Abstract
Bioenergetic dysfunction occurs in Alzheimer's disease (AD) and mild cognitive impairment (MCI), a clinical syndrome that frequently precedes symptomatic AD. In this study, we modeled AD and MCI bioenergetic dysfunction by transferring mitochondria from MCI, AD and control subject platelets to mtDNA-depleted SH-SY5Y cells. Bioenergetic fluxes and bioenergetics-related infrastructures were characterized in the resulting cytoplasmic hybrid (cybrid) cell lines. Relative to control cybrids, AD and MCI cybrids showed changes in oxygen consumption, respiratory coupling and glucose utilization. AD and MCI cybrids had higher ADP/ATP and lower NAD+/NADH ratios. AD and MCI cybrids exhibited differences in proteins that monitor, respond to or regulate cell bioenergetic fluxes including HIF1α, PGC1α, SIRT1, AMPK, p38 MAPK and mTOR. Several endpoints suggested mitochondrial mass increased in the AD cybrid group and probably to a lesser extent in the MCI cybrid group, and that the mitochondrial fission–fusion balance shifted towards increased fission in the AD and MCI cybrids. As many of the changes we observed in AD and MCI cybrid models are also seen in AD subject brains, we conclude reduced bioenergetic function is present during very early AD, is not brain-limited and induces protean retrograde responses that likely have both adaptive and mal-adaptive consequences.
INTRODUCTION
Perturbed bioenergetic function, and especially mitochondrial dysfunction, is observed in brains and peripheral tissues of subjects with Alzheimer's disease (AD) and mild cognitive impairment (MCI) (1,2), a clinical syndrome that frequently represents a transition between normal cognition and AD dementia (3). Neurons are vulnerable to mitochondrial dysfunction due to their high energy demands and dependence on respiration to generate ATP (4). Mitochondrial dysfunction may, therefore, drive or mediate various AD pathologies (5).
While AD hippocampal neurons have reduced numbers of morphologically preserved mitochondria (6), other changes in overall neuron mitochondrial mass are less straightforward. Dystrophic neurites concentrate mitochondria within autophagic vesicles, a phenomenon that could reflect defective retrograde transport (7–9). While PCR-amplifiable mitochondrial DNA (mtDNA) levels are reduced (10), approaches that detect mtDNA within autophagosomes in addition to mtDNA within intact mitochondria suggest hippocampal neuron mtDNA levels may actually increase (6). Other studies report mitochondrial mass increases in healthy-appearing neurons while markedly declining in tangle-bearing neurons from AD subject hippocampi (11).
Mitochondrial fission–fusion dynamics are also altered in AD and could impact organelle proliferation, movement and distribution (12–14). Tissue from AD patient hippocampi shows increased Fis1 as well as reduced Drp1, Opa1, Mfn1 and Mfn2. These proteins further redistribute away from neuron processes and towards cell bodies (15). In AD subject frontal cortex, expression of the pro-fission Drp1 and Fis1 genes increases, while Mfn1, Mnf2 and Opa1 pro-fusion gene expression decreases (16). In AD fibroblasts, H2O2 and Aβ (amyloid beta) treatment reduces Drp1 and disrupts mitochondrial networks (17). Perturbed mitochondrial fission–fusion in AD may relate to increased mitochondrial autophagy (mitophagy), since altering fusion and fission protein levels induces mitophagy (18).
In order to study AD and MCI bioenergetic fluxes, intermediates and proteins that relate to those fluxes, and mitochondrial mass/morphology-related phenomena, we generated cytoplasmic hybrid (cybrid) cell lines using platelet mitochondria from AD, MCI and age-matched control subjects. We found that bioenergetic flux and flux-associated parameters were altered in MCI and AD cybrid lines. Many of the changes we observed either mediate or represent retrograde responses that are probably compensatory in nature. These compensatory changes, though, may have mal-adaptive as well as adaptive consequences.
RESULTS
COX Vmax activities
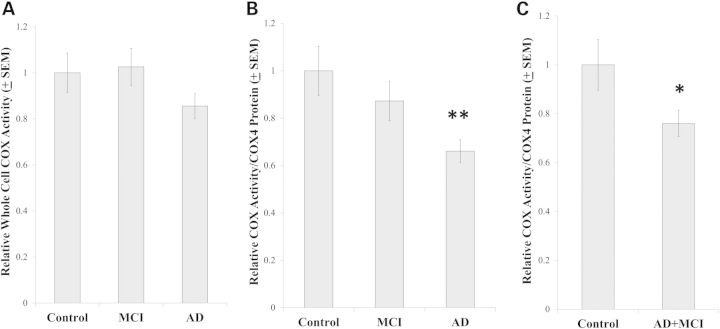
Numerous studies have found that following a 6-week selection process, relative to control cybrid cell lines, AD cybrid cell lines have reduced COX Vmax activities (1). One previous study reported MCI cybrid COX Vmax activities are also reduced relative to that of control cybrids (19), and another study found that after correcting for mtDNA synthesis rates AD cybrid COX Vmax activity reductions persist for at least 1–2 months after the 6-week cybrid selection process is complete (20). Our COX Vmax data, which come from cybrid cell lines maintained in continuous culture for two additional months after the 6-week selection process was completed, are for the most part consistent with the existing literature. Although reporting whole cell COX Vmax activities only as pseudo-first-order rate constants normalized to mg protein (sec−1/mg protein) did not reveal differences (Fig. 1A), further normalizing to a marker of COX quantity, in this case COX4I1 protein, found that the COX activity mean per COX subunit protein was lower in the AD cybrid lines than it was in the control cybrid lines (Fig. 1B). The MCI cybrid COX Vmax activity mean was intermediate, as it was not statistically different from either the control or AD group means (Fig. 1B). The possibility that COX function is perturbed in MCI cybrids is further supported by an analysis in which the MCI and AD cybrids were combined to create a single group. In this case, the AD+MCI cybrid COX Vmax mean, when normalized to COX4I1 protein levels, was lower than that of the control cybrids (Fig. 1C). Conversely, adding the MCI and control cybrids together created a group whose COX Vmax mean was statistically comparable to that of the AD cybrids (data not shown).
