Abstract
During treatment, mutations in HIV-1 protease (PR) are selected rapidly that confer resistance by decreasing affinity to clinical protease inhibitors (PIs). As these unique drug resistance mutations can compromise the fitness of the virus to replicate, mutations that restore conformational stability and activity while retaining drug resistance are selected on further evolution. Here we identify several compensating mechanisms by which an extreme drug-resistant mutant bearing 20 mutations (PR20) with >5-fold increased Kd and >4000-fold decreased affinity to the PI darunavir functions. 1) PR20 cleaves, albeit poorly, Gag polyprotein substrates essential for viral maturation. 2) PR20 dimer, which exhibits distinctly enhanced thermal stability, has highly attenuated autoproteolysis, thus likely prolonging its lifetime in vivo. 3) The enhanced stability of PR20 results from stabilization of the monomer fold. Both monomeric PR20T26A and dimeric PR20 exhibit Tm values 6-7.5 °C higher than their PR counterparts. Two specific mutations in PR20, L33F and L63P at sites of autoproteolysis, increase the Tm of monomeric PRT26A by ~8 °C, similar to PR20T26A. However, without other compensatory mutations as seen in PR20, L33F and L63P substitutions, together, neither restrict autoproteolysis nor significantly reduce binding affinity to darunavir. To determine whether dimer stability contributes to binding affinity for inhibitors, we examined single-chain dimers of PR and PRD25N in which the corresponding identical monomer units were covalently linked by GGSSG sequence. Linking of the subunits did not appreciably change the ΔTm on inhibitor binding; thus stabilization by tethering appears to have little direct effect on enhancing inhibitor affinity.
Keywords: HIV protease, drug resistance, structural stability, calorimetry, enzyme kinetics
The protease (PR) of HIV-1 is indispensable for processing the retroviral Gag and Gag-Pol polyproteins to release the structural and functional proteins essential for maturation and propagation of infectious virus. PR is one of the primary targets for antiviral therapy and several potent protease inhibitors (PIs) have been developed for clinical use. Encoded as part of the viral Gag-Pol polyprotein, PR mediates its own release by a multi-step autocatalytic process (autoprocessing).1-4 A crucial step in this process is the cleavage at the N-terminus of the PR domain, accompanied by appearance of catalytic activity and stable dimer formation.3 The mature PR is a homodimer consisting of two 99-amino acid monomer units each of which contributes one aspartyl residue to the active site, part of the conserved active site interface triad Asp-Thr-Gly.5 Other regions contributing to the dimer interface are the flaps, which adopt a closed conformation upon binding of a substrate or inhibitor, and the four-stranded antiparallel β-sheet, comprising the four N- and C-terminal strands of the two monomers which play a major role in holding the subunits together.6 Several PIs, 8-10 of which are in widespread current use,7 have been designed to bind to the active site of the PR dimer at low nM-pM affinities. However, the noteworthy success of these drugs is accompanied by a serious drawback, since error-prone replication by the virally encoded reverse transcriptase facilitates the rapid emergence of drug resistant mutants (DRMs), selected in the presence of PIs as functionally competent enzymes. Some of these highly evolved PR variants contain multiple mutations and are resistant to most if not all PIs.8,9
In addition to major DRMs, which directly affect substrate and/or inhibitor binding, so-called accessory or compensatory mutations remote from the active site, some of which correspond to naturally occurring variants,10 are frequently observed to co-evolve with the major mutations. These polymorphisms may pre-exist or they may evolve after emergence of major DRM(s) under drug pressure, and they are selected because they confer functional advantages to the drug resistant PR. Structural studies to date have focused on mechanisms whereby long-range effects of distal mutations alter the conformation of the binding site and/or flaps, and consequently the energetics of inhibitor binding.11-13 Some of these mutations in PR also partially restore the biological fitness of the mutant virus to replicate, which is adversely affected by major DRMs.14-16 Accessory mutations such as L10I, L63P and A71V have been shown to increase the thermal stability and pH tolerance of a drug resistant PR bearing the destabilizing active-site mutation I84V.17 Several drug resistant PRs bearing both active-site and remote mutations exhibit thermal stability exceeding that of the wild type dimer. These include ANAM-11,11 which bears 11 mutations including L63P, and mutant I50L/A71V showing Tm increases of 4.8 °C and ~2 °C, respectively, relative to wild type.18 In the present study we describe the basis for enhanced thermal stabilization of a highly drug resistant mutant protease, PR20, which to our knowledge has the highest such thermal stabilization (as well as the largest number of mutations) for a drug resistant PR observed to date.
PR20, a clinical isolate first reported by Dierynck et al.,19 bears 20 amino acid substitutions both near and remote from the active site. Mature PR20 exhibits a 3-4 orders of magnitude decreased affinity for PIs darunavir (DRV) and saquinavir, relative to the wild type PR. Furthermore a model precursor (TFR-PR20) in which the protease is flanked at its N-terminus by the transframe region (TFR) of Gag-Pol, undergoes efficient autoprocessing to release the TFR and PR20, in the presence of every clinical PI including darunavir (DRV) up to 250 μM concentration.9 By contrast the processing of the corresponding wild-type precursor is inhibited by DRV with an IC50 of ~1 μM. These observations account for the viability of HIV-1 with mutant PR20, which successfully evades inhibition even by highly effective second generation PIs. In the present study we examine the kinetic parameters of PR20 using a chromogenic substrate, its monomer-dimer equilibrium, and urea induced unfolding. Next, using differential scanning calorimetry, we investigate several properties relating to the stability of PR20 such as 1) whether the enhanced stability of PR20 relative to PR results from inter-monomer interactions or a more stable monomer fold, 2) if specific mutations L33F and L63P at sites of autoproteolysis selected under drug pressure in PR20 affect both autoproteolysis and stability and 3) whether an entropically stabilized PR dimer as a single-chain construct influences binding affinity to inhibitors. Finally, we explore the relative catalytic efficiencies of PR20- and PR-mediated hydrolysis of peptide substrates corresponding to natural sites in Gag polyprotein to help understand if hyper resistance is associated with selection for cleavage of some substrates different in their order of efficiencies as compared to wild-type PR.
MATERIALS AND METHODS
Protein expression and purification
Previously reported constructs used in this study are PR,20 PR20,9,19 ANAM-11,9,11 PRD25N,21 and PRT26A.22 Constructs used for the first time are PR20K7Q, PR20T26A, PRL33F/L63P, PRT26A/L33F/L63P, PRL33F/E34D/L63P, ScPR and ScPRD25N. Genes were synthesized and cloned in pET11a vector between Nde1 and BamH1 sites and transformed into E. coli BL-21(DE3). Substitution mutations were introduced using the Quik-Change mutagenesis kit reagents and protocol (Agilent Technologies) and verified by DNA sequencing. Cells bearing the corresponding plasmid were grown at 37 °C either in Luria-Bertani medium or in a modified minimal medium with 15N ammonium chloride and 13C glucose as the sole nitrogen and carbon sources. Induction for protein expression, isolation of inclusion bodies and purification were carried out as described previously.23,24 Proteins were subjected to electrospray ionization mass spectrometry (ESI-MS) to verify composition prior to use. Proteins were folded freshly as described, either by the dialysis or the quench protocol25 for enzyme kinetics, calorimetric and NMR studies.
Spectrophotometric enzyme assays
Enzymatic activity was measured at 28 °C with chromogenic substrate IV [Lys-Ala-Arg-Val-Nle-(4-NO2Phe)-Glu-Ala-Nle-NH2, California Peptide Research, Napa, CA] by following the decrease in absorbance at 310 nm in 50 mM sodium acetate buffer, pH 5, containing 250 mM sodium chloride. The protease was folded by the quench protocol to a final concentration of 0.5 μM as described25 and reactions were initiated by addition of substrate. Absorbance change was converted to molarity by use of Δε = 1797 M−1 cm−1, and the data at substrate concentrations from 72-430 μM were analyzed with the enzyme kinetics module of SigmaPlot 10 (Systat Software, Inc.). The dimer dissociation constant, Kd, was measured under the same conditions at a substrate concentration of 430 μM and varying concentrations of PR20 from 0.026 to 1.9 μM (as monomers). An equation previously described24 was used to fit the data. The effect of up to 2.4 M urea on the rate of substrate (400 μM) hydrolysis by 0.53 μM PR20 was measured at the same pH and NaCl concentration as above (pH of urea stock solution adjusted to 5.07).
