Abstract
The past several decades have seen tremendous advances in the field of medical genetics. The application of genetic technologies to the field of reproductive medicine has ushered in a new era of medicine that is likely to greatly expand in the coming years. Concurrent with an in vitro fertilization (IVF) cycle, it is now possible to obtain a cellular biopsy from a developing embryo and genetically evaluate this sample with increasing sophistication and detail. Preimplantation genetic screening (PGS) is the practice of determining the presence of aneuploidy (either too many or too few chromosomes) in a developing embryo. However, how and in whom PGS should be offered is a topic of much debate.
Keywords: preimplantation genetic screening, PGS, embryo, recurrent pregnancy loss, microarray, FISH, PGD
Introduction
Over the past several decades the world has witnessed incredible advances in many medical fields. No field has seen more dramatic change, however, than the field of genetics. The mammoth explosion of technologies surrounding genetics is staggering. Sequencing the human genome was only a short time ago considered the “Mount Everest” of medical achievement. Today, sequencing an entire individual’s genome is widely available and can be performed in a day (rather than years) for thousands (rather than millions) of dollars.1 In a similar manner, technology surrounding preimplantation genetic screening (PGS) has advanced greatly in recent years. In the following review we will outline PGS, current applications for this technology, and the limitations of the procedure.
An in vitro fertilization (IVF) cycle consists of administrating injectable gonadotropins to women and induce controlled ovarian hyperstimulation in which more than the usual number of ovarian follicles are recruited and matured.2,3 The oocytes within these follicles are then surgically harvested and inseminated with sperm. Typically, resultant embryos are then grown in vitro until either 3 or 5 days of development, at which time the 1 or 2 “best” embryos are placed into the uterus and the remaining embryos are cryopreserved. Determining which embryos are the “best” has been a subject of much debate since the birth of the technology in the late 1970’s. Traditionally the use of morphology, the visual appearance of embryos, has been the principal modality of choosing optimal embryos for uterine transfer.4 However, the implantation rate per transferred embryo in most clinics rarely exceeds 40%.5 Therefore, many investigators have for some time been searching for other diagnostic methods capable of more accurately determining embryo quality than morphology alone. This effort has produced several promising technologies including metabolomics, real-time videography, and PGS.
The Evolution of PGS
PGS is the practice of taking a biopsy either from the polar body of a mature oocyte or from cells taken from developing embryos and genetically analyzing the composition of these cells. Results of this genetic analysis direct the embryologist in choosing embryos for uterine transfer. Because PGS can only be performed using cells obtained at embryo biopsy, this technology is only possible in conjunction with an in vitro fertilization (IVF) cycle. PGS is the practice of evaluating embryos for chromosomal aneuploidy, the presence of either too many or too few chromosomes, in chromosomally normal parents. In contrast, preimplantation genetics diagnosis (PGD) is the practice of evaluating embryos for specific genetic abnormalities, such as sickle cell disease or cystic fibrosis, where carrier status has been documented in each of the parents.
Certain patient populations, including couples with advanced maternal age, recurrent miscarriage, repeated implantation failure, and severe male factor, are thought to have a predisposition for producing aneuploid embryos.3,6,7 Many have suggested that these patient populations may be benefit from PGS.3,7 However, the indications for using PGS in many centers is constantly expanding.3 As reported by the European Society of Human Reproduction and Embryology (ESHRE), over the past 10 years, 61% of all preimplantation genetic testing cycles were performed for PGS.8 This paper will focus exclusively on the clinical applications associated with PGS rather than PGD.