Figure 1.
COX Vmax activities. The pseudo-first-order rate constant in sec−1 was spectrophotometrically determined by following the conversion of reduced cytochrome c to oxidized cytochrome c. In (A), each cybrid line's rate constant was normalized to the amount of whole cell protein in its assay cuvette to provide a rate in sec−1/mg protein (A). As immunochemical measurements of COX4I1 protein revealed the amount of COX4I1 protein differed between the groups (see Fig. 8), we also normalized the sec−1/mg protein rate for each line to its relative amount of COX4I1 protein (B and C). Asterisk indicates a P < 0.05 difference from the control group; Double asterisk indicates a P < 0.01 difference from the control group.
Oxygen and glucose fluxes
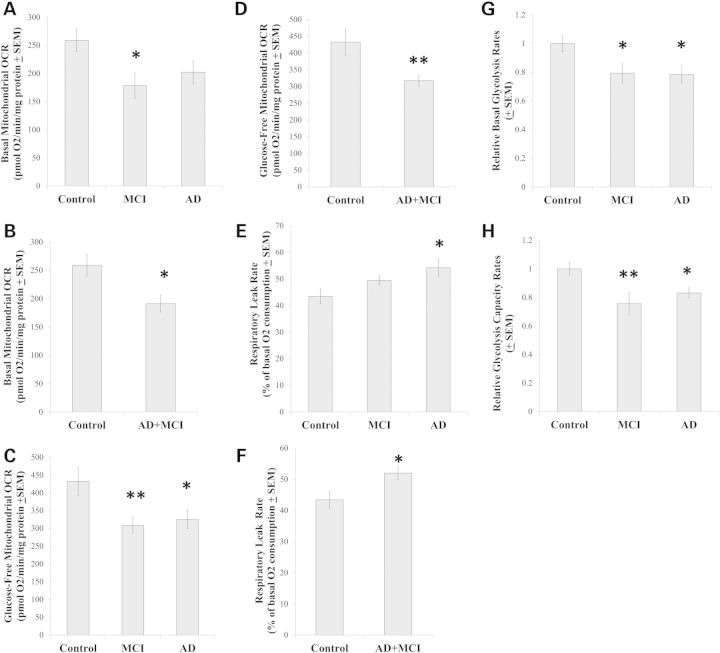
We used a Seahorse XF24 Analyzer to assess cybrid line respiration-related fluxes. To quantify mitochondrial OCRs under unstressed conditions, the total OCR for each cell line was determined in the presence of 25 mm glucose before and after the addition of rotenone and antimycin A to the cells. Rotenone (a complex I inhibitor) and antimycin A (a complex III inhibitor) blocks mitochondrial respiration, and reveals the cell non-mitochondrial OCR contribution. Subtracting the non-mitochondrial OCR from the total OCR yields the mitochondrial OCR. The mitochondrial OCRs were then normalized to the amount of total protein that was present in the well that was being read. In the presence of 25 mm glucose, the MCI group OCR mean was lower than the control group OCR mean (Fig. 2A). On post-hoc testing, the AD group was statistically comparable to both groups (P = 0.07 compared with the control group, and P = 0.42 compared with the MCI group). Combining the MCI and AD cybrids generated a single group whose OCR was lower than that of the controls (Fig. 2B), while a group formed by combining the MCI with the control group was not statistically different from the AD group (data not shown).
Figure 2.
Mitochondrial respiration and ECAR values. Mitochondrial OCR values in the presence of non-limiting amounts of glucose (A and B) and in the absence of glucose (C and D) were determined for each separate cybrid group and for the combined AD+MCI group. Mitochondrial OCR values for each cybrid group (E) and for the AD+MCI group (F) were determined following exposure to oligomycin. The basal glycolysis rate for each cybrid line was estimated by determining its ECAR in the presence of non-limiting amounts of glucose (G). The maximal glycolysis rate (the glycolysis capacity) for each cybrid line was estimated by determining the ECAR in the presence of glucose and oligomycin (H). Asterisk indicates a P < 0.05 difference from the control group; Double asterisk indicates a P < 0.01 difference from the control group.
Glucose deprivation prevents glycolysis, which renders cells increasingly dependent on oxidative phosphorylation to make energy. This manifests as an increased OCR. Relative to total cell OCR measurements taken under 25 mm glucose conditions, under glucose deprivation conditions the control group OCR increased by 67%, the MCI group OCR increased by 74% and the AD group OCR increased by 61%. When no glucose was present in the assay medium, the mitochondrial OCR rate was lower in the MCI and AD groups than it was in the control group (Fig. 2C). Combining the MCI and AD cybrids generated a single group whose glucose-deprived OCR was lower than that of the controls (Fig. 2D), while a group formed by combining the MCI with the control group was not statistically different from the AD group (data not shown).
To evaluate the mitochondrial proton leak rate, we determined each cell line's OCR following oligomycin treatment. Oligomycin inhibits the mitochondrial ATP synthase, at which point all mitochondrial oxygen consumption is due to the ATP synthase-independent leakage of protons from the intermembrane space to the mitochondrial matrix. The respiratory leak rate was higher in the AD cybrids than it was in the control cybrids. The MCI leak rate was intermediate, as its value was not significantly different from the other two groups (Fig. 2E). Combining the MCI and AD groups in order to increase statistical power yielded a group whose leak rate exceeded that of the control group (Fig. 2F). Combining the control and MCI groups yielded a group whose leak rate did not differ from the AD group (data not shown).
