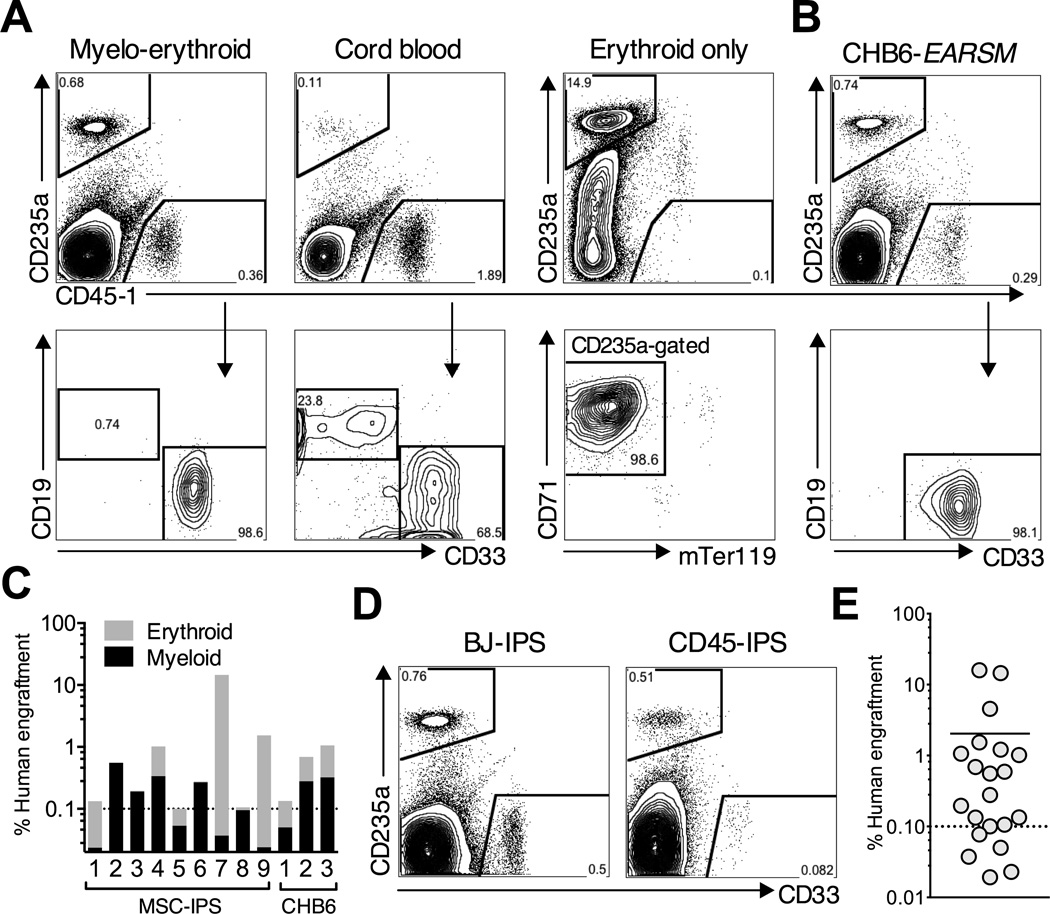
Figure 6. Short-term engraftment of re-specified progenitors.
EB progenitors derived from MSC-IPS (A) or CHB6-ESC (B) were re-specified with inducible EARSM in serum free culture with Dox for 7–16 days, and 0.8 – 1.2 × 106 cells were transplanted into the right femur of adult NSG mice. As a control, 1 × 106 CB mononuclear cells were transplanted. All mice were kept on Dox in the drinking water. Human engraftment was analyzed 4–5 weeks post-transplantation using two human CD45 (myeloid, lymphoid) and CD235a (erythroid) antibodies. Only engraftment the in injected femur is shown. (A) Representative myelo-erythroid engraftment in EARSM- (left) and cord blood- (middle) transplanted mice. Cells positive for CD45 are sub-gated on CD19 and CD33 (bottom row). Right: a representative mouse engrafted only with human erythroid cells. CD235a–positive cells are sub-gated on CD71 and mouse Ter119. (B) Representative engraftment in CHB6-ESC mice. (C) Myeloid and erythroid lineage distribution in engrafted (>0.1% of human chimerism) MSC-IPS and CHB6-ESC mice. The height of the bar indicates total engraftment level calculated as %CD235a plus %CD45+. (D) Representative engraftment in mice transplanted with independently-derived IPSC lines, BJ-IPS and CD45-IPS. (E) Summary of engraftment level in all transplanted mice with four different hPSC lines. See also Figure S6 and S7.