PGS, unlike PGD, has been and continues to be a controversial technology. Recent studies indicate that greater than 60%–90% of all first trimester miscarriages may be the result of chromosomal aneuploidy.3 Because so many early miscarriages are due to aneuploidy, PGS seems to be a reasonable intervention to improve the efficiency with which euploid (chromosomally normal) embryos are selected for uterine transfer in IVF cycles. Classic studies have reported that miscarriages that are caused by aneuploidy are disproportionally concentrated on select chromosomes.9,10 These data are based on karyotype analysis of failed pregnancies that developed far enough along to have tissue available for genetic analysis.9,10 Consequently, clinics performing PGS in the early days of the technology focused on detecting aneuploidy on only select chromosomes using fluorescence in situ hybridization (FISH), which typically evaluates between 5–14 chromosome pairs rather than all 23 chromosome pairs.11,12 Traditionally, PGS biopsy was exclusively performed at approximately 3 days of embryonic development following fertilization.11,12 Initial data using PGS in the context of cleavage stage biopsy with FISH showed promising results and generated much excitement for this new technology.3,13–15 Unfortunately the results from this approach failed to result in improvements in clinical pregnancy rates and this lack of efficacy was widely referenced following a landmark paper by Mastenbroek in the New England Journal of Medicine.16 Subsequently, similar papers cast further doubt on the benefits of PGS and position statements from major medical societies formally discouraged its use.17–19
Further research, however, elucidated several biologic limitations that could explain the prior shortcomings of clinically applied PGS. The practice of polar body biopsy to determine the genetic composition of a fertilized oocyte is a commonly utilized modality for performing preimplantation genetic testing.3,8,20 A critical component of oocyte development is meiotic division in which a haploid set of unused maternal DNA is marginalized into what is termed a polar body.3,8 Genetic evaluation of this polar body was initially quite popular, as this process obtained a diagnosis without disturbing the developing embryo and could be performed prior to fertilization.3 However, this approach is incapable of detecting paternally derived genetic errors, or any errors introduced after or during fertilization. Due to these limitations, polar body biopsy is now principally performed in countries where strict legislation limits the practice of embryo biopsy.3,21
However, PGS using biopsied cells from developing embryos also presents challenges. Studies have repeatedly documented that embryos at day 3 of development have high levels of mosaicism.22,23 Mosaicism is a condition in which a single developing embryo is comprised of more than one distinct genetic cell line. In other words, mosaic embryos may have euploid (normal) and aneuploid (abnormal) cell lines within a single embryo. Studies evaluating this phenomenon have concluded the majority of all embryos may be mosaic at day 3 of development.22–24 Consequently, a biopsy performed at day 3 of development may produce a result that is not representative of the entire embryo.3 Mosaicism has been shown to also exist at day 5 of embryo development.25 However, recent data suggests that mosaicism may be much reduced by day 5 of development.3,26
Another limitation of traditionally performed PGS was the use of FISH for determination of chromosomal abnormalities. FISH typically evaluates between 5–14 rather than all 23 chromosome pairs.27 Recent studies have indicated that embryonic aneuploidy occurs in clinically significant amounts in all 23 chromosome pairs.28 Therefore, FISH is incapable of diagnosing many of the chromosomal abnormalities commonly found in developing embryos.
Realization of these two principal limitations has led many genetic laboratories to offer PGS using technologies evaluating the chromosomal status of all 23 chromosome pairs using an embryonic biopsy performed at the blastocyst stage, typically reached by day 5 or 6 of development. The clinical pregnancy rates using this approach have been reported to be markedly superior to the traditional approach of performing PGS.29,30 For example, a recent study evaluating more than 4,500 embryos using 23 chromosome pair determinations found clinical pregnancy rates in women suffering from recurrent pregnancy loss (RPL) to be significantly improved above similar studies using FISH PGS.29 Additionally, pregnancy rates were further improved when 23 chromosome evaluation PGS was performed on blastocyst stage embryos (day 5/6 of development) as compared to when the biopsy was performed on embryos at day 3 of development. 29,31,32 Similar results have been reported consistently by many clinics in the United States and around the world.31,32 This has generated a renewed interest in PGS, although it still remains to be determined whether PGS is an efficacious technology, and which patient populations are best served by PGS.
Evaluation of all 23 chromosomes in the context of PGS possesses inherent complexities that potentially may compromise the integrity of data if not properly performed. There are multiple approaches that are used to perform 23 chromosome pair evaluation. The two modalities that are most commonly used today utilize microarray technology, either by using single nucleotide polymorphism (SNP) or comparative genomic hybridization (CGH) technology.3 Both of these technologies rely on obtaining embryonic DNA, fragmenting and then amplifying this DNA, and evaluating this amplified product using microarrays. This amplification process is a potential source of error, as failure to amplify the entire embryonic DNA product could produce a false result. Additionally, because the DNA product being initially amplified is taken from only 1 to several cells, any external DNA contamination can produce a spurious result.