Fluorescence measurements
The effect of urea on the intrinsic fluorescence of PR20 was measured by use of a Fluoromax-3 spectrofluorimeter (Horiba Jobin Yvon, Edison, NJ) with excitation and emission wavelengths of 285 and 350 nm, respectively, and a bandwidth of 2 nm. The method, based on a previously described protocol25 consisted of replacing successive 70-μL aliquots of the measured solution (1 mL of 1 μM folded PR20 in 50 mM sodium acetate, pH 5; initially containing no urea) with an equal volume of a solution of the same composition containing 3.6 M urea to reach a final urea concentration of 2.2 M. The fluorescence signal was recorded upon reaching a stable endpoint after each successive replacement. A second experiment followed the same procedure with 2 μM DRV in both solutions.
NMR experiments
1H-15N TROSY-HSQC spectra were recorded at 293 K on a uniformly 15N and 13C labeled sample at 100 μM protein concentration in 20 mM sodium acetate buffer, pH 5.8, using a 600 MHz Bruker Avance II spectrometer equipped with a z-axis TCI cryogenic probe. 512* × 1024* complex points in the indirect and direct dimensions were collected for acquisition times of 266 (t1) and 122 (t2) ms. The interscan delay was set to 1.5 s. The spectra were processed using NMRPipe26 and displayed with SPARKY.27
Differential scanning calorimetry
Proteins were folded from 7 or 12 mM HCl by a quench protocol as previously described, 25 to give a final pH of 5.0 in 50 mM acetate or 3.6 in 25 mM formate buffers (see Table 1). To avoid possible precipitation at higher buffer concentrations, scPRD25N (with and without inhibitor) was folded by addition of 5.66 volumes of 5 mM sodium acetate buffer, pH 6.0, to one volume of protein in 12 mM HCl (final pH 4.7). For experiments in the presence of DRV or RPB, the inhibitor (~2-fold molar excess relative to dimers) was added in the folding buffer. DSC measurements were performed using a MicroCal VP-DSC microcalorimeter (GE Healthcare), and data were analyzed with the instrument's Origin software.21
Table 1.
DSC dataa and dimer dissociation constants (Kd) for selected PR constructs
| (A) | |||
|---|---|---|---|
| Protein | Tm (−DRV) | Tm (+DRV) | ΔTm (I)b |
| PR | 65.7c | 88.1c | 22.4 |
| PRL33F/E34D/L63P | nd | 85.4 | -- |
| PR20d | 71.7 | 77.0 | 5.3 |
| ANAM-11d | 70.5 | 81.9 | 11.4 |
| PRT26A | 60.0 | 63.0 | 3.0 |
| PR20T26A | 67.5 | 67.5 | 0 |
| PRT26A/L33F/L63P | 68.1 | 67.1 | −1.0 |
| (B) | |||||
|---|---|---|---|---|---|
| Protein | T m | T m | ΔTm (I)b | ΔTm (Sc)e | Kd (μM) |
| (− RPB) | (+ RPB) | ||||
| PRf | 56.6 | 65.8 | 9.2 | 11 (− RPB) | <0.01c |
| ScPRf | 67.6 | 75.4 | 7.8 | 9.6 (+ RPB) | -- |
| (− DRV) | (+ DRV) | ||||
| PRD25N | 58.4c | 61.5c | 3.1 | 9.0 (− DRV) | 1.3c |
| ScPRD25N | 67.4 | 70.3 | 2.9 | 8.8 (+ DRV) | -- |
At pH 4.7-5.0 in sodium acetate buffer unless noted otherwise
Tm(+inhibitor)-Tm(−inhibitor)
cited from reference21; an approximately 100-fold increase in Kd observed based on the active site mutation D25N in PR accompanies ~106-fold weaker binding of DRV
reference9
Tm(single chain)-Tm(homodimer)
in 25 mM sodium formate, pH 3.6
nd (not determined) due to more rapid autoproteolysis than PR, see Figure 2
Kinetics using peptides representing natural sites of polyprotein processing
Assays with peptides incorporating natural cleavage sites in Gag were initiated by mixing 5 μl (50 - 14000 nM) purified PR20 or 5 μl (9 – 350 nM) purified PR with 10 μl 2x incubation buffer (0.5 M potassium phosphate buffer, pH 5.6, containing 10% glycerol, 2 mM EDTA, 10 mM dithiothreitol, 4 M NaCl) and 5 μl 0.1 - 6 mM substrate dissolved in water (or in the case of Met-containing peptides in 10 mM DTT). After incubation at 37 °C for 1 hour, the reaction was terminated by the addition of 180 μl 1% trifluoroacetic acid. Substrates and cleavage products were separated using a reverse-phase HPLC method described previously.28 Kinetic parameters were determined by fitting data obtained at less than 20% substrate hydrolysis to the Michaelis-Menten equation by use of SigmaPlot software. The experimental errors of the determined parameters were smaller than 25 %. The amount of active proteases used in the assays was determined by active site titration using the potent HIV-1 PR inhibitor amprenavir. Active site titration was performed using the HPLC method, except 0.2 μl aliquots of the inhibitor (0 - 10 μM) were added to the reaction mixture.
RESULTS AND DISCUSSION
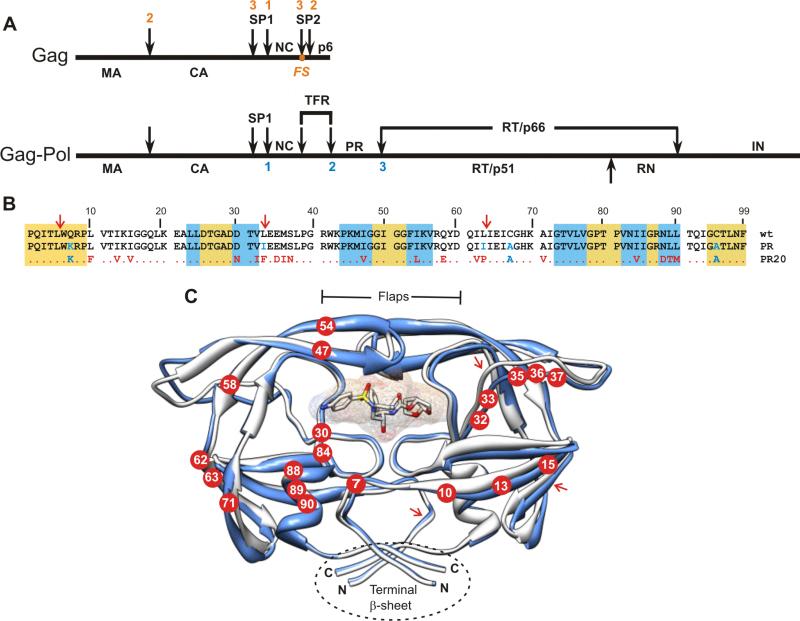
Figure 1A shows a schematic representation of the Gag and Gag-Pol polyproteins and the order in which the protease-catalyzed cleavage steps in their processing occur. In Figure 1B, the amino acid sequence of PR2019 is aligned with the wild type enzyme (PRwt) and with an optimized construct, designated as PR.4,20,29 We have shown previously that substituting the Cys residues to Ala does not alter the conformational stability or the kinetic parameters of PR and facilitates sample handling for NMR experiments.4,20 Mutations defined as major DRMs associated with DRV and saqunavir (SQV) resistance are 147V, I54L, I84V (DRV), and L90M (SQV).7 A recent crystallographic study has revealed novel flap conformations and an expanded inhibitor binding site for PR2013 and relates its reduced inhibitor binding affinity to propagation of structural changes resulting from the distal “accessory” mutations. The structure shown in Figure 1C represents a superimposition of PR20 (3UCB13) on PR (2IEN30), showing the location of the 20 mutations present in PR20. The present study focuses on the mechanism(s) by which such mutations also contribute to structural stabilization and function of the PR.
Figure 1.