The presence of erythroid engraftment was notable since immunodeficient mice do not robustly support human erythropoiesis (Hu et al., 2011). We observed that several mice with symptoms of radiation sickness - low weight, enlarged spleens, and peripheral lymphocytopenia, had high levels of erythroid-only engraftment suggesting that intense myeloablation promotes erythropoiesis in this model (Figure 6C). Erythroid development in vivo was not inhibited by exposure to Dox possibly due to lower effective levels in tissues. Engraftment levels declined during extended transplantation. Low-level myeloid engraftment persisted 10 – 12 weeks post-transplant; however erythroid cells were not detectable beyond 6 weeks (Figure S6C). These findings demonstrate that progenitors re-specified from multiple hPSC lines mediate reproducible short-term engraftment.
To determine the basis for the beneficial effect of SOX4 and MYB on engraftment, we carried out expression profiling of CD34+CD38- cells induced with the five factors. These samples clustered closely to EAR indicating an overall similarity between all re-specified progenitors (Figure S7A). Gene ontology categories for hematopoietic development and immune function were enriched among genes differentially regulated by EARSM relative to EAR (Figure S7B). EAR was associated with aberrant induction of a number of genes, such as heat-shock proteins (HSPA6) and Fcγ receptors (FCGR2A, B, C) (Figure S7C). Repression of these transcripts by SOX4 and MYB may restore engraftment capacity of in vitro-specified cells.
Globin switching during in vivo human erythropoiesis
A pervasive feature of erythroid differentiation from hPSCs is the retention of embryonic and fetal patterns of globin expression (Chang et al., 2006; Dias et al, 2011). This property has been reported using independent protocols suggesting that erythroid precursors fail to receive cues that promote proper developmental maturation reflected by globin gene switching (Anstee et al, 2012). Consistent with reported findings, BFU-Es isolated from CD34+CD45+ EBs expressed a combination of embryonic ε globin and fetal γ globin, but little or no adult β globin (Figure 7A). Re-specification with EARSM did not alter this pattern of globin expression in vitro (Figure 7A). An important question that we sought to address is whether the adult bone marrow microenvironment could provide the cues to alter globin preference. To this end, we analyzed erythroid engraftment in EARSM-transplanted mice. Mouse erythrocytes were gated out with anti-mouse Ter119, showing that the majority of human erythrocytes were CD235a+CD71+ (Figure 7B). The marrow of cytopenic mice contained more Ter119+ cells reflective of stress erythropoiesis, accompanied by progressive maturation of human erythrocytes (Figure 7B). The majority of sorted human CD235a+CD71+ cells were at the late erythroblast – normoblast stage of differentiation by morphology (Figure 7C), and marked by high levels of Band3 and low levels of CD49d (Figure 7D). Human erythrocytes do not readily enucleate in vivo; however we detected some enucleated cells both by morphology and nuclear dye uptake (Figure 7C and 7D). Thus, hPSC-derived progenitors undergo erythroid maturation in vivo.