SNP arrays directly evaluate ploidy status using a dense array of approximately 300,000 genetic markers.3 CGH arrays, in contrast, evaluate far fewer genetic markers and compare this result to a known normal DNA sample.3 Each of these microarray platforms have advantages and disadvantages. A purported advantage of SNP arrays is their ability to detect relatively small genetic duplications or deletions, though the value of this information is, at the present time, generally unclear. An advantage of CGH arrays is that they may be performed in 12–16 hours as opposed to several days for most SNP arrays.
Evidence for the Clinical Application of PGS
Studies from centers using PGS on day 5 and 6 blastocysts show promising results with transferred embryos generating pregnancy rates greater than 75%.29,31,32 However, a central criticism of the widespread use of PGS is a lack of randomized controlled trials that conclusively show the procedure to be beneficial. To our knowledge, at the time of writing this article, there is only one randomized trial to show a pregnancy benefit using PGS versus IVF using morphology alone in the setting of exclusive single embryo uterine transfer.33 This study was relatively small and had several significant limitations. Therefore, further larger and more rigorously studies are required before PGS will be more broadly accepted.3 Several large randomized controlled clinical trials are currently underway that will hopefully provide such data in the near future.
Despite the lack of support from professional societies and lack of large randomized controlled trials definitively demonstrating the benefits of the technology, PGS comprises the majority of all preimplantation genetic testing internationally and is being increasingly utilized.8 However, the patient population in which PGS may be appropriate is unclear at the present time.3 Many PGS clinics have traditionally recommended PGS for couples with risk factors believed to be associated with embryonic aneuploidy such as unexplained RPL, severe male factor, and advanced maternal age. However, in recent years many clinics have liberally expanded the use of PGS to many women without such risk factors. In fact, some clinics broadly recommend PGS to virtually all IVF patients as a strategy to improve pregnancy rates in couples battling infertility. The debate surrounding the appropriate patient populations for PGS is currently in flux and will likely be a source of debate for years to come.
Limitations of PGS
Despite the positive data that is emerging within the field of PGS, there are tangible technical and biological limitations to the technology. The limitations of FISH PGS evaluation and the use of biopsy taken from day 3 embryos are significant and have been discussed previously. Additionally, technical limitations surrounding the use of both SNP and CGH arrays may produce spurious results if not properly executed.3 Furthermore, while automated results are generated, the raw data is also interpreted by a trained geneticist. Therefore, the interpretation of results may be subjectively interpreted and this leaves inherent room for human error in addition to inaccurate automated resulting.
Perhaps the most significant source of error from PGS using 23 chromosome pair evaluation and blastocyst biopsy (day 5/6 of development) is the presence of cellular discordance within the developing embryo. A blastocyst embryo is comprised of 2 components, an inner cell mass (ICM) and trophectoderm (TE).3 The ICM possesses cells that are destined to form fetal tissue and the TE possesses cells that will form the placenta. Blastocyst biopsy utilizes cells taken from the TE to minimize potential deleterious effects that may be caused from biopsy of the ICM, cells destined to form the fetus. Though there is clearly a high degree of correspondence between the genetic composition of the ICM and the TE, some data suggests that in up to 10% of developing blastocysts, there may be aneuploidy present in the TE but not ICM or vice versa.26 Therefore, TE biopsy taken at the blastocyst stage, from a biological standpoint, may not be universally predictive of the chromosomal status of the developing embryo even if no technical error exists in the performance of genetic analysis.3 Mosaicism may also exist within a given TE cell population. Array technology, however, is capable of detecting all but very low levels of mosaicism within TE samples analyzed provided.26,34 The above limitations of PGS demand that patients be adequately counseled on the risks and limitations of PGS, preferably with the aid of a specialized physician, geneticist, or genetic counselor. Furthermore, antenatal genetic testing is still recommended in all patients undergoing PGS.3
Conclusion
New technologies associated with PGS are producing data that suggest that the procedure could be a valuable adjunct to assisted reproductive technologies in the future to enhance pregnancy success in many patients. Defining the exact benefit conferred by PGS and determining exactly which patient populations could be best served by PGS, however, is currently controversial. In the near future, several large and high quality studies will attempt to answer these questions. Despite the current lack of definitive data, PGS is increasingly being applied to patients with ever-expanding clinical indications. While there are few large randomized controlled studies defining the benefits of PGS, clinical data emerging from many PGS laboratories around the world is encouraging. Therefore, the judicious use of PGS seems reasonable at the present time, as the preponderance of available current evidence suggests that some couples may see a benefit from this technology. Only with time will the role of PGS be clearly defined.