(A) Organization of the Gag and Gag-Pol polyproteins of HIV-1. Nomenclature of domains in Gag:2 MA, matrix; CA, capsid; SP1, spacer peptide 1, NC, nucleocapsid; SP2, spacer peptide 2. In Gag-Pol: TFR, transframe region; PR, protease; RT, reverse transcriptase; RN, Rnase H; IN, integrase. FS designates the location of the - 1 frame shift during translation that results in production of Gag-Pol. Numbers shown above (red) and below (blue) the schematic lines designate the order in which sequential cleavages occur in Gag and in Gag-Pol respectively.2,9 (B) Sequence alignment of PR20 with wild type HIV-1 protease (PRwt) and its optimized mutant (termed PR). Residues in PR and PR20 shown in blue were introduced to limit autoproteolysis and to preclude cysteine-thiol oxidation. Residues shown in red are natural mutations in the clinical isolate from which PR20 is derived.19 The gold and white backgrounds indicate highly conserved regions in the protease domain in drug treated patients,41 and areas of greatest variability among isolates, respectively. The blue background designates regions where major DRMs (as defined in http:/hivdb.stanford.edu/cgi-bin/PIResiNote.cgi) occur. (C) Ribbon representation (in blue) of PR20 in complex with DRV (pdb accession code 3UCB13) superimposed on PR (in white, pdb accession code 2IEN30). Locations of the mutations in PR20 are shown as red circles with residue numbers in white. The DRV inhibitor is shown as a stick and surface representation in the active site cavity. Major sites of autoproteolysis are indicated in B and C with red arrows. Molecular representations were prepared using Chimera.42
Kinetic and biochemical characterization of PR20
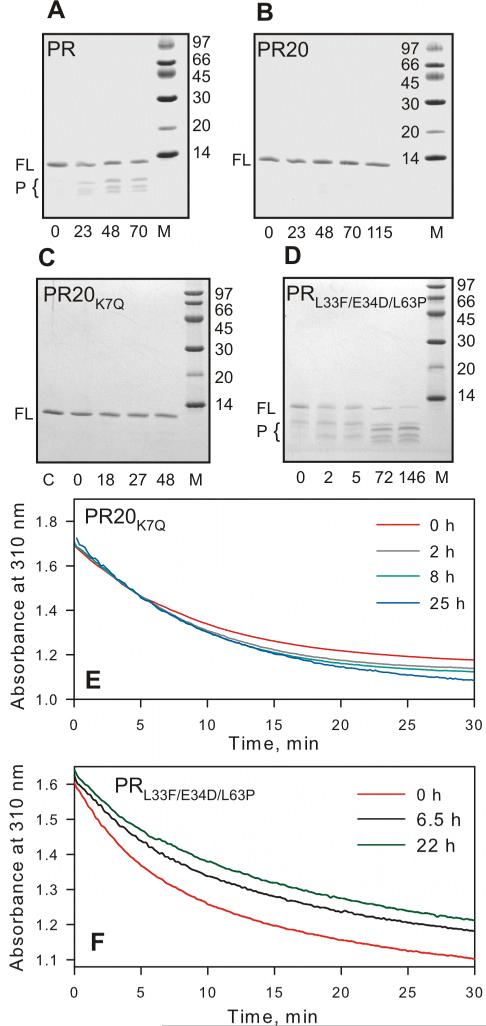
PRwt (Figure 1B) undergoes rapid and substantial self-cleavage (autoproteolysis) concomitant with loss of activity upon folding and incubation under optimal conditions, which may constitute a mechanism for down-regulation of PR activity.20 To overcome this problem a construct was designed bearing mutations Q7K, L331 and L63I at residues at or near the three major sites of autoproteolysis.20,29 Autoproteolysis of this construct was markedly reduced even after incubation for 11 days at 5-10 μM, pH 5.5 and 25 °C.29 Incubation of 13 μM PR (which also includes C67A and C95A substitutions, Figure 1B) for 23-70 h at room temperature in 50 mM sodium acetate buffer, pH 5, resulted in a small, time dependent appearance of products of autoproteolysis (Figure 2A, bands marked P). By contrast, incubation of PR20 for up to 115 h under the same conditions revealed essentially no lower-mass cleavage products (Figure 2B). We speculate that attenuation of autoproteolysis may contribute to the longevity of PR20 in vivo, thus possibly compensating for its decreased catalytic efficiency (kcat/Km, see below). It is likely that attenuated autoproteolysis of PR20 is favored by specific mutations influencing the catalytic machinery of PR20.
Figure 2.
Time course of autoproteolysis of (A) PR, (B) PR20, (C) PR20K7Q and (D) PRL33F/E34D/L63P at room temperature in 50 mM sodium acetate buffer, pH 5. Proteins were folded from 12 mM HCl by a quench protocol as previously described,25 to give a final concentration of 13 μM (as dimer) in 50 mM acetate, pH 5. Samples were withdrawn at the indicated times (in hours), mixed with gel loading buffer and analyzed by SDS-PAGE (20% homogeneous PhastGel, GE Healthcare). FL and P designate full length protease and products of autoproteolysis, respectively. M denotes molecular weight markers in kDa. Numbers below the gels denote hours of incubation at room temperature. (C and E) show that PR20 bearing the wild-type residue in position 7 (K7Q) is equally resistant to autoproteolysis as PR20. Sample PR20K7Q prior to initiating the assay is shown in lane C (panel C). (E and F) Time dependent decrease in protease mediated hydrolysis of substrate IV assayed at the indicated times at a final enzyme concentration of 440 nM for PR20K7Q and 330 nM PRL33F/E34D/L63P and final substrate concentration 380 μM, 28 °C. PRL33F/E34D/L63P shows a significant decrease in catalytic activity with increasing time of incubation, consistent with appearance of products of autoproteolysis, unlike PR20 or PR20K7Q (compare B and C with D). (D and F) demonstrate that mutations L33F and L63P are not responsible for the reduced rate of autoproteolysis seen with PR20 consistent with a rate comparable to wild-type PR.
It is noteworthy that the major site of autoproteolysis between Leu5/Trp64,29 in PR20 does not contain naturally selected mutations flanking 4 residues on either side (P/P’) of this site including K719 unlike the other 2 sites (L33/E34 and L63/I64, see Figure 1B). To ascertain that Leu5/Trp6 site is not a preferred site for autoproteolysis, we created a revertant bearing K7Q mutation (wild-type site) in PR20. Incubation of folded PR20K7Q showed no loss in catalytic activity over a period of 25 hours (Figure 2E) and analysis by SDS-PAGE even after 48 hours also failed to show significant products corresponding to cleavage of this site (Figure 2C). However, by ESI-MS we were able to detect a very small fraction of the protein (<5%) corresponding only to the product spanning protease residues 6-99 after 48 hours. Products corresponding to the cleavage at the other 2 sites are absent in PR20K7Q similar to PR20. These results clearly indicate that the 19 mutations selected in PR20 under drug pressure naturally suppress the Leu5/Trp6 cleavage without additional substitutions at P/P’ positions, different from L33/E34 and L63/I64 sites. We chose to retain the Q7K mutation as all our previous studies were carried out with this mutation, thus enabling a strict comparison with our present PR20 data on its characterization. Notably biochemical and structural studies are unreliable to carry out without this substitution in the wild-type protease because of rapid autoproteloysis leading to dimer dissociation and loss of catalytic activity4,20 and the mutation Q7K is not expected to influence autoproteolysis at L33/E34 and L63/I64 sites, which is indeed our focus.