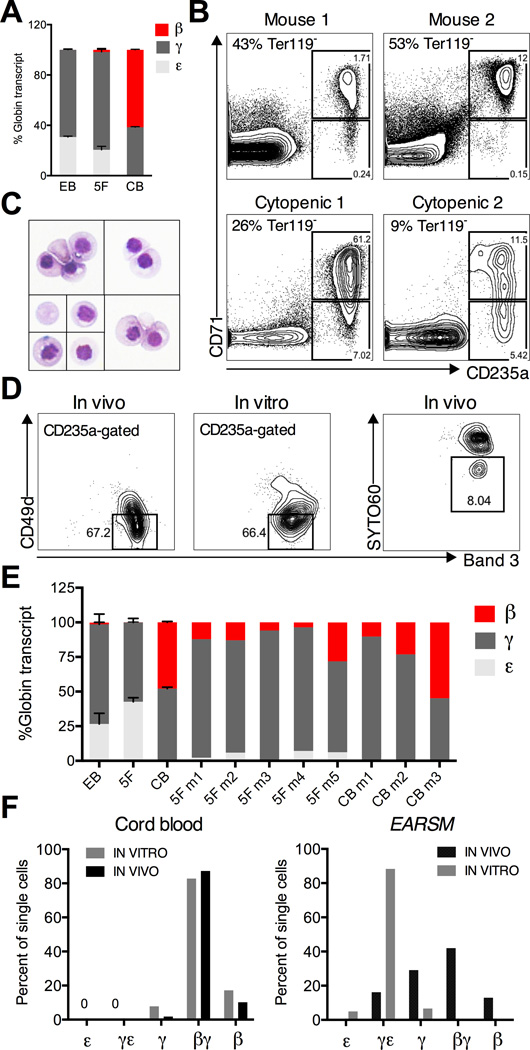
Figure 7. In vivo globin switching in embryonic erythrocytes.
(A) Quantitative PCR for the expression of adult HBB (β), fetal HBG (γ), and embryonic HBE (ε) hemoglobin genes in BFU-E colonies derived from EBs, EARSM (5F), and CB. Expression of each gene was calculated using absolute quantification and displayed as percent of total globin transcript. (B) Analysis of human erythroid graft in two normopenic (Mouse 1 and 2) and two cytopenic EARSM mice using human CD71 and CD235a. Plots are sub-gated on Ter119-negative mouse cells, and percent of mouse erythrocytes is indicated. (C) Giemsa staining of Ter119-CD71+CD235+ human erythrocytes. Representative single cells and clusters are shown. (D) Erythroid maturation in vivo and in vitro determined by expression of Band3 and CD49d (integrin α4). Progressive maturation is marked by high levels of Band3 and loss of CD49d (gated population). Only CD235a+ cells are shown. (Right panel) The proportion of enucleated erythrocytes in vivo using cell-permeable nuclear dye SYTO60 (gated SYTO60-negative population). (E) Hemoglobin expression in Ter119-CD71+CD235+ human erythroblasts isolated from the marrow of multiple mice transplanted with CB or EARSM progenitors, in comparison with BFU-Es from EBs, 5F, and CB. (E) Hemoglobin expression in single Ter119-CD71+CD235+ erythroblasts isolated from transplanted mice (IN VIVO) or from cultured BFU-Es (IN VITRO). The cut-off for co-expression was set at 10% (i.e. a cell with 90/10% ratio of β/γ transcript is classified as βγ, while 95/5% as β-only). Single cell expression was validated by comparing against wells sorted with 50 cells. Data in (A) and (E) is shown as the mean ± SEM of at least 2 samples. Data in (F) represents >80 sorted single cells.
We next examined the pattern of hemoglobin expression in CD235a+CD71+ erythroblasts isolated from engrafted mice. Remarkably, ε globin was suppressed (4.0% of total globin, range 0.9 –7.1%), and β globin (12.6% of total globin, range 3.6 – 28.1%) was induced, compared to cultured erythroblasts (Figure 7E). As expected, mice engrafted with CB expressed a mixture of γ and β globins. These findings were supported by analysis in single sorted CD235a+CD71+ cells. Single CB cells isolated from colonies or engrafted mice almost uniformly co-expressed γ and β globins (Figure 7F). Remarkably, 41.9% of EARSM-engrafted cells co-expressed γ and β globin, and a significant proportion (12.9%) expressed only β globin (Figure 7F). By comparison, adult globin-expressing cells were never detected in vitro (Figure 7F). The total levels of globin transcripts, and the degree of erythroid maturation as evidenced by Band3 and CD49d were similar between cells isolated in vitro or in vivo (Figure 7D). These findings demonstrate that human IPSC-derived erythroid precursors re-specified with transcription factors undergo hemoglobin switching in vivo, giving rise to definitive erythrocytes.