Figure 1.
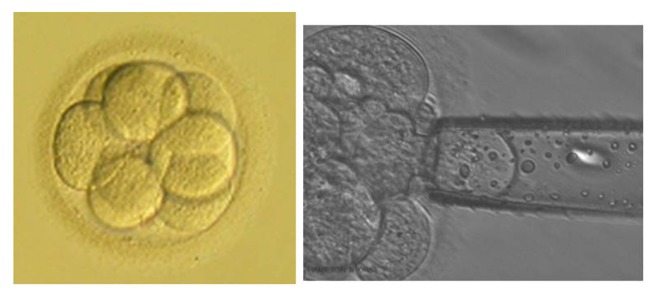
Cleavage stage embryo.
Notes: These photographs show an embryo at the cleavage stage. On the right is a photograph of a cleavage stage biopsy.
Figure 2.
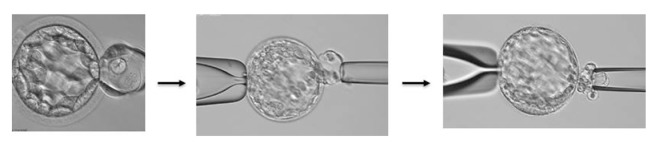
Blastocyst stage embryo.
Notes: These photographs show an embryo at the Blastocyst stage. The leftmost photo shows the herniation of TE cells after the application of a laser to breach the zona pellucida. The next 2 photographs show the process of obtaining a sheet of trophectoderm (TE) cells that will be analyzed for PGS.
Acknowledgments
We thank Dr. Jianchi Ding for help with the writing of this manuscript.
Footnotes
Contributors
PRB and WHK primarily searched the literature. RWK performed literature review. PRB wrote the first draft of the manuscript. RWK participated in writing and provided review assistance. WHK advised on the content of the manuscript regarding generalist physicians, assisted in the literature search, and contributed to the writing of the manuscript. PRB is guarantor.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest. Provenance: the authors were invited to submit this paper.
Funding
Author(s) disclose no funding sources.
References
- 1.Rizzo JM, Buck MJ. Key principles and clinical applications of “next-generation” DNA sequencing. Cancer Prev Res (Phila) 2012;5:887–900. doi: 10.1158/1940-6207.CAPR-11-0432. [DOI] [PubMed] [Google Scholar]
- 2.Brezina PR, Benner A, Rechitsky S, et al. Single-gene testing combined with single nucleotide polymorphism microarray preimplantation genetic diagnosis for aneuploidy: a novel approach in optimizing pregnancy outcome. Fertil Steril. 2011;95:1786. doi: 10.1016/j.fertnstert.2010.11.025. [DOI] [PubMed] [Google Scholar]
- 3.Brezina PR, Brezina DS, Kearns WG. Preimplantation genetic testing. BMJ. 2012;345:e5908. doi: 10.1136/bmj.e5908. [DOI] [PubMed] [Google Scholar]
- 4.Brezina PR, Zhao Y. The ethical, legal, and social issues impacted by modern assisted reproductive technologies. Obstet Gynecol Int. 2012;2012:686253. doi: 10.1155/2012/686253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ajduk A, Zernicka-Goetz M. Advances in embryo selection methods. F1000. Biol Rep. 2012;4:11. doi: 10.3410/B4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fragouli E, Wells D, Whalley KM, Mills JA, Faed MJ, Delhanty JD. Increased susceptibility to maternal aneuploidy demonstrated by comparative genomic hybridization analysis of human MII oocytes and first polar bodies. Cytogenet Genome Res. 2006;114(1):30–8. doi: 10.1159/000091925. [DOI] [PubMed] [Google Scholar]
- 7.Vialard F, Boitrelle F, Molina-Gomes D, Selva J. Predisposition to aneuploidy in the oocyte. Cytogenet Genome Res. 2011;133(2–4):127–35. doi: 10.1159/000324231. [DOI] [PubMed] [Google Scholar]
- 8.Harper JC, Wilton L, Traeger-Synodinos J, et al. The ESHRE PGD Consortium: 10 years of data collection. Hum Reprod Update. 2012;18:234–47. doi: 10.1093/humupd/dmr052. [DOI] [PubMed] [Google Scholar]
- 9.Monni G, Ibba RM, Zoppi MA. Prenatal Genetic Diagnosis through Chorionic Villus Sampling. In: Milunsky A, Milunsky J, editors. Genetic Disorders and the Fetus. 6th ed. Oxford: Wiley-Blackwell; 2010. [Google Scholar]