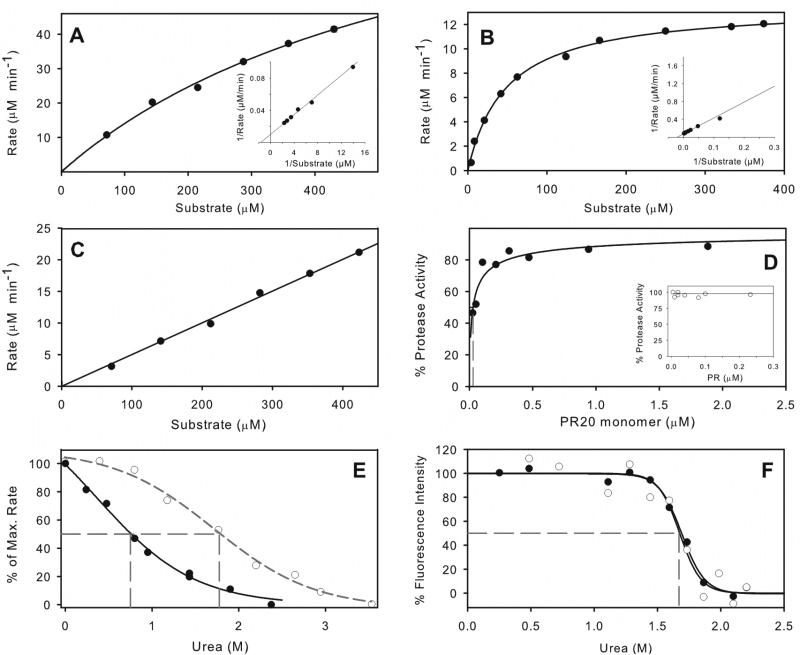
Hydrolysis by PR20 of a chromogenic substrate based on the Gag CA/SP1 cleavage site, substrate IV, exhibits Michaelis-Menten kinetics, with an estimated Km higher than concentrations that can be used, because of substrate inhibition above 500 μM (Figure 3A). Kinetic parameters Km = 617 ± 84 μM and kcat = 215 ± 19 μM min−1 at pH 5 and 250 mM NaCl were estimated by curve fitting to the acquired kinetic data (see also inset Lineweaver-Burke plot). A low value of kcat/Km for PR20, roughly an order of magnitude smaller than for PR under the same conditions (Figure 3B),31 thus appears to be largely a consequence of the high Km rather than a decreased kcat. Compromised substrate binding correlates with the open flap conformations evident in PR20 crystal structures.13 In particular, PR20 has 5 mutations between residues 32 and 37 in or near the flap hinge region that can account for different flap orientations. Another multi-drug resistant mutant, MDR769 (PDB accession code ITW7)12 which exhibits a pronounced “wide-open” conformation of both flaps, also contains several of the substitutions seen in PR20: namely, S37N, I62V, I63P, A71V, I84V, and L90M.
Figure 3.
Kinetic properties of PR20 and comparison with PR. Michaelis-Menten kinetics for hydrolysis of substrate IV by 0.5 μM PR20 in 50 mM sodium acetate buffer, pH 5, containing 250 mM sodium chloride (A), and its hydrolysis under the same conditions31 catalyzed by 0.078 μM PR (B). Kinetic parameters for PR20 are kcat ≥196 min−1 and Km ≥533 μM. For PR, kcat = 174 ± 3 min−1 and Km = 48.2 ± 3.1 μM. Solid lines are based on curve fitting of the Michaelis-Menten equation to the data. Lineweaver-Burke plots are shown as insets. (C) shows the linear dependence of reaction rate for PR20 hydrolysis on substrate concentration, resulting from an increased Km in the presence of 0.8 M urea under the same conditions as (A). (D) Dependence of catalytic activity on the concentration of PR20 in the same buffer gives a Kd of 29 ± 10 nM (dashed gray line). For comparison the inset shows that Kd for PR is < 5 nM under similar conditions20 (E) Effect of increasing urea concentration on hydrolysis of substrate IV by PR20 (solid symbols) and by PR20 (open symbols). (F) Effect of urea concentration on the intrinsic fluorescence of PR20 in 50 mM sodium acetate buffer, pH 5. Solid circles, data in the absence of inhibitor; open circles, in the presence of 2 μM DRV. The urea concentrations at which 50% of the maximum effect (UC50) is observed are indicated by dashed gray lines in (E) and (F).
A dimer dissociation constant of ~30 nM measured for PR20 (Figure 3D) is at least 5-fold higher than that for PR, which is too small to measure accurately by enzyme kinetics (<5 nM, see inset4). At a relatively low concentration, urea substantially decreases the catalytic activity as measured kinetically for PR20. The mid-point of maximum effect is observed at ~0.75 M urea (UC50) even in the presence of added sodium chloride, which is expected to stabilize the protease32 (Figure 3E). This observation is due at least in part to an increase in Km in the presence of urea, resulting in a strictly linear Michaelis-Menten plot (compare Figure 3A and 3C). A similar effect of urea on Km was previously observed for HIV-2 protease23 but not for the protease of group N.24 The effect of urea on PR20 as measured by the decrease in intrinsic fluorescence of PR20 in 50 mM sodium acetate buffer (Figure 3F) was qualitatively consistent with a significantly higher UC50 of ~1.7M, which is comparable to 1.8 M measured for PR (shown in 3E20) in the same buffer, and presumably corresponds to a true unfolding process. The dependence of fluorescence on urea concentration was unchanged in the presence of 2 μM DRV, indicating no stabilization, in spite of an estimated KL of 41 nM (see Table 2).
Table 2.
Thermodynamic parameters for DRV binding determined by ITC
| Protein | KA (M−1) | KL (nM)a | ΔH (kcal/mol) | −TΔS (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|
| PRb | 2.2 × 1011 | 0.005 | −12.1 | −3.1 | −15.2 |
| PR20c | 2.45 ± 0.59 × 107 | 41 ± 10 | −7.6 ± 0.1 | −2.6 | −10.2 |
| PRD25Nd | 3.17 ± 0.29 × 105 | 3,200 ± 293 | −5.8 ± 0.2 | −1.8 | −7.6 |
| ANAM-11c | 6.2 ± 3.4 × 108 | 1.6 ± 0.8 | −10.0 ± 0.07 | −2.1 | −12.1 |
All titrations, unless noted otherwise, were performed in 50 mM sodium acetate buffer at pH 5.0 and 28 °C as described.9 The thermodynamic parameters for PR20 and ANAM-11 are reported here for the first time. Other values incorporated in the table are solely for comparison.
Dissociation constant equal to 1/KA
For wild-type PR, in 10 mM sodium acetate buffer, pH 5, at 20 °C38
reference9
reference21
Thermal stability of PR20 and its monomeric T26A mutant
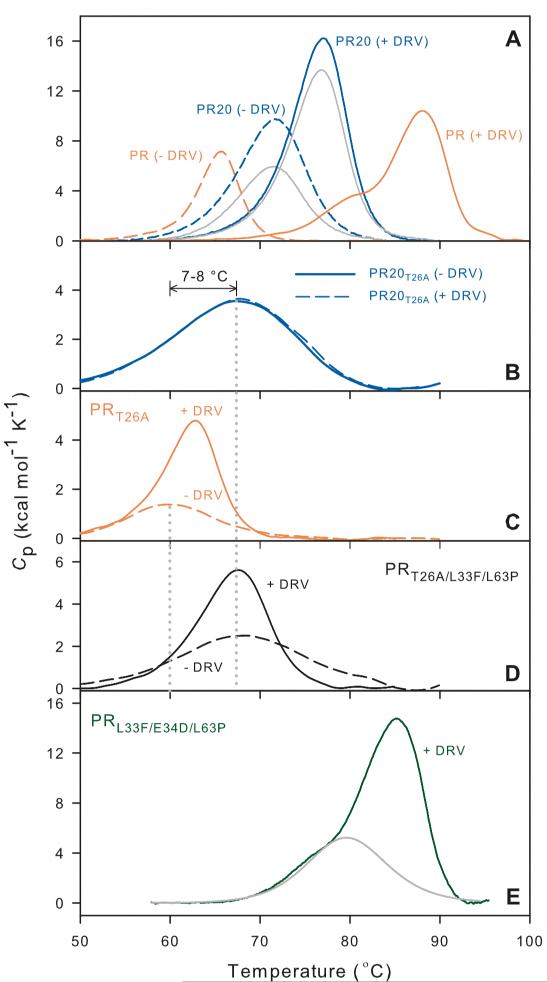
Although the effect of urea concentration on denaturation is similar for PR and PR20, their sensitivity to thermal denaturation differs, such that the Tm for thermal denaturation of dimeric PR20, as measured by differential scanning calorimetry (DSC), is 6 °C higher than for PR. In contrast, the Tm for the ternary PR20 dimer/inhibitor DRV complex is 11.1 °C lower than that of the corresponding PR dimer/DRV complex. This is a reflection of the drastically decreased binding affinity of PR20 for DRV (by a factor of at least 4000, Table 2).9 We previously used the increase in Tm upon inhibitor binding to dimeric PR (ΔTm = Tm of PR/PI complex – Tm of PR without PI) as a semi-quantitative indication of relative binding affinity;21 thus for PR20 ΔTm upon DRV binding is only 5.3 °C as compared to 22.4 °C for PR/DRV (Table 1A). Thermal denaturation of PR at pH ~5 is essentially irreversible,21 whereas denaturation of PR20 and its DRV complex exhibited partial reversibility as shown by repeated DSC scans, which is likely a result of its resistance to autoproteolysis.