Discussion
Generation of engraftable HSPCs from human pluripotent cells is a major goal in regenerative medicine. Here we report a significant advance towards this goal using a novel transcription factor-mediated approach. In our system hPSCs are differentiated to hematopoietic progenitors and then transduced with a combination of HSC transcription factors that aim to “re-specify” these progenitors to a stem cell fate. Using an in vitro screen to identify relevant combinations of factors, we demonstrate that HOXA9, ERG, and RORA convert myeloid-restricted precursors into self-renewing multi-lineage HSPCs with erythroid and lymphoid potential. These factors are sufficient to expand progenitors while maintaining a hierarchy with CD34+CD38− “stem cells”. HOXA9 and ERG can be further modified by SOX4 and MYB to enable short-term myelo-erythroid engraftment. These findings establish the first transcription factor-based system for engraftment from hPSCs, which will serve as a platform for further improvements in transplantation potential.
The advent of IPSC technology has enabled the generation of pluripotent lines from patients with hematologic diseases. The hope has been that these lines can serve as models to gain insight into the molecular basis of these diseases and as platforms for drug screens (Cherry and Daley, 2013). However, the inability to generate large numbers of engraftable cells has precluded disease modeling in vivo, thereby limiting the scope of experiments and drug screens that can be performed. Our system described here may be suitable for modeling anemias and other red blood cell disorders, due to the ability to generate large numbers of erythroid precursors in vitro, robust erythroid engraftment that can be enhanced by intensive myeloablation, and a high degree of hemoglobin maturation in vivo. Due to the near-unlimited capacity to produce erythroid cells, this transcription factor approach may also be suitable for transfusion-related applications (Migliaccio et al., 2012).
Erythroid precursors differentiated from hPSCs express embryonic and fetal but not adult hemoglobin (Chang et al., 2006). Induction of a switch to adult globin, critical for modeling thalassemias and sickle cell anemia caused by mutations in the HbB locus, has been the subject of intense investigation. Silencing of embryonic and fetal globins is mediated by BCL11A, in tandem with KLF1, MYB, and others (Sankaran and Orkin, 2013; Sankaran et al., 2009). One possibility is that the expression or activity of these factors is not induced during hPSC differentiation in vitro resulting in the persistence of embryonic and fetal globins. The finding that erythrocytes in transplanted mice silence embryonic and induce adult globins supports the long-standing hypothesis that the bone marrow microenvironment can promote globin switching. This raises the possibility that globin switching is initiated by specific signaling pathways, which activate BCL11A and other regulators following the migration of HSCs from fetal liver to bone marrow after birth. The nature of these signals remains completely unknown. Since globin switching is regulated in a species-specific manner, transplantation of hPSC progenitors can serve as a model to interrogate the nature of the signaling interactions that drive globin switching (Sankaran et al., 2009).
Transcription factors that control stem cell state act as part of complex gene regulatory networks, and while these networks have been extensively modeled, functional studies have largely focused on the role of individual factors in controlling HSC fate (Orkin and Zon, 2008; Wilson et al., 2010). The re-specification approach presents an opportunity to identify functional networks of cooperating factors. HOXA9 and ERG have been known to be required for HSC specification and expansion (Lawrence et al., 2005; Loughran et al., 2008). Both factors are also known oncogenes, and it is now well-established that normal self renewal programs, if constitutively activated or dysregulated, can promote aberrant expansion of leukemic cells (Eppert et al., 2011). Our findings demonstrate that HOXA9, ERG, RORA, SOX4 and MYB are part of the same functional gene regulatory network. HOXA9 occupies ERG, SOX4, and MYB promoters linking them on a molecular level (Huang et al., 2012). We propose that this network may have an important role in adult HSCs and leukemia-initiating cells. Our approach may provide deeper insights into the principles underlying the organization of HSC networks.