In addition to measuring medium oxygen levels and calculating the rate at which oxygen is consumed by cultured cells, Seahorse Analyzers also measure medium pH and calculate the rate at which cells acidify their surrounding medium. Since cell lactic acid excretion is primarily responsible for medium acidification, and lactic acid is produced by anaerobic glycolysis, ECARs can be used to estimate glycolysis fluxes.
We initially placed cells from each of our cybrid lines in a glucose-free medium, at which point ECARs were quite low and equivalent between the groups; at the conclusion of each experiment 2-deoxyglucose was added to each well, and revealed baseline ECAR measurements were stable throughout. The post-2-deoxyglucose ECARs were used to provide non-glycolysis acidification rates. An initial injection introduced 10 mm glucose to the cells. The non-glycolysis, baseline ECAR for each line was subtracted from the rate in 10 mm glucose to yield a true glycolysis-dependent ECAR. The glucose injection was followed by an injection of oligomycin, which eliminates mitochondrial ATP production by inhibiting the mitochondrial ATP synthase and forces a compensatory increase in glycolysis-related ATP production. The difference between the post-oligomycin ECAR and the post-2-deoxyglucose baseline ECAR indicates the glycolysis capacity. The difference between the post-glucose, pre-oligomycin ECAR and the post-glucose, post-oligomycin ECAR indicates the glycolysis spare reserve capacity. The ECARs from each line were normalized to cell protein, organized by groups and further normalized to the control group mean.
The post-glucose, pre-oligomycin ECARs in the MCI and AD cybrids, as a group, were lower than the control cybrid ECARs (Fig. 2G). This suggests that glycolysis rates were lower in the MCI and AD cybrids. The post-glucose, post-oligomycin ECARs were also lower in the MCI and AD cybrids, suggesting their glycolysis capacity was also lower than that of the control cybrid lines (Fig. 2H). The calculated glycolysis spare capacities were lower in the MCI and AD cybrid groups (data not shown).
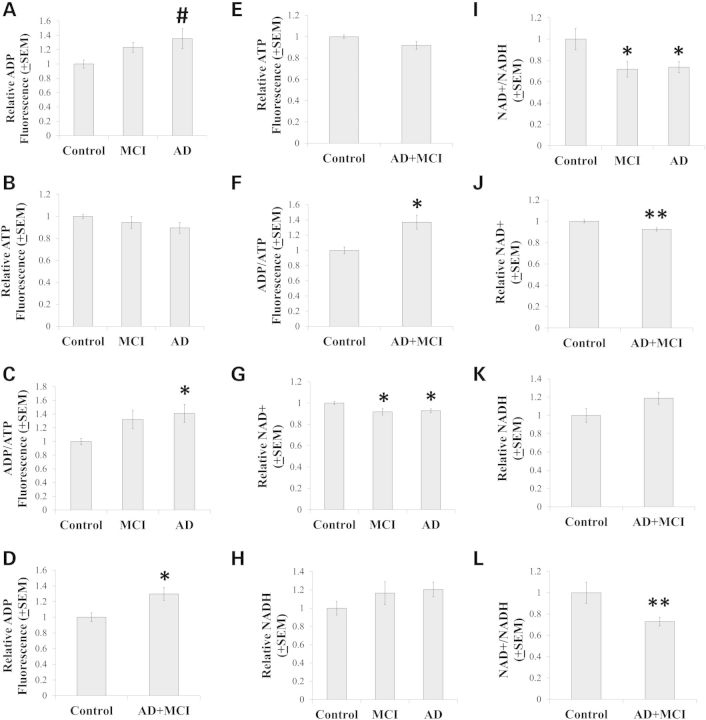
Energy and redox intermediates
Similar to what was found in a prior study of AD and control cybrid cell lines prepared on an NT2 cell nuclear background (21), inter-group differences in energy substrate levels were observed. These differences were subtle but distinct. Mean ADP levels between AD, MCI and control cybrid lines were comparable on ANOVA, although a post-hoc test suggested a potential increase in the AD cybrid ADP mean when compared with the control cybrid ADP mean [P < 0.05 on least significant difference (LSD) test] (Fig. 3A). ATP levels were comparable between the three groups (Fig. 3B). The ADP/ATP ratio, though, was elevated in the AD group when compared with the control group, while the MCI group ADP/ATP ratio was intermediate between the AD and control groups as it did not differ from either of those groups (Fig. 3C). Since MCI is often an AD precursor state, we attempted to increase power by generating a combined AD+MCI group. In this analysis, the AD+MCI group ADP concentration and ADP/ATP ratio exceeded the control ADP concentration and ADP/ATP ratio (Fig. 3D and F). No differences were seen when the MCI and control cybrid values were combined and compared with the AD cybrid values (data not shown).
Figure 3.
Energy and redox intermediates. Relative ADP levels, ATP levels and ADP/ATP ratios for each separate group are shown in (A–C). Relative ADP levels, ATP levels and ADP/ATP ratios are shown for the combined AD+MCI group in (D–F). Relative NAD+ levels, NADH levels and NAD+/NADH ratios for each separate group are shown in (G–I). Relative NAD+ levels, NADH levels and NAD+/NADH ratios are shown for the combined AD+MCI group in (J–L). Asterisk indicates a P < 0.05 difference from the control group; Double asterisk indicates a P < 0.01 difference from the control group; # indicates the ANOVA calculation itself was not statistically significant, but the LSD post-hoc comparison with the control group showed a P-value of < 0.05.
MCI and AD cybrid whole cell NAD+/NADH ratios were lower than the control cybrid NAD+/NADH ratio (Fig. 3I). This difference was driven by statistically significant reductions in MCI and AD cybrid NAD+ concentrations (Fig. 3G). Increases in MCI and AD cybrid NADH concentrations may also have accentuated the inter-group ratio differences (see Fig. 3H), although NADH levels were statistically equivalent between the groups. Combining the MCI and AD cell lines into a single group did not alter this analysis; the combined group showed decreased NAD+, statistically unchanged NADH and a decreased NAD+/NADH ratio (Fig. 3J–L). Combining the MCI and control groups to form a single group, which was then compared with the AD group, also revealed no quantitative differences in NAD+ concentration (data not shown).