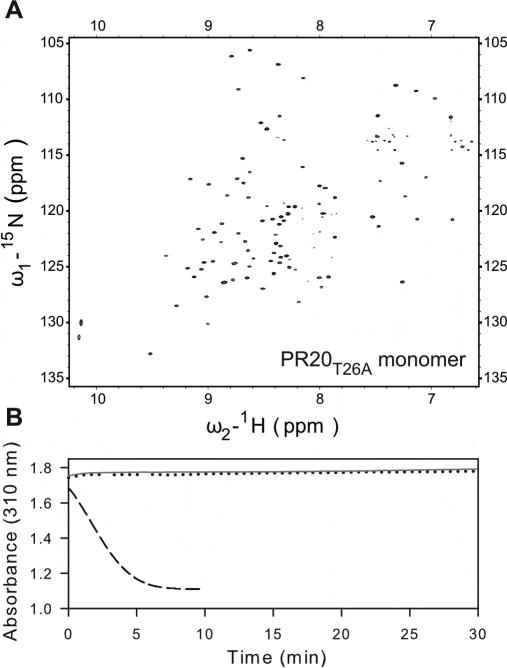
In order to determine whether the thermal stability of PR20 is related to its tertiary fold together with interactions at its interface as a dimer or simply an effect of enhanced stability of the monomer fold, we created a PR20 monomer construct. The PR dimer in the absence of inhibitor is held together mainly through interactions of its conserved active-site residues Asp25-Thr26-Gly27 (fireman's grip) and the terminal 4 residues 1-4 and 96-99. Based on our earlier observation with PR that a single substitution mutation T26A dramatically shifts the monomer-dimer equilibrium towards the monomer (Kd >500 μM),4 we introduced the same mutation in PR20. Figure 4A shows a 1H-15N TROSY-HSQC spectrum of PR20T26A acquired at a concentration of 100 μM (as monomer) at pH 5.8. Well dispersed signals characteristic of a folded structure were observed, analogous to that observed for PRT26A monomer.22 Similar to PR or its analogues, PR20 or PR20T26A could not be subjected to column fractionation, coupled to light-scattering and RI detectors to determine its molecular status (dimer or monomer), because of their non-specific interaction with column matrices. We estimate that Kd for PR20T26A is likely well above 500 μM, since Kd for PR20 is at least 5-fold higher than for PR (30 nM versus <5 nM).4 Several lines of evidence support the conclusion that PR20T26A is monomeric at the concentrations used in our experiments. 1) A spectrum of PR20T26A reacquired at a 4-fold lower protein concentration exhibited peaks that clearly superimpose on those observed at 100 μM indicating that PR20T26A is monomeric even at 100 μM. 2) PR20T26A is catalytically inactive when assayed at both 1 and 5 μM, consistent with its being a monomer (Figure 4B). 3) The DSC trace for PR20T26A in the absence of inhibitor exhibits a low, broad thermal transition and decreased Tm relative to PR20, similar to observations with PRT26A and other PR monomers.21
Figure 4.
(A) 600 MHz 1H-15N TROSY-HSQC spectrum of a freshly prepared uniformly 15N- and 13C-labeled PR20T26A (100 μM as monomer) in 20 mM sodium phosphate buffer, pH 5.8, 20 °C. A spectrum acquired using a 4-fold diluted sample (not shown) clearly superimposes on the one shown, indicating no changes in the monomer-dimer or tertiary fold status, and that the protein is all monomeric in conjunction with data shown in (B). (B) Activity assays with 360 μM substrate IV in 50 mM sodium acetate buffer, pH 5, containing 250 mM NaCl. Dotted black and solid gray lines are for 1 and 5 μM PR20T26A (as dimer), respectively. For comparison, the dashed line is for 300 nM active PR dimer shows depletion of substrate within 5 min.
Having established that PR20T26A is a monomer under the conditions of DSC (~ 14 μM as dimer) allowed us to compare the Tm of both PR and PR20 monomers under identical conditions. In the absence of an inhibitor PR20T26A exhibits a Tm that is 7.5 °C higher than that for PRT26A. This is comparable to the 6 °C stabilization of the PR20 dimer, and suggests that stabilization of the monomer fold may be a major factor contributing to the enhanced overall thermal stability of PR20 dimer. Interestingly, in the presence of excess DRV, the thermal transition for PRT26A becomes more pronounced (ΔH) and its Tm is increased by 3 °C. This suggests at least partial formation of a weak ternary PRT26A dimer/DRV complex promoted by DRV binding (Figure 5C). By contrast, no evidence for interaction of PR20T26A with DRV or ternary complex formation was detected (Figure 5B and Table 1A). This observation is consistent with both the lower affinity of PR20 for DRV and its somewhat higher dimer dissociation constant relative to PR. The nearly 4 orders of magnitude difference in affinity of DRV to PR20 relative to PR as measured by ITC (Table 2) corresponds to a ~5 kcal/mol less favorable free energy, much of which is due to a less favorable ΔH (−5.8 as compared to −12 kcal/mol) for binding, consistent with fewer and/or weaker polar interactions with the inhibitor that can result from its expanded binding cavity and more open flap conformation.13
Figure 5.
Differential scanning calorimetry of PR, PR20 and PRL33F/E34D/L63P dimers (A, E) and its corresponding monomers (B, C, D). Dashed and solid lines are thermal denaturation curves in the absence and presence of DRV, respectively. (A) Orange lines correspond to PR and blue to PR20. Traces shown in gray for PR20, obtained on a second scan of the DSC sample after completion of the initial scan, indicate partial reversibility of thermal melting in both the presence and absence of DRV. Data for PR, shown for comparison, are from.21 Panels B, C, and D show the effect of DRV on the thermal denaturation of PR20, PR, and PRL33F/L63P monomers bearing the T26A mutation. The absence of any effect of DRV on PR20T26A (B) indicates that this monomer likely does not form a complex with DRV. (E) Thermal denaturation of PRL33F/E34D/L63P dimer as its DRV complex (green). The curve in gray represents deconvolution of a transition with Tm 79.7 °C, analogous to that seen in the biphasic DSC plot for PR (panel A). For values of Tm and ΔTm derived from these plots see Table 1A.
Effect of mutations at sites of autoproteolysis in PR20
Natural mutations L33F and L63P in PR2019 occur at positions that are mutated (L33I, L63I) from the wild type to limit autoproteolysis of PR, and the presence of L63P provided a reasonable hypothesis to account for diminished autoproteolytic activity, since Pro occurs very rarely at the P1 position of substrates cleaved by PR.33 To examine the effects of these mutations we introduced L33F and L63P into a PR background (termed PRL33F/L63P). The resultant construct failed to accumulate, likely due to rapid degradation in the E. coli expression system. Thus we conclude that these two mutations by themselves do not impede autoproteolysis at their respective sites, likely requiring additional contributions from mutations at more-remote sites as seen in PR20 (Figure 1B). As it was not possible even after repeated attempts to isolate full length active PRL33F/L63P, we introduced a conservative E34D substitution, which occurs rarely if at all at the P1’ position of a variety of PR substrates33 and thus was thought unlikely to promote cleavage. This modification resulted in barely visible accumulation of PRL33F/E34D/L63P, sufficient to purify and examine its thermal stability and inhibitor binding. This mutant undergoes time-dependent autoproteolysis comparable to or more than that of PR, concomitant with a decrease in catalytic activity within few hours (Figure 2D and 2F). Major degradation products identified by ESI-MS correspond to well characterized self-cleavage sites for PR, between residues 33/34 and 63/64. Notably, thermal denaturation of PRL33F/E34D/L63P in the presence of a two-fold excess of DRV gives a biphasic transition curve that closely resembles that of PR with a large ΔH and a Tm for the major transition that is only 2.7 °C lower than that for PR/DRV, and 10 °C higher than PR20/DRV complex (Figure 5E), indicative of tight binding to the inhibitor. Thus L33F and L63P are also unlikely to play a significant role in the >4000-fold diminished DRV binding affinity of PR20 relative to PR, and mutations at other sites must be involved in drug resistance.
We also examined monomeric PRT26A/L33F/L63P to assess the effect of L33F and L63P on thermal stability of the PRT26A monomer fold. In the absence of inhibitors PRT26A/L33F//L63P exhibits a typically weak thermal transition, but surprisingly, with a Tm of ~68 °C that is very similar to that of PR20T26A and 7-8 °C higher than that of PRT26A (Figure 5D). The Tm of PRT26A/L33F/L63P, like that of PR20, is essentially unaffected in the presence of a 2-fold molar excess of DRV; however, ΔH for this transition is increased by DRV, possibly due to weak interaction between the protease monomer and the inhibitor. Taken together the present observations suggest that L33F and L63P in the wild-type context contribute significantly to stabilize the momomer fold while retaining the very high affinity for DRV. Thus when extrapolated, the increased monomer stability of PR20 likely does not directly correlate with its drastically diminished affinity for DRV.