- 10.Kearns WG, Pen R, Graham J, et al. Preimplantation genetic diagnosis and screening. Semin Reprod Med. 2005;23(4):336–47. doi: 10.1055/s-2005-923391. [DOI] [PubMed] [Google Scholar]
- 11.Schoolcraft WB, Fragouli E, Stevens J, Munne S, Katz-Jaffe MG, Wells D. Clinical application of comprehensive chromosomal screening at the blastocyst stage. Fertil Steril. 2010;94:1700–6. doi: 10.1016/j.fertnstert.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Brezina PR, Kearns WG. Preimplantation genetic screening in the age of 23-chromosome evaluation; why FISH is no longer an acceptable technology. J Fertiliz In Vitro. 2011;1:e103. [Google Scholar]
- 13.Munné S, Chen S, Fischer J, et al. Preimplantation genetic diagnosis reduces pregnancy loss in women aged 35 years and older with a history of recurrent miscarriages. Fertil Steril. 2005;84(2):331–5. doi: 10.1016/j.fertnstert.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 14.Platteau P, Staessen C, Michiels A, Van Steirteghem A, Liebaers I, Devroey P. Preimplantation genetic diagnosis for aneuploidy screening in women older than 37 years. Fertil Steril. 2005;84(2):319–24. doi: 10.1016/j.fertnstert.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 15.Mantzouratou A, Mania A, Fragouli E, et al. Variable aneuploidy mechanisms in embryos from couples with poor reproductive histories undergoing preimplantation genetic screening. Hum Reprod. 2007;22(7):1844–53. doi: 10.1093/humrep/dem102. [DOI] [PubMed] [Google Scholar]
- 16.Mastenbroek S, Twisk M, Echten-Arends J, Sikkema-Raddatz B, Korevaar JC, Verhoeve HR, et al. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;357:9–17. doi: 10.1056/NEJMoa067744. [DOI] [PubMed] [Google Scholar]
- 17.Checa MA, Alonso-Coello P, Solà I, Robles A, Carreras R, Balasch J. IVF/ICSI with or without preimplantation genetic screening for aneuploidy in couples without genetic disorders: a systematic review and meta-analysis. J Assist Reprod Genet. 2009;26:273–83. doi: 10.1007/s10815-009-9328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mastenbroek S, Twisk M, van der Veen F, Repping S. Preimplantation genetic screening: a systematic review and meta-analysis of RCTs. Hum Reprod Update. 2011;17:454–66. doi: 10.1093/humupd/dmr003. [DOI] [PubMed] [Google Scholar]
- 19.Practice Committee of Society for Assisted Reproductive Technology; Practice Committee of American Society for Reproductive Medicine. Preimplantation genetic testing: a Practice Committee opinion. Fertil Steril. 2008;90:S136–43. doi: 10.1016/j.fertnstert.2008.08.062. [DOI] [PubMed] [Google Scholar]
- 20.Verlinsky Y, Rechitsky S, Evsikov S, et al. Preconception and preimplantation diagnosis for cystic fibrosis. Prenat Diagn. 1992;12(2):103–10. doi: 10.1002/pd.1970120205. [DOI] [PubMed] [Google Scholar]
- 21.Tuffs A. Germany allows restricted access to preimplantation genetic testing. BMJ. 2011;343:d4425. doi: 10.1136/bmj.d4425. [DOI] [PubMed] [Google Scholar]