Flux modifying genes and proteins
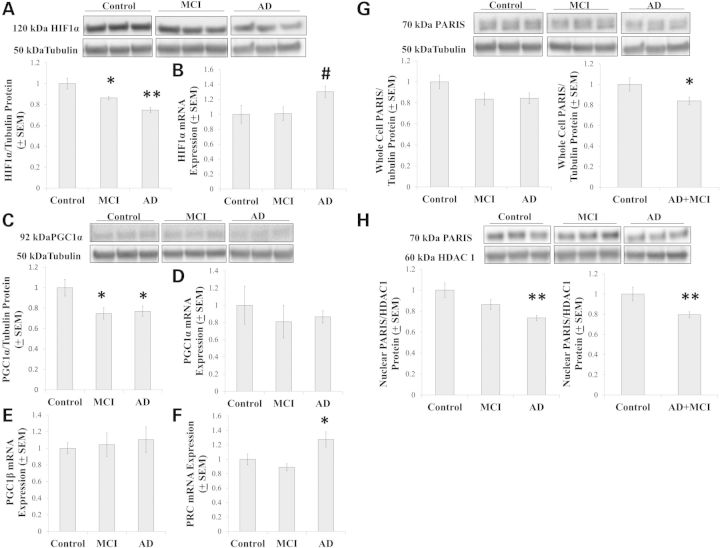
We assessed the status of genes and proteins that regulate or are regulated by bioenergetic fluxes. HIF1α, a protein that senses and responds to cell oxygenation status (22), was lower in MCI and AD cybrids than it was in control cybrids (Fig. 4A). HIF1α mRNA expression levels were comparable between the groups (P = 0.054 on ANOVA), although a post-hoc analysis suggested HIF1α mRNA levels in the AD cybrid group could have exceeded that of the MCI and control cybrid groups (P < 0.05 on LSD) (Fig. 4B).
Figure 4.
Flux regulatory pathways. Relative HIF1α protein levels are shown in (A). Relative HIF1α mRNA levels are shown in (B). Relative PGC1α protein levels are shown in (C) and relative PGC1α mRNA levels are shown in (D). Relative PGC1β (E) and PRC (F) mRNA levels were also determined. Relative whole cell PARIS protein levels are shown for all three groups and for the combined AD+MCI group (G). Relative nuclear PARIS protein levels are shown for all three groups and for the combined AD+MCI group (H). Asterisk indicates a P < 0.05 difference from the control group; Double asterisk indicates a P < 0.01 difference from the control group; # indicates the ANOVA calculation itself was not statistically significant, but the LSD post-hoc comparison with the control group showed a P-value of < 0.05.
Peroxisome proliferator-activated receptor γ coactivator α (PGC1α) mediates changes in mitochondrial mass and coupling efficiency (23). Total cell PGC1α protein levels were lower in MCI and AD cybrids than they were in control cybrids (Fig. 4C). No differences in PGC1α mRNA expression were observed, perhaps because of an unexpectedly large variation in individual cell line values (Fig. 4D). We also assessed the mRNA levels of two other PGC family members, peroxisome proliferator-activated receptor γ coactivator β (PGC1β) and PGC1α-related coactivator (PRC). PGC1β expression was comparable between the groups, while PRC expression was higher in the AD cybrid group than it was in the control and MCI groups (Fig. 4E and F).
It was recently shown that PGC1α expression is also regulated by PARIS, a protein that binds the PGC1α promoter and blocks PGC1α gene transcription (24). While ANOVA between the control, MCI and AD cybrids showed no intergroup differences in whole cell PARIS, whole cell PARIS levels in the combined AD+MCI group were reduced relative to the control group level (Fig. 4G). When just nuclear PARIS was considered, protein levels were lower in the AD group than they were in the control group and also in the combined AD+MCI group (Fig. 4H).
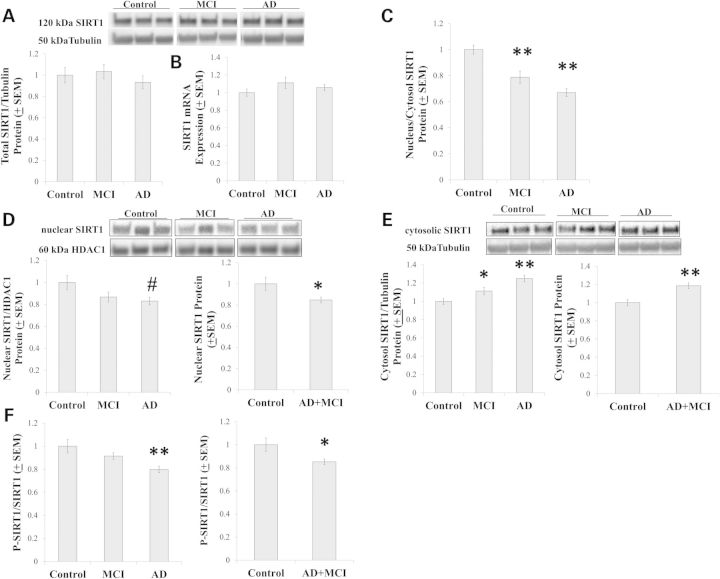
PGC1α activity is post-translationally regulated. Acetylation reduces its activity while de-acetylation, which is SIRT1-mediated, increases activity (25,26). Whole-cell SIRT1 protein and expression levels were comparable between the groups (Fig. 5A and B). SIRT1 compartmentalization, though, changed and this manifested as a decrease in the protein's nucleus to cytoplasm ratio (Fig. 5C). Compared with the control group, on a post-hoc analysis nuclear SIRT1 trended towards a lower level, and in the combined AD+MCI group nuclear SIRT1 levels were significantly lower (Fig. 5D). Cytosolic SIRT1 protein levels were increased in both the MCI and AD groups, as well as in the combined AD+MCI group (Fig. 5E). The ratio of phosphorylated to total SIRT1 was also reduced in the AD and AD+MCI groups (Fig. 5F).