Relationship between stability and inhibitor binding of single chain PR dimer
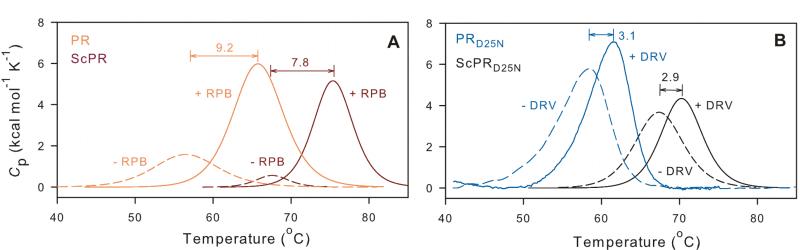
Since the monomer to dimer equilibrium is slightly less favorable for PR20 than for PR we assessed whether dimer stabilization exerts a significant effect on inhibitor binding. Thus we examined two single-chain (Sc) dimers consisting of (1) two PR monomers both with active-site Asp25 residues, linked by a GGSSG sequence (ScPR) between the C terminus of one monomer and the N terminus of the second monomer, and (2) the corresponding single chain dimer with both Asp25 residues mutated to Asn (ScPRD25N). Since the affinity of inhibitors to PR20 is relatively low, we used conditions that would correspond to similar (nM to low μM) affinities, i.e., ScPR with a substrate (CA/SP1) analogue inhibitor (RPB, H-Arg-Val-Leu-(®)-Phe-Glu-Ala-Nle-NH2, where (®) denotes a reduced peptide bond) at pH 3.6, and ScPRD25N with DRV, which binds very poorly to PRD25N, at pH 5.0. The Tm value (Table 1B) is 11 °C higher for ScPR than for homodimeric PR due to the stabilization provided by covalently linking two PR monomers. A comparable thermal stabilization of ~10 °C was observed for ScPRD25N. Inhibitors designed to bind with high affinity to the stable mature PR dimer were shown to promote dimerization of protease precursors34 and monomeric constructs4 which exhibit high Kds. However, our present models with moderate to weakly bound inhibitors exhibit ΔTm values on inhibitor binding (Table 1B and Figure 6) to the single chain and homodimeric proteins that are the same within the accuracy of the DSC experiments (several of which exhibit low, broad thermal responses). Binding of RPB to both ScPR and PR at pH 3.6 gave ΔTm(I) values of 8-9 °C, and binding of DRV to ScPRD25N and homodimeric PRD25N gave ΔTm(I) values of ~3 °C. It is interesting to note that stabilization due to tethering (ΔTm(Sc), Table 1B) is similar in the presence and absence of inhibitors. We conclude that the ΔTm values observed with inhibitors that are not tightly bound may primarily reflect the disruption of direct interactions of the inhibitor with the active site and contiguous residues, which are essentially the same for both dimeric and single-chain constructs and do not induce significant remote changes that affect dimer stability. Since all currently available competitive inhibitors of PR bind only weakly to PR20, this lack of cooperative effects suggests that dimer stability may have, at best, only a limited role in PR20-inhibitor interactions.
Figure 6.
Effect of dimer stability on inhibitor binding and thermal denaturation of single-chain PR constructs, ScPR (dark red) and ScPRD25N (black) in comparison with PR (orange) and PRD25N (blue) homodimers. Dashed and solid lines designate scans without inhibitor and with ~2-fold molar excess of inhibitor RPB or DRV relative to the respective proteases (as dimers). ΔTm values shown for each pair of traces show the increase in Tm of each protein upon inhibitor binding. For experimental details see Materials and Methods. Actual Tm values are given in Table 1B.
Cleavage of natural substrates by PR20
Having defined some of the physical properties of PR20, and measured its kinetic parameters by using a chromogenic substrate, it was of interest to examine the ability of PR20, in comparison with PR, to catalyze cleavages in the Gag polyprotein ultimately required for the assembly of the virus particle. Catalytically competent, dimeric PR is released in a stepwise process (Figure 1A) involving intramolecular cleavages of the Gag-Pol (via transient dimers) (1) at the SP1/NC site followed by (2) between the TFR and PR domains at the N-terminus of PR, and subsequent intermolecular cleavage between PR and RT. Mature PR thus becomes available to catalyze release of structural proteins from the more prevalent Gag polyprotein by cleavages at SP1/NC followed by MA/CA and SP2/p6, and finally at the CA/SP1 and NC/SP2 sites.2 Highly drug resistant protease mutants are compromised in their fitness to replicate, due to reduced ability to cleave the polyprotein at these sites. PR20 exhibits significantly reduced catalytic efficiency for peptides corresponding to several of these cleavage sites at the high ionic strength optimal for oligopeptide substrate processing (Table 3), with a wide range of differences in kcat/Km between 0.2% and 7% relative to PR. Km values for PR and PR20 are similar for SP1/NC, NC/SP2 and SP2/P6, such that differences in catalytic efficiency (kcat/Km) on these substrates largely reflect differences in kcat. The catalytic efficiency decreases in the order shown in the last column and the trend for PR and PR20 remains the same. The substrates cluster into two groups: SP1/NC, MA/CA, and CA/SP1 (group 1) are hydrolyzed with relative efficiency, whereas NC/SP2 and SP2/P6 (group 2) are processed much less efficiently. Differences between the two groups are not clearly related to the temporal order of Gag processing. Thus in group 1 the two best substrates for both enzymes, MA/CA and SP1/NC, represent early Gag cleavages, whereas cleavage at CA/SP1 (efficiency comparable to MA/CA) occurs later. Similarly the early SP2/p6 cleavage is relatively inefficient. This leads us to speculate that the full-length Gag conformation(s) and the microenvironment in vivo also likely account for some of these differences.
Table 3.
Kinetic data for PR-catalyzed hydrolysis of synthetic peptides corresponding to natural cleavage sites in HIV-1 Gag polyprotein
| Substrate | Sequence | Enzyme | Km (mM) | kcat (s−1) | kcat/Km (mM−1s−1) | Activty (%) relative to PR | Order of cleavagea |
|---|---|---|---|---|---|---|---|
| MA/CA | VSQNY↓PIVQ | PR | 0.15 | 6.9 | 46.0 | 2 | |
| PR20 | 0.91 | 3.0 | 3.2 | 7 | 1 | ||
| CA/SP1 | KARVL↓AEAMS | PR | 0.02 | 0.9 | 42.4b | 3 | |
| PR20 | 0.11 | 0.1 | 1.2 | 2.8 | 3 | ||
| SP1/NC | TATIM↓MQRGN | PR | 0.10 | 6.7 | 67.0 | 1 | |
| PR20 | 0.10 | 0.2 | 2.0 | 3.0 | 2 | ||
| NC/SP2 | ERQAN↓FLGKI | PR | 0.17 | 0.15 | 0.9c | 5 | |
| PR20 | 0.23 | 0.0005 | 0.002 | 0.2 | 6 | ||
| NC/SP2 mutant | ERRVN↓FLGKI | PR | nd | nd | 20.1c | 4 | |
| PR20 | 0.23 | 0.01 | 0.04 | 0.2 | 4 | ||
| SP2/p6 | RPQNF↓LQSRP | PR | 0.48 | 0.4 | 0.8 | 6 | |
| PR20 | 0.43 | 0.01 | 0.02 | 2.5 | 5 |
The MA/CA cleavage in Gag is critical in that it must proceed almost to completion in order to ensure proper assembly of infectious virions.2 In accordance, MA/CA peptide is the best substrate examined for PR20, although not for PR. It exhibits the largest kcat (only 2.3-fold slower than PR), which is offset by a high Km. However, the local concentration of Gag is estimated to be ~4.4 mM within the immature virus particle2 during or shortly after budding, when this cleavage probably occurs.2,35 Thus the high Km for MA/CA cleavage by mature PR20 may not present a serious disadvantage. In contrast to MA/CA, cleavage of the SP1/NC substrate by PR20 exhibits a moderately low kcat and a low Km, resulting in overall catalytic efficiency that is close to that for MA/CA. The NC/SP2 substrate is cleaved by PR20 with the poorest efficiency. A double mutant containing polymorphisms QA→RV associated with drug resistance exhibited kcat/Km ~20-fold greater than for the wild type substrate. Interestingly, A→V mutation at P2 of the NC/SP2 site was shown to accompany the pair of PR mutations L33F/L63P in three HIV sequences derived from patients exhibiting multiple drug resistance.36 However, the efficiency of PR20 relative to PR was unchanged for the mutant substrate. This suggests that the same pair of mutations present in Gag would confer equal advantages to PR and PR20. We speculate that substrate polymorphisms at other, as yet unidentified, sites in Gag and Gag-Pol may contribute to enhancing the catalytic efficiency of PR20 and viability of the virus.