Current strategies for deriving HSPCs from heterologous cell types are based on directed differentiation from hPSCs, or reprogramming of somatic cells (Pereira et al., 2013). We propose a hybrid strategy aiming to re-specify HSCs from closely related progenitors by activating stem cell gene regulatory networks. However, re-specification with our current factors has not achieved the major goal of generating a cell with long-term and multi-lineage repopulation potential. HSC state is controlled by a large number of transcription factors and epigenetic modifiers (Doulatov et al., 2012; Orkin and Zon, 2008). Powerful screening strategies are thus needed to identify rare combinations of cooperating factors. Our approach is based on detecting clonogenic progenitors arising as a result of self-renewal divisions in culture. However, these cell states may be incompatible with the in vivo requirements for HSC function. The alternative is directly to re-specify in vivo to take advantage of the cues that normally support HSC self-renewal. However, only very rare combinations can likely be detected in this setting. The strategy we describe here provides a logical platform for pursuing both in vitro and in vivo screens aiming to reveal improved sets of factors that extend stem cell properties of re-specified progenitors.
Experimental Procedures
Details on colony assays, flow cytometry, microarray profiling, mouse transplantation, assessment of human cell engraftment, and quantitative and single cell PCR are described in the Supplemental Experimental Procedures available online.
hPSC culture
All experiments were performed with CHB6 ESC (NIH #0006) and MSC-IPS1 (Park et al., 2008). Human ES and IPS cells were maintained on mouse embryonic fibroblasts (GlobalStem) feeders in DMEM/F12 + 20% KnockOut-Serum Replacement (Invitrogen), 1 mM L-glutamine, 1 mM NEAA, 0.1 mM β-mercaptoethanol, and 10 ng/ml bFGF. Media was changed daily, and cells were passaged 1:4 onto fresh feeders every 7 days using standard clump passaging with collagenase IV.
EB differentiation
EB differentiation was performed as previously described (Chadwick et al., 2003). Briefly, hPSC colonies were scraped into non-adherent rotating 10 cm plates. EB media was KO-DMEM + 20% FBS (Stem Cell Technologies), 1 mM L-glutamine, 1 mM NEAA, penicillin/streptomycin, 0.1 mM β-mercaptoethanol, 200 µg/ml h-transferrin, and 50 µg/ml ascorbic acid. After 24 hrs, media was changed by allowing EBs to settle by gravity, and replaced with EB media supplemented with growth factors: 50 ng/ml BMP4 (R&D Systems), 300 ng/ml SCF, 300 ng/ml FLT3, 50 ng/ml G- CSF, 20 ng/ml IL-6, 10 ng/ml IL-3 (all Peprotech). Media was changed on day 5, and day 10. EBs were dissociated on day 14 by digesting with collagenase B (Roche) for 2 hrs, followed by treatment with enzyme-free dissociation buffer (Gibco), and filtered through an 80 µ m filter. Dissociated EBs were frozen in 10% DMSO, 40% FBS freezing solution.
Lentivirus production
Plasmids for the 9F library were obtained as Gateway plasmids (GeneCopoeia). ORFs were cloned into lentiviral vectors using LR Clonase (Invitrogen). Two vectors were used: pSMAL-GFP (constitutive) and pINDUCER-21 (Meerbrey et al., 2011). Lentiviral particles were produced by transfecting 293T-17 cells (ATCC) with the 3rd-generation packaging plasmids. Virus was harvested 12 and 36 hrs after transfection and concentrated by ultracentrifugation at 23,000 rpm for 2 hrs. Constructs were titered by serial dilution on 293T cells.
Progenitor sorting
Dissociated EB cells were thawed following the Lonza Poietics protocol(http://bio.lonza.com/uploads/tx_mwaxmarketingmaterial/Lonza_ManualsProductInstructions_Procedure_for_Thawing_Poietics_Cells.pdf) and resuspended at 1×106 per 100 µl staining buffer (PBS + 2% FBS). Cells were stained with a 1:50 dilution of CD45 PE-Cy5 (Immu19.2; Clontech), CD34 PE-Cy7 (8G12; BD), and DAPI for 20 min at RT. All sorting was performed on a BD FACS Aria II cell sorter using a 70 µm nozzle.