Figure 5.
SIRT1 analyses. Relative SIRT1 and whole-cell protein levels (A) and mRNA levels (B) were similar between groups. However, the AD and MCI cybrid nucleus/cytosolic SIRT1 protein ratio decreased (C). This was due to a decrease in nuclear SIRT1 protein in the AD+MCI group and perhaps the AD group (D), as well as increased amounts of cytosolic SIRT1 protein in the AD, MCI and AD+MCI groups (E). SIRT1 phosphorylation was reduced in the AD and AD+MCI groups (F). Asterisk indicates a P < 0.05 difference from the control group; Double asterisk indicates a P < 0.01 difference from the control group; # indicates the ANOVA calculation itself was not statistically significant, but the LSD post-hoc comparison with the control group showed a P-value of < 0.05.
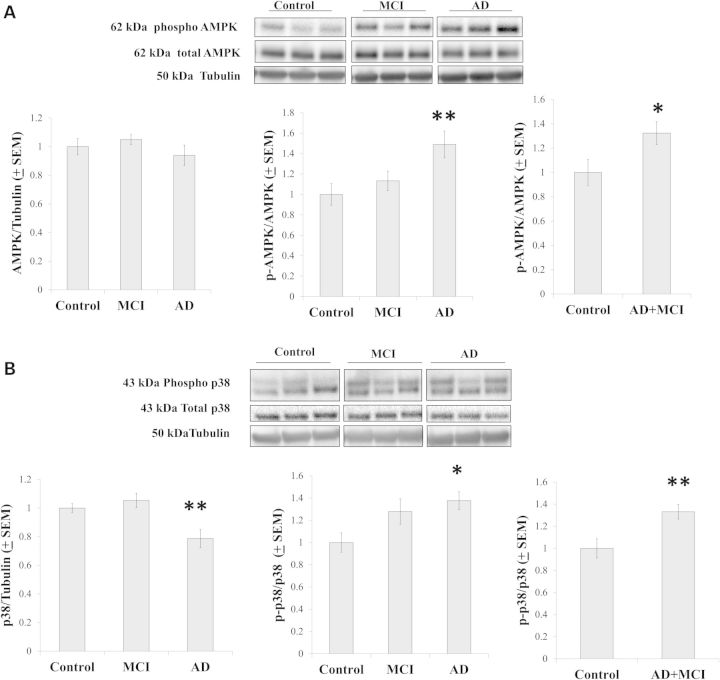
Phosphorylation provides another layer of PGC1α post-translational modification; increased phosphorylation associates with increased activity (25). AMPK is one enzyme that phosphorylates PGC1α (27). While total AMPK protein levels were comparable between groups, relative to the control group AMPK phosphorylation increased in the AD and AD+MCI groups (Fig. 6A). We measured levels of a second PGC1α-phosphorylating enzyme, the p38 mitogen-associated protein kinase (28), and found that relative to the control group, total p38 protein levels were decreased in the AD group. Similar to what was previously found in other studies of AD cybrids (29,30), though, the ratio of phosphorylated to non-phosphorylated p38 was increased in the AD cybrids and this ratio was also increased in the AD+MCI group (Fig. 6B).
Figure 6.
AMPK and p38 analyses. (A) Total cell AMPK protein levels were equivalent between the groups, while AMPK phosphorylation was increased in the AD and AD+MCI groups. (B) Total cell p38 protein levels decreased in the AD group, while p38 phosphorylation increased in the AD and AD+MCI groups. Asterisk indicates a P < 0.05 difference from the control group; Double asterisk indicates a P < 0.01 difference from the control group.
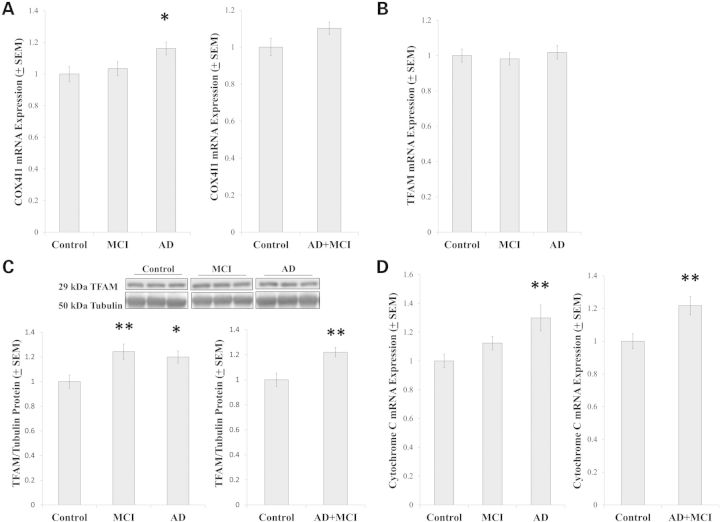
As various results were consistent with either increased or decreased PGC1α activation, we measured COX4I1 mRNA levels since COX4I1 expression is induced by PGC1α-nuclear respiratory factor (NRF) complexes (31). COX4I1 expression was higher in the AD group than it was in the control group, although the control and AD+MCI groups were comparable (Fig. 7A). We further assessed mRNA and protein levels of transcription factor A of the mitochondria (TFAM), which is also induced by PGC1α-NRF complexes (31). Although TFAM mRNA levels were comparable between groups (Fig. 7B), relative to the control group, TFAM protein was increased in the MCI, AD and AD+MCI groups (Fig. 7C). Cytochrome c is transcribed from a nuclear gene, translated in the cytoplasm and mostly resides in the mitochondrial intermembrane space. We did not measure cytochrome c protein levels in this study, but relative to the control group cytochrome c expression was increased in the AD and AD+MCI groups (Fig. 7D).