- 22.Harton GL, Magli MC, Lundin K, Montag M, Lemmen J, Harper JC. ESHRE PGD Consortium/Embryology Special Interest Group—best practice guidelines for polar body and embryo biopsy for preimplantation genetic diagnosis/screening (PGD/PGS) Hum Reprod. 2011;26:41–6. doi: 10.1093/humrep/deq265. [DOI] [PubMed] [Google Scholar]
- 23.Munné S, Weier HU, Grifo J, Cohen J. Chromosome mosaicism in human embryos. Biol Reprod. 1994;51:373–9. doi: 10.1095/biolreprod51.3.373. [DOI] [PubMed] [Google Scholar]
- 24.van Echten-Arends J, Mastenbroek S, Sikkema-Raddatz B, et al. Chromosomal mosaicism in human preimplantation embryos: a systematic review. Hum Reprod Update. 2011;17(5):620–7. doi: 10.1093/humupd/dmr014. [DOI] [PubMed] [Google Scholar]
- 25.Bielanska M, Tan SL, Ao A. High rate of mixoploidy among human blastocysts cultured in vitro. Fertil Steril. 2002;78(6):1248–53. doi: 10.1016/s0015-0282(02)04393-5. [DOI] [PubMed] [Google Scholar]
- 26.Brezina PR, Sun Y, Anchan RM, Li G, Zhao Y, Kearns WG. Aneuploid embryos as determined by 23 single nucleotide polymorphism (SNP) microarray preimplantation genetic screening (PGS) possess the potential to genetically normalize during early development. Fertil Steril. 2012;98(3):S108. [Google Scholar]
- 27.Harper JC, Sengupta SB. Preimplantation genetic diagnosis: state of the art 2011. Hum Genet. 2012;131:175–86. doi: 10.1007/s00439-011-1056-z. [DOI] [PubMed] [Google Scholar]
- 28.Brezina PR, Tobler K, Benner AT, Du L, Xu X, Kearns WG. All 23 Chromosomes Have Significant Levels of Aneuploidy in Recurrent Pregnancy Loss Couples. Fertil Steril. 2012;97(3):S7. [Google Scholar]
- 29.Brezina PR, Tobler K, Benner AT, Du L, Boyd B, Kearns WG. In vitro fertilization (IVF) cycles and 4,873 embryos using 23-chromosome single nucleotide polymorphism (SNP) microarray preimplantation genetic screening (PGS) Fertil Steril. 2012;97:S23–4. [Google Scholar]
- 30.Wells D, Alfarawati S, Fragouli E. Use of comprehensive chromosomal screening for embryo assessment: microarrays and CGH. Mol Hum Reprod. 2008;14:703–10. doi: 10.1093/molehr/gan062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forman EJ, Tao X, Ferry KM, Taylor D, Treff NR, Scott RT., Jr Single embryo transfer with comprehensive chromosome screening results in improved ongoing pregnancy rates and decreased miscarriage rates. Hum Reprod. 2012;27:1217–22. doi: 10.1093/humrep/des020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoolcraft WB, Fragouli E, Stevens J, Munne S, Katz-Jaffe MG, Wells D. Clinical application of comprehensive chromosomal screening at the blastocyst stage. Fertil Steril. 2010;94:1700–6. doi: 10.1016/j.fertnstert.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 33.Yang Z, Liu J, Collins GS, Salem SA, et al. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Mol Cytogenet. 2012;5:24. doi: 10.1186/1755-8166-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mamas T, Gordon A, Brown A, Harper J, Sengupta S. Detection of aneuploidy by array comparative genomic hybridization using cell lines to mimic a mosaic trophectoderm biopsy. Fertil Steril. 2012;97(4):943–7. doi: 10.1016/j.fertnstert.2011.12.048. [DOI] [PubMed] [Google Scholar]