Lentiviral gene transfer
Sorted CD34+CD45+ progenitors were seeded on fibronectin-coated (10 ug/cm2) 96 well plates at a density of 2 – 5 ×104 cells per well. The infection media was IMDM + 20% BIT (StemCell Technologies), 1 mM L-glutamine, and 0.1 mM P-mercaptoethanol, with 300 ng/ml SCF, 300 ng/ml FLT3, 50 ng/ml G- CSF, 20 ng/ml IL6, 10 ng/ml IL3 (all Peprotech). Lentiviral infections were carried out in a total volume of 150 ul. The multiplicity of infection for the factors was as follows: 9F 20, ERG 5.0, HOXA9 5.0, RORA 3.0, SOX4 3.0, MYB 3.0. Virus was concentrated onto cells by centrifuging the plate at 2500 rpm for 30 min at RT. Infections were carried out for 24 –36 hrs.
Progenitor culture
Following gene transfer, progenitors were cultured in suspension in infection media supplemented with 50 ng/ml SCF, 50 ng/ml FLT3, 50 ng/ml TPO, 50 ng/ml IL6, and 10 ng/ml IL3 (all R&D Systems). To standardize these conditions, for all experiments with inducible constructs (including all transplantation experiments), infection media was replaced with StemSpan SFEM (StemCell Technologies). Dox was added at 2 ng/ml (Sigma). For culturing on immobilized ligands, non-tissue culture plates were coated with 2.5 µg/ml Delta1 extIgG and 5 µg/ml recombinant human retronectin (CH296, Takara Bio) for 2 hrs at 37°C. Culture media was same as above. Cultures were maintained at a density of <1 ×106 cells/ml, and media was changed every 3–4 days.
Supplementary Material
Highlights.
Re-specification is a novel strategy to generate HSCs from committed progenitors
ERG, HOXA9, and RORA induce multipotent progenitors from human IPS cells
Addition of SOX4 and MYB enables robust short-term erythro-myeloid transplant
Engrafted human erythroblasts mature and undergo hemoglobin switching
Acknowledgements
The authors would like to thank Thorsten Schlaeger and the BCH ESC core, Ronald Mathieu from the flow cytometry core, and Norman Gerry from the Coriell Genotyping and Microarray Center, for assistance. Andrea Ditadi and Gordon Keller provided reagents and advice. Band3 antibody was a gift from the Narla Mohandas lab. GQD is supported by grants from the US National Institute of Diabetes and Digestive and Kidney Diseases (R24-DK092760) and the National Heart, Lung, Blood Institute Progenitor Cell Biology Consortium (UO1-HL100001); Alex’s Lemonade Stand; and the Doris Duke Medical Foundation. GQD is an associate member of the Broad Institute, and an investigator of the Howard Hughes Medical Institute and the Manton Center for Orphan Disease Research. SD is supported by a fellowship from Helen Hay Whitney Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Microarray data from this study has been deposited to GEO database under accession number GSE49938.
Supplemental Information
The online Supplemental Information that accompanies this article includes Experimental Procedures, Figures S1-S7, and Tables S1-S4.