Figure 7.
Expression of PGC1α-coactivated genes. COX4I1 mRNA levels were increased in the AD group but not the AD+MCI group (A). TFAM mRNA levels were equivalent between groups (B), while TFAM protein levels were increased in the MCI, AD and AD+MCI groups (C). Cytochrome c mRNA levels were increased in the AD and AD+MCI groups (D). Asterisk indicates a P < 0.05 difference from the control group; Double asterisk indicates a P < 0.01 difference from the control group.
Mitochondrial mass and morphology
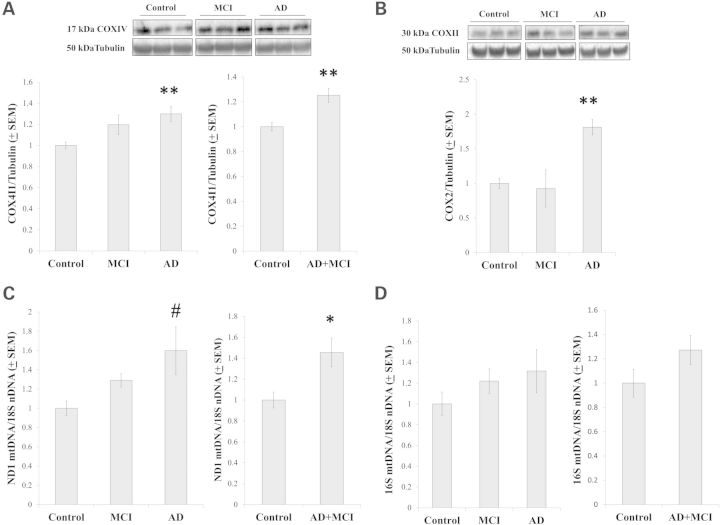
We evaluated several markers of mitochondrial mass. COX4I1, a COX subunit, is encoded by a nuclear gene and translocates to the inner mitochondrial membrane. On the ANOVA comparison, COX4I1 protein was higher in the AD group than it was in the control group. COX4I1 protein levels were not statistically different between the control and MCI groups, with the P-value being 0.067 (Fig. 8A). COX4I1 protein levels were greater in the AD+MCI group than they were in the control group (Fig. 8A). COX2, another COX subunit, is encoded by an mtDNA gene and localizes to the inner mitochondrial membrane. On the ANOVA comparison, COX2 protein was higher in the AD group than it was in either the control or MCI group (Fig. 8B); the control and AD+MCI group COX2 protein levels were comparable (data not shown). Lastly, we determined relative mtDNA to nDNA ratios in our cybrid cell lines. When we normalized the copy number of an mtDNA gene, ND1, to the copy number of a nuclear gene, the 18S RNA, the ratio was higher in the AD and AD+MCI groups than it was in the control group (Fig. 8C). When the amount of the 16S RNA mtDNA gene was normalized to the amount of the 18S gene no statistically significant quantitative differences were seen between the groups, although qualitatively the 16S:18S data resembled the ND1:18S data (Fig. 8D).
Figure 8.
Mitochondrial mass markers. COX4I1 protein levels were increased in the AD and MCI+AD groups (A). COX2 protein was increased in the AD group (B). When relative mtDNA levels were determined using primers to the mtDNA ND1 gene, mtDNA levels trended higher in the AD group and were elevated in the AD+MCI group (C). When relative mtDNA levels were determined using primers to the mtDNA 16S gene, no inter-group differences were seen although qualitatively results obtained with the 16 s primers resembled those obtained with the ND1 primers (D). Asterisk indicates a P < 0.05 difference from the control group; Double asterisk indicates a P < 0.01 difference from the control group; # indicates the ANOVA calculation itself was not statistically significant, but the LSD post-hoc comparison with the control group showed a P-value of < 0.05.
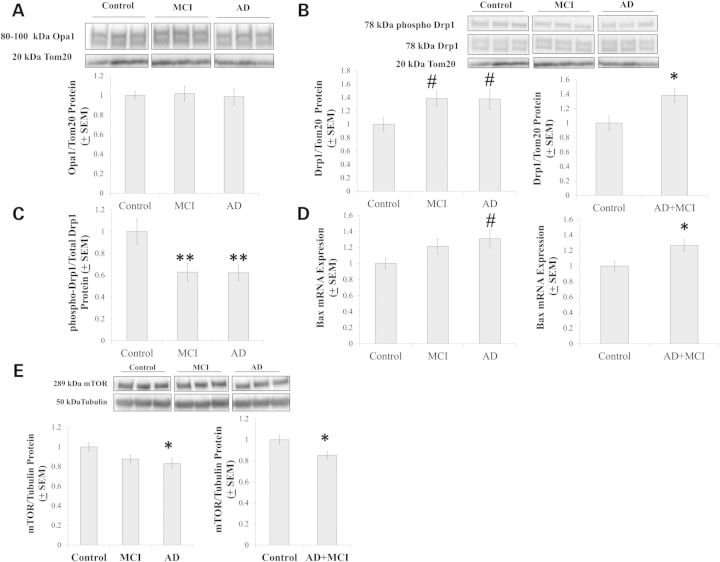
Mitochondrial morphology is partly determined by fission–fusion dynamics (32,33). Mitochondrial fission and fusion are enzyme-mediated processes that facilitate bioenergetic adaptation. In purified mitochondrial fractions, no differences in Opa1, a protein that drives mitochondrial fusion, were observed between our cybrid groups (Fig. 9A). Levels of Drp1 protein, which drives mitochondrial fission, were comparable on ANOVA (P = 0.076), but a post-hoc analysis suggested Drp1 trended higher in the MCI and AD groups (both less than P < 0.05 on LSD comparison) than it was in the control group (Fig. 9B). Drp1 was higher in the combined AD+MCI group than it was in the control group (Fig. 9B). Drp1 activity is also controlled through post-translational modification, and Drp1 phosphorylation at serine 637 inversely correlates with Drp1 activity (34–36). We found that Drp1 phosphorylation, when normalized to total mitochondrial Drp1 protein, was lower in the MCI and AD groups than it was in the control group (Fig. 9C).
Figure 9.
Mitochondrial fission, mitochondrial fusion, and mTOR analyses. Opa1 protein levels were equivalent between groups (A), while Drp1 protein levels trended higher in the MCI and AD groups and were higher in the AD+MCI group (B). Drp1 serine 637 phosphorylation was reduced in the MCI and AD groups (C). Bax expression trended higher in the AD group, and was higher in the AD+MCI group (D). mTOR protein levels were reduced in the AD and AD+MCI groups (E). *indicates a P < 0.05 difference from the control group; **indicates a P < 0.01 difference from the control group; # indicates the ANOVA calculation itself was not statistically significant, but the LSD post-hoc comparison with the control group showed a P value of < 0.05.
Since the pro-apoptotic protein Bax reportedly enhances mitochondrial fission and fragmentation, we quantified Bax expression (37,38). The inter-group ANOVA was negative (P = 0.099), although on post-hoc analysis there was a trend for higher expression in the AD group when compared with the control group (P = 0.036 on LSD) (Fig. 9D). Bax expression was higher in the combined AD+MCI group than it was in the control group (Fig. 9D). Finally, we quantified cell mTOR protein levels, as mTOR is sensitive to bioenergetic states and counters autophagy, a process that might predictably increase under conditions that favor increased mitochondrial fission (39,40). Relative to the control group, mTOR levels were reduced in the AD group and in the AD+MCI groups (Fig. 9E).
DISCUSSION
Cybrid cell lines created through transfer of AD and MCI subject mitochondria show altered bioenergetic function and bioenergetics-associated infrastructures. COX holoenzyme function and respiratory fluxes are less efficient, glucose utilization is less robust and these changes impact cell energy and redox states. Proteins that sense, respond to and regulate bioenergetic fluxes are altered. While the majority of these retrograde responses are certainly compensatory, they appear to represent a mix of adaptive and mal-adaptive changes.
COX activity in AD or MCI groups is lower than it is in age-matched control groups (1,2,41,42). Initial studies reported COX activity is also lower in AD and MCI cybrid cell lines than it is in control cybrid lines (1,19,43), despite apparent attempts by AD cybrids to increase their mitochondrial mass (20). We again found COX in AD and, to a lesser extent, MCI cybrid cell lines are less efficient than it is in control cybrid lines despite the presence of seemingly adaptive compensations.
In AD and MCI cybrid cell lines, mitochondrial oxygen consumption is altered under non-stress conditions. Respiration differences are accentuated when cells are forced to increase their mitochondrial ATP production. Decreased respiratory coupling in AD and, to a lesser extent, MCI cybrids further suggests respiration in AD and MCI cybrids is less efficient than it is in control cybrids.
Despite this, glycolysis fluxes are reduced, rather than enhanced, in AD and MCI cybrids. Glycolysis flux is partly determined by NAD+/NADH ratios and NAD+ availability. We found AD and MCI cybrids have less NAD+ and lower NAD+/NADH ratios than control cybrids. Such changes would predictably reduce glycolysis flux. While activities of some glycolysis enzymes are increased in AD brain neurons (44), glucose utilization is reduced in brains from AD and MCI subjects (45,46). Cybrid modeling recapitulates this well-recognized bioenergetic phenomenon.
Reduced glucose utilization would constitute a mal-adaptive retrograde response because it predictably exacerbates rather than alleviates an energy shortfall. In AD and MCI cybrids, this manifests as an elevated ADP/ATP ratio. Addressing respiration-induced glycolysis flux reductions in AD and MCI cybrids and, ideally, subjects might favorably influence cell energy stores.
HIF1α protein was reduced in AD and MCI cybrids. HIF1α enhances glycolysis by promoting glycolysis enzyme gene expression (22,47). Reduced HIF1α levels would seem to represent an ultimately mal-adaptive compensation (44). One study previously reported HIF1α protein levels are reduced in AD brains (48). Our cybrid data support the view that increasing HIF1α protein levels is a justifiable AD therapeutic strategy (44).
While HIF1α protein decreases in AD cybrids, HIF1expression increases. HIF1α is regulated more by protein degradation rates than gene expression rates, so dissociations should be possible. Interestingly, one study found HIF1α gene expression was increased in AD subject brain microvessels (49).
PGC1α expression is reduced in AD brains (50,51). We did not replicate this finding in our AD and MCI cybrids. Levels of PARIS, a protein that blocks PGC1α expression (24), were low in AD and MCI cybrids and this also would tend to favor increased rather than decreased PGC1α expression. Expression of a related gene, PRC (52), increased in AD cybrids. The status of PRC in AD brains is unknown.
AD subject brains, AD cybrid cell lines and MCI cybrid cell lines all show reduced PGC1α protein levels (51,53). If this exacerbates a respiratory chain defect, then PGC1α protein reduction constitutes a mal-adaptive compensation. Alternatively, reducing PGC1α protein levels could conceivably benefit AD and MCI cybrid cells by limiting exposure to impaired mitochondria.
Reductions in nuclear SIRT1 and SIRT1 phosphorylation, in conjunction with or perhaps due to low NAD+, would predictably increase PGC1α acetylation and decrease PGC1α activity (54,55). On the other hand, increased AMPK and p38 phosphorylation, which we presume occurred in response to energy compromise and oxidative stress, should increase PGC1α phosphorylation and increase PGC1α activity (25,27,28). These results indicate respiratory chain dysfunction alters cell milieus in ways that permit the coincident presence of contradictory responses. Similar contradictory profiles are also seen in AD brains, where SIRT1 protein is reportedly reduced while AMPK and p38 are reportedly activated (56–60).
COX4I1 expression, which is co-activated by PGC1α (31), and COX4I1 protein levels were increased in AD and, to a lesser extent, MCI cybrids. TFAM expression, which is also co-activated by PGC1α (31), was unchanged in AD and MCI cybrids but TFAM protein levels were increased. Cytochrome c, a gene co-activated by both PGC1α and PRC (31), showed increased expression in AD and MCI cybrids. Therefore, despite the fact that PGC1α protein levels are reduced in AD and MCI cybrids, we cannot conclude overall PGC1α activity is reduced.
Whether it occurs due to PGC1α activation or despite reduced PGC1a protein, mitochondrial mass markers increase in AD and MCI cybrids. AD cybrids have more COX2 protein, COX4I1 protein, COX4I1 expression, cytochrome c expression, TFAM protein and mtDNA than control cybrids. If we combine the AD and MCI cybrids to form one group, then the MCI cybrids also showed evidence of increased mitochondrial mass.
Less mTOR protein was present in the AD and AD+MCI groups. Since mTOR counters autophagy (61,62), decreased mTOR should predict increased autophagy. While total mTOR levels were unchanged in one study of AD brains (63), autophagy markers do increase in AD (7,64–66). AMPK, which was activated in our AD and MCI cybrids, also inhibits mTOR and through this action promotes autophagy (60).
For our mitochondrial fission–fusion protein analysis, in order to more directly test protein–mitochondria relationships, we measured Opa1 and Drp1 in mitochondrial fractions rather than whole cell lysates. Data obtained via this approach are consistent with our mTOR and AMPK data, in that AD and MCI cybrids show enhanced mitochondrial fission infrastructure. An increase in fission infrastructure, which is seen in AD subject brains (12,15,16), would predictably facilitate mitochondrial autophagy (18,67).
Autophagy differences may complicate mtDNA copy number measurements. While levels of PCR-amplifiable mtDNA are consistently reduced in AD brain (6,10,68–70) one study found, using an immunochemical rather than PCR approach, large amounts of mtDNA within an expanded autophagosome pool (6). Since less mtDNA amplification was seen in AD brains in that study, this autophagosome-localized mtDNA may have eluded amplification (6).
While overall numbers of normal-appearing mitochondria are reduced in AD (6,10,71,72), between individual neurons mitochondrial mass estimations range from markedly increased to profoundly decreased (10,11,73). The balance between mitochondrial biogenesis and mitochondrial autophagy may, therefore, vary between different neurons at any particular time, and vary over time within a single neuron.
Several factors may explain the small magnitude of most inter-group changes observed in this study. Differences could truly be subtle, cybrids may minimize differences or diagnostic inaccuracy could converge means and inflate variations. Increasing statistical power would address such limitations. We accomplished this by treating the AD and MCI groups as a single group. We believe this was justified because in most cases the MCI clinical syndrome occurs in association with, or is due to, AD (41,74).
Our data suggest AD-specific bioenergetic profiles are present when clinical symptoms manifest and longitudinally evolve. This chronology is consistent with the mitochondrial cascade hypothesis, which postulates AD histopathology in sporadic, late-onset AD is initiated when an individual's mitochondrial function declines below a critical threshold (5,75,76). If, as has previously been assumed, mtDNA accounts for differences between cybrid lines created from cytoplasts of different origins (77), then mtDNA differences are already present in subjects when clinical changes begin (78).
In this cybrid study, correlative inference was occasionally used to deduce functional and, in particular cases, causal relationships. Some interpretive points, therefore, should be considered within that context. For example, we assumed changes observed in cytosolic or nuclear compartments arise due to changes in mitochondrial function. While this seems reasonable given our understanding of cybrid cell lines, the degree to which cytosolic or nuclear compartment changes in turn alter mitochondrial function is difficult to quantify. We further assumed bioenergetic flux changes impact cell signaling pathways more than cell signaling pathways impact bioenergetic fluxes. In a study like ours ascertaining the effect, and especially the magnitude of effect, of one parameter on another is certainly challenging since many of the parameters we studied are exceedingly intertwined. Future experiments designed to address this issue are indicated.
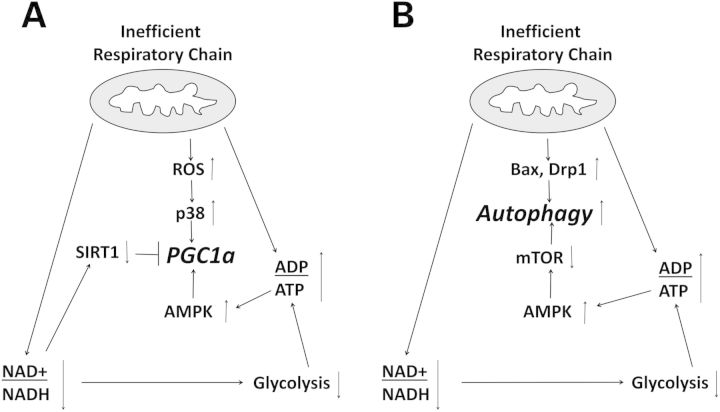
In summary (Fig. 10), our data suggest respiratory chain changes affect respiratory fluxes and cell redox states. Redox changes, in turn, reduce glycolysis flux and further stress cell energy supplies. Mitochondrial biogenesis and autophagy are up-regulated, at least initially, but contradictions within pathways that regulate mitochondrial mass occur. The balance between mitochondrial biogenesis and mitochondrial autophagy is lost, and over time the functional mitochondrial pool shrinks