Concluding remarks
The mutant HIV protease PR20 bears multiple mutations that render it extremely resistant to PIs due to its weak binding to these inhibitors.9,13 Although some of these mutations are known to compromise fitness, the virus is able to propagate. In fact PR20 is a reasonably robust enzyme due to many secondary mutations that preserve its overall stability and function. (1) Mutations at or near sites of autoproteolysis acting in conjunction with other mutations in PR20 presumably increase the longevity of active PR20 in vivo by limiting degradation; (2) PR20 exhibits enhanced thermal stability relative to PR, which contributes to its functionality and viability of the virus (primarily through selection of mutations L33F and L63P) and (3) PR20 cleaves peptides corresponding to sites in the Gag polyprotein essential for viral maturation. However, this catalysis is highly inefficient relative to PR. PR20 hydrolyzes a co-evolved NC/SP2 substrate with ~20-fold increased efficiency relative to the wild-type site although not with the same efficiency as PR cleaving its natural NC/SP2 substrate. This is consistent with observations37 that mutations affecting cleavage sites in the Gag and Gag-Pol can co-evolve with a highly drug resistant PR bearing multiple mutations, and provide a mechanism for partially circumventing inefficient catalysis. Lack of available sequence information spanning the Gag of the PR20 isolate precludes the identification of the role of such mutations in preserving the viability of this virus.
Our observation of a slightly compromised dimer dissociation constant for PR20 relative to PR, as well as comparable thermal stabilization of the PR20 monomer and dimer (6-7.5 °C) suggests that the enhanced thermal stability of this mutant protease over PR is predominantly or entirely the result of a more stable monomer fold. Thus, the enlarged binding cavity13 and moderate weakening of interactions in regions contiguous to the active site and flaps may limit tight binding of inhibitors, while compensating changes elsewhere preserve the protein's overall structural integrity. The DRM, ANAM-11,11 bears six mutations identical or similar to PR20 (L10I/F, M36I, L63P, A71V, I84V, L90M) and exhibits similar properties of increased dimer dissociation (Kd = 0.1 ± 0.04 μM)9 along with moderately enhanced thermal stability (Table 1A). We speculate that monomer stabilization similar to that observed in the present study may be a characteristic of other DRMs, and possibly constitutes an “evolutionary necessity” to permit the viability of highly drug resistant mutants. Since the protease precursor is monomeric and undergoes autoprocessing only via a transient dimer, mutations that structurally stabilize the monomer may also shift the equilibrium to favor precursor monomers over transient dimers that can be trapped as inactive dimer/PI complexes. This could account in part for the failure of PIs to interact with PR20 precursor to inhibit autoprocessing, thus making it impossible to isolate the precursor TFR-PR20, unlike TFR-PR where extensive studies have been performed.9 A monomer construct, PR20T26A, shows no evidence for binding or partial dimerization in the presence of DRV under conditions where its (pseudo)-wild type counterpart, PRT26A, interacts weakly with this PI (Figure 5B and C). Substitutions of two PR20 residues, L33F and L63P, in PR or its monomeric model, PRT26A, increase the monomer stability to an extent comparable with PR20, but permit effective DRV binding to a PR dimer. Thus, other mutations must contribute to the remarkable resistance to PIs exhibited by PR20. It is tempting to suggest that identification and targeting of such combinations of mutations could provide a potential strategy to overcome PI resistance in highly evolved PR variants or their precursors bearing multiple mutations.
ACKNOWLEDGMENTS
We thank Drs. Adriaan Bax and Irene T. Weber for discussions. PIs used in this study were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This research was supported by the Intramural Research Program of the NIDDK, National Institutes of Health and the Intramural AIDS-Targeted Program of the Office of the Director, NIH, and in part by the TÁMOP 4.2.2.A-11/1/KONV-2012-0023 “VÉD-ELEM” and by the Hungarian Science and Research Fund (OTKA 101591).
Abbreviations
- HIV-1
human immunodeficiency virus type 1
- PR
mature HIV-1 protease containing the mutations Q7K, L33I, L63I, C67A, C95A
- PI
clinical protease inhibitor
- DRM
drug resistance mutation (or drug resistant mutant), PR20, wild-type protease bearing 19 DRMs, Q7K, C67A and C95A
- DRV
darunavir
- DSC
differential scanning calorimetry
- ITC
isothermal titration calorimetry
- Kd
protease dimer dissociation constant
- KL
ligand (inhibitor) dissociation constant
- ScPR
single chain PR
- HSQC
heteronuclear single quantum coherence
- TROSY
transverse relaxation optimized spectroscopy
REFERENCES
- 1.Oroszlan S, Luftig RB. Retroviral proteinases. Curr. Top. Microbiol. Immunol. 1990;157:153–185. doi: 10.1007/978-3-642-75218-6_6. [DOI] [PubMed] [Google Scholar]
- 2.Lee SK, Potempa M, Swanstrom R. The choreography of HIV-1 proteolytic processing and virion assembly. J. Biol. Chem. 2012;287:40867–40874. doi: 10.1074/jbc.R112.399444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis JM, Weber IT, Tozser J, Clore GM, Gronenborn AM. HIV-1 protease: maturation, enzyme specificity, and drug resistance. Adv. Pharmacol. 2000;49:111–146. doi: 10.1016/s1054-3589(00)49025-3. [DOI] [PubMed] [Google Scholar]
- 4.Louis JM, Ishima R, Torchia DA, Weber IT. HIV-1 Protease: Structure, Dynamics, and Inhibition. Adv. Pharmacol. 2007;55:261–298. doi: 10.1016/S1054-3589(07)55008-8. [DOI] [PubMed] [Google Scholar]
- 5.Pearl LH, Taylor WR. A structural model for the retroviral proteases. Nature. 1987;329:351–354. doi: 10.1038/329351a0. [DOI] [PubMed] [Google Scholar]
- 6.Wlodawer A, Erickson J. Structure-based inhibitors of HIV-1 protease. Annu. Rev. Biochem. 1993;62:543–585. doi: 10.1146/annurev.bi.62.070193.002551. [DOI] [PubMed] [Google Scholar]
- 7.Johnson VA, Calvez V, Gunthard HF, Paredes R, Pillay D, Shafer RW, Wensing AM, Richman DD. Update of the drug resistance mutations in HIV-1: March 2013. Top. Antivir. Med. 2013;21:6–14. [PMC free article] [PubMed] [Google Scholar]
- 8.Agniswamy J, Weber IT. HIV-1 Protease: Structural perspectives on drug resistance. Viruses. 2009;1:1110–1136. doi: 10.3390/v1031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louis JM, Aniana A, Weber IT, Sayer JM. Inhibition of autoprocessing of natural variants and multidrug resistant mutant precursors of HIV-1 protease by clinical inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9072–9077. doi: 10.1073/pnas.1102278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pieniazek D, Rayfield M, Hu DJ, Nkengasong J, Wiktor SZ, Downing R, Biryahwaho B, Mastro T, Tanuri A, Soriano V, Lal R, Dondero T. Protease sequences from HIV-1 group M subtypes A-H reveal distinct amino acid mutation patterns associated with protease resistance in protease inhibitor-naive individuals worldwide. HIV Variant Working Group. AIDS. 2000;14:1489–1495. doi: 10.1097/00002030-200007280-00004. [DOI] [PubMed] [Google Scholar]
- 11.Muzammil S, Ross P, Freire E. A major role for a set of non-active site mutations in the development of HIV-1 protease drug resistance. Biochemistry. 2003;42:631–638. doi: 10.1021/bi027019u. [DOI] [PubMed] [Google Scholar]