References
- Agarwal S, Loh YH, McLoughlin EM, Huang J, Park IH, Miller JD, Huo H, Okuka M, Dos Reis RM, Loewer S, et al. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature. 2010;464:292–296. doi: 10.1038/nature08792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsellem S, Pflumio F, Bardinet D, Izac B, Charneau P, Romeo PH, Dubart-Kupperschmitt A, Fichelson S. Ex vivo expansion of human hematopoietic stem cells by direct delivery of the HOXB4 homeoprotein. Nature medicine. 2003;9:1423–1427. doi: 10.1038/nm953. [DOI] [PubMed] [Google Scholar]
- Anstee DJ, Gampel A, Toye AM. Ex-vivo generation of human red cells for transfusion. Current opinion in hematology. 2012;19:163–169. doi: 10.1097/MOH.0b013e328352240a. [DOI] [PubMed] [Google Scholar]
- Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- Chadwick K, Wang L, Li L, Menendez P, Murdoch B, Rouleau A, Bhatia M. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood. 2003;102:906–915. doi: 10.1182/blood-2003-03-0832. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Boles NC, Lin KY, Tierney MP, Bowman TV, Bradfute SB, Chen AJ, Merchant AA, Sirin O, Weksberg DC, et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell stem cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KH, Nelson AM, Cao H, Wang L, Nakamoto B, Ware CB, Papayannopoulou T. Definitive-like erythroid cells derived from human embryonic stem cells coexpress high levels of embryonic and fetal globins with little or no adult globin. Blood. 2006;108:1515–1523. doi: 10.1182/blood-2005-11-011874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry AB, Daley GQ. Reprogrammed cells for disease modeling and regenerative medicine. Annual review of medicine. 2013;64:277–290. doi: 10.1146/annurev-med-050311-163324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry AB, Gagne KE, McLoughlin EM, Baccei A, Gorman B, Hartung O, Miller JD, Zhang J, Zon RL, Ince TA, et al. Induced Pluripotent Stem Cells with a Pathological Mitochondrial DNA Deletion. Stem cells. 2013 doi: 10.1002/stem.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KD, Vodyanik MA, Togarrati PP, Suknuntha K, Kumar A, Samarjeet F, Probasco MD, Tian S, Stewart R, Thomson JA, et al. Identification of the hemogenic endothelial progenitor and its direct precursor in human pluripotent stem cell differentiation cultures. Cell reports. 2012;2:553–567. doi: 10.1016/j.celrep.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney C, Varnum-Finney B, Aoyama K, Brashem-Stein C, Bernstein ID. Dose-dependent effects of the Notch ligand Delta1 on ex vivo differentiation and in vivo marrow repopulating ability of cord blood cells. Blood. 2005;106:2693–2699. doi: 10.1182/blood-2005-03-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di-Poi N, Koch U, Radtke F, Duboule D. Additive and global functions of HoxA cluster genes in mesoderm derivatives. Developmental biology. 2010;341:488–498. doi: 10.1016/j.ydbio.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Dias J, Gumenyuk M, Kang H, Vodyanik M, Yu J, Thomson JA, Slukvin II. Generation of red blood cells from human induced pluripotent stem cells. Stem cells and development. 2011;20:1639–1647. doi: 10.1089/scd.2011.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nature immunology. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell stem cell. 2012;10:120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, Metzeler KH, Poeppl A, Ling V, Beyene J, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nature medicine. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- Garcon L, Ge J, Manjunath SH, Mills JA, Apicella M, Parikh S, Sullivan LM, Podsakoff GM, Gadue P, French DL, et al. Ribosomal and hematopoietic defects in induced pluripotent stem cells derived from Diamond Blackfan anemia patients. Blood. 2013 doi: 10.1182/blood-2013-01-478321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Van Rooijen N, Yang YG. Macrophages prevent human red blood cell reconstitution in immunodeficient mice. Blood. 2011;118:5938–5946. doi: 10.1182/blood-2010-11-321414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Sitwala K, Bronstein J, Sanders D, Dandekar M, Collins C, Robertson G, MacDonald J, Cezard T, Bilenky M, et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood. 2012;119:388–398. doi: 10.1182/blood-2011-03-341081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Kennedy M, Awong G, Sturgeon CM, Ditadi A, LaMotte-Mohs R, Zuniga-Pflucker JC, Keller G. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell reports. 2012;2:1722–1735. doi: 10.1016/j.celrep.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Kumano K, Arai S, Hosoi M, Taoka K, Takayama N, Otsu M, Nagae G, Ueda K, Nakazaki K, Kamikubo Y, et al. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood. 2012;119:6234–6242. doi: 10.1182/blood-2011-07-367441. [DOI] [PubMed] [Google Scholar]