- 12.Martin P, Vickrey JF, Proteasa G, Jimenez YL, Wawrzak Z, Winters MA, Merigan TC, Kovari LC. “Wide-open” 1.3 A structure of a multidrug-resistant HIV-1 protease as a drug target. Structure. 2005;13:1887–1895. doi: 10.1016/j.str.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Agniswamy J, Shen CH, Aniana A, Louis JM, Sayer JM, Weber IT. HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements. Biochemistry. 2012;51:2819–2828. doi: 10.1021/bi2018317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nijhuis M, Schuurman R, de Jong JD, Erickson J, Gustchina E, Albert J, Schipper P, Gulnik S, Boucher CA. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS. 1999;13:2349–2359. doi: 10.1097/00002030-199912030-00006. [DOI] [PubMed] [Google Scholar]
- 15.Quiñones-Mateu ME, Weber J, Rangel HR, Chakraborty B. HIV-1 Fitness and Antiviral Drug Resistance. AIDS Rev. 2001;3:223–234. [Google Scholar]
- 16.van Maarseveen NM, de JD, Boucher CA, Nijhuis M. An increase in viral replicative capacity drives the evolution of protease inhibitor-resistant human immunodeficiency virus type 1 in the absence of drugs. J. Acquir. Immune. Defic. Syndr. 2006;42:162–168. doi: 10.1097/01.qai.0000219787.65915.56. [DOI] [PubMed] [Google Scholar]
- 17.Chang MW, Torbett BE. Accessory mutations maintain stability in drug-resistant HIV-1 protease. J. Mol. Biol. 2011;410:756–760. doi: 10.1016/j.jmb.2011.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanchunas J, Jr., Langley DR, Tao L, Rose RE, Friborg J, Colonno RJ, Doyle ML. Molecular basis for increased susceptibility of isolates with atazanavir resistance-conferring substitution I50L to other protease inhibitors. Antimicrob. Agents Chemother. 2005;49:3825–3832. doi: 10.1128/AAC.49.9.3825-3832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dierynck I, De Wit M, Gustin E, Keuleers I, Vandersmissen J, Hallenberger S, Hertogs K. Binding kinetics of darunavir to human immunodeficiency virus type 1 protease explain the potent antiviral activity and high genetic barrier. J. Virol. 2007;81:13845–13851. doi: 10.1128/JVI.01184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louis JM, Clore GM, Gronenborn AM. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat. Struct. Biol. 1999;6:868–875. doi: 10.1038/12327. [DOI] [PubMed] [Google Scholar]
- 21.Sayer JM, Liu F, Ishima R, Weber IT, Louis JM. Effect of the active-site D25N mutation on the structure, stability and ligand binding of the mature HIV-1 protease. J. Biol. Chem. 2008;283:13459–13470. doi: 10.1074/jbc.M708506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louis JM, Ishima R, Nesheiwat I, Pannell LK, Lynch SM, Torchia DA, Gronenborn AM. Revisiting monomeric HIV-1 protease. Characterization and redesign for improved properties. J. Biol. Chem. 2003;278:6085–6092. doi: 10.1074/jbc.M209726200. [DOI] [PubMed] [Google Scholar]
- 23.Louis JM, Ishima R, Aniana A, Sayer JM. Revealing the dimer dissociation and existence of a folded monomer of the mature HIV-2 protease. Protein Sci. 2009;18:2442–2453. doi: 10.1002/pro.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sayer JM, Agniswamy J, Weber IT, Louis JM. Autocatalytic maturation, physical/chemical properties, and crystal structure of group N HIV-1 protease: relevance to drug resistance. Protein Sci. 2010;19:2055–2072. doi: 10.1002/pro.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishima R, Torchia DA, Louis JM. Mutational and structural studies aimed at characterizing the monomer of HIV-1 protease and its precursor. J. Biol. Chem. 2007;282:17190–17199. doi: 10.1074/jbc.M701304200. [DOI] [PubMed] [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Goddard TD, Kneller DG. Sparky 3. University of California; San Francisco: 2010. Sparky 3. [Google Scholar]
- 28.Tozser J, Blaha I, Copeland TD, Wondrak EM, Oroszlan S. Comparison of the HIV-1 and HIV-2 proteinases using oligopeptide substrates representing cleavage sites in Gag and Gag-Pol polyproteins. FEBS Lett. 1991;281:77–80. doi: 10.1016/0014-5793(91)80362-7. [DOI] [PubMed] [Google Scholar]
- 29.Mildner AM, Rothrock DJ, Leone JW, Bannow CA, Lull JM, Reardon IM, Sarcich JL, Howe WJ, Tomich CS, Smith CW. The HIV-1 protease as enzyme and substrate: mutagenesis of autolysis sites and generation of a stable mutant with retained kinetic properties. Biochemistry. 1994;33:9405–9413. doi: 10.1021/bi00198a005. [DOI] [PubMed] [Google Scholar]
- 30.Tie Y, Boross PI, Wang YF, Gaddis L, Hussain AK, Leshchenko S, Ghosh AK, Louis JM, Harrison RW, Weber IT. High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multi-drug-resistant clinical strains. J. Mol. Biol. 2004;338:341–352. doi: 10.1016/j.jmb.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 31.Ishima R, Torchia DA, Lynch SM, Gronenborn AM, Louis JM. Solution structure of the mature HIV-1 protease monomer: insight into the tertiary fold and stability of a precursor. J. Biol. Chem. 2003;278:43311–43319. doi: 10.1074/jbc.M307549200. [DOI] [PubMed] [Google Scholar]
- 32.Szeltner Z, Polgar L. Conformational stability and catalytic activity of HIV-1 protease are both enhanced at high salt concentration. J. Biol. Chem. 1996;271:5458–5463. doi: 10.1074/jbc.271.10.5458. [DOI] [PubMed] [Google Scholar]
- 33.Chou KC, Tomasselli AG, Reardon IM, Heinrikson RL. Predicting human immunodeficiency virus protease cleavage sites in proteins by a discriminant function method. Proteins. 1996;24:51–72. doi: 10.1002/(SICI)1097-0134(199601)24:1<51::AID-PROT4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 34.Agniswamy J, Sayer JM, Weber IT, Louis JM. Terminal Interface Conformations Modulate Dimer Stability Prior to Amino Terminal Autoprocessing of HIV-1 Protease. Biochemistry. 2012;51:1041–1050. doi: 10.1021/bi201809s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adamson CS, Freed EO. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv. Pharmacol. 2007;55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- 36.Dam E, Quercia R, Glass B, Descamps D, Launay O, Duval X, Krausslich HG, Hance AJ, Clavel F. Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS. Pathog. 2009;5:e1000345. doi: 10.1371/journal.ppat.1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozisek M, Henke S, Saskova KG, Jacobs GB, Schuch A, Buchholz B, Muller V, Krausslich HG, Rezacova P, Konvalinka J, Bodem J. Mutations in HIV-1 gag and pol compensate for the loss of viral fitness caused by a highly mutated protease. Antimicrob. Agents Chemother. 2012;56:4320–4330. doi: 10.1128/AAC.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King NM, Prabu-Jeyabalan M, Nalivaika EA, Wigerinck P, de Bethune MP, Schiffer CA. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J. Virol. 2004;78:12012–12021. doi: 10.1128/JVI.78.21.12012-12021.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feher A, Weber IT, Bagossi P, Boross P, Mahalingam B, Louis JM, Copeland TD, Torshin IY, Harrison RW, Tozser J. Effect of sequence polymorphism and drug resistance on two HIV-1 Gag processing sites. Eur. J. Biochem. 2002;269:4114–4120. doi: 10.1046/j.1432-1033.2002.03105.x. [DOI] [PubMed] [Google Scholar]
- 40.Ishima R, Ghirlando R, Tozser J, Gronenborn AM, Torchia DA, Louis JM. Folded monomer of HIV-1 protease. J. Biol. Chem. 2001;276:49110–49116. doi: 10.1074/jbc.M108136200. [DOI] [PubMed] [Google Scholar]
- 41.Ceccherini-Silberstein F, Erba F, Gago F, Bertoli A, Forbici F, Bellocchi MC, Gori C, D'Arrigo R, Marcon L, Balotta C, Antinori A, Monforte AD, Perno CF. Identification of the minimal conserved structure of HIV-1 protease in the presence and absence of drug pressure. AIDS. 2004;18:F11–F19. doi: 10.1097/01.aids.0000131394.76221.02. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]