- Kyba M, Perlingeiro RC, Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- Laurenti E, Doulatov S, Zandi S, Plumb I, Chen J, April C, Fan JB, Dick JE. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nature immunology. 2013 doi: 10.1038/ni.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, Humphries RK, Largman C. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledran MH, Krassowska A, Armstrong L, Dimmick I, Renstrom J, Lang R, Yung S, Santibanez-Coref M, Dzierzak E, Stojkovic M, et al. Efficient hematopoietic differentiation of human embryonic stem cells on stromal cells derived from hematopoietic niches. Cell stem cell. 2008;3:85–98. doi: 10.1016/j.stem.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Lee GS, Kim BS, Sheih JH, Moore M. Forced expression of HoxB4 enhances hematopoietic differentiation by human embryonic stem cells. Molecules and cells. 2008;25:487–493. [PubMed] [Google Scholar]
- Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA, Henley KJ, Ellis S, Voss AK, Metcalf D, et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nature immunology. 2008;9:810–819. doi: 10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]
- Martin CH, Woll PS, Ni Z, Zuniga-Pflucker JC, Kaufman DS. Differences in lymphocyte developmental potential between human embryonic stem cell and umbilical cord blood-derived hematopoietic progenitor cells. Blood. 2008;112:2730–2737. doi: 10.1182/blood-2008-01-133801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurier F, Doedens M, Gan OI, Dick JE. Rapid myelo-erythroid repopulation after intrafemoral transplantation of NOD-SCID mice reveals a new class of human stem cells. Nature medicine. 2003;9:959–963. doi: 10.1038/nm886. [DOI] [PubMed] [Google Scholar]
- McKinney-Freeman S, Cahan P, Li H, Lacadie SA, Huang HT, Curran M, Loewer S, Naveiras O, Kathrein KL, Konantz M, et al. The transcriptional landscape of hematopoietic stem cell ontogeny. Cell stem cell. 2012;11:701–714. doi: 10.1016/j.stem.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaccio AR, Whitsett C, Papayannopoulou T, Sadelain M. The potential of stem cells as an in vitro source of red blood cells for transfusion. Cell stem cell. 2012;10:115–119. doi: 10.1016/j.stem.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller LU, Milsom MD, Harris CE, Vyas R, Brumme KM, Parmar K, Moreau LA, Schambach A, Park IH, London WB, et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood. 2012;119:5449–5457. doi: 10.1182/blood-2012-02-408674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Pereira CF, Chang B, Qiu J, Niu X, Papatsenko D, Hendry CE, Clark NR, Nomura-Kitabayashi A, Kovacic JC, Ma'ayan A, et al. Induction of a Hemogenic Program in Mouse Fibroblasts. Cell stem cell. 2013 doi: 10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Orkin SH. The switch from fetal to adult hemoglobin. Cold Spring Harbor perspectives in medicine. 2013;3 doi: 10.1101/cshperspect.a011643. a011643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Xu J, Ragoczy T, Ippolito GC, Walkley CR, Maika SD, Fujiwara Y, Ito M, Groudine M, Bender MA, et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460:1093–1097. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulpule A, Kelley JM, Lensch MW, McPherson J, Park IH, Hartung O, Nakamura T, Schlaeger TM, Shimamura A, Daley GQ. Pluripotent stem cell models of shwachman-diamond syndrome reveal a common mechanism for pancreatic and hematopoietic dysfunction. Cell stem cell. 2013;12:727–736. doi: 10.1016/j.stem.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Menendez P, Shojaei F, Li L, Mazurier F, Dick JE, Cerdan C, Levac K, Bhatia M. Generation of hematopoietic repopulating cells from human embryonic stem cells independent of ectopic HOXB4 expression. The Journal of experimental medicine. 2005;201:1603–1614. doi: 10.1084/jem.20041888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell stem cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Ye Z, Zhan H, Mali P, Dowey S, Williams DM, Jang YY, Dang CV, Spivak JL, Moliterno AR, Cheng L. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009;114:5473–5480. doi: 10.1182/blood-2009-04-217406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Mali P, Huang X, Dowey SN, Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011;118:4599–4608. doi: 10.1182/blood-2011-02-335554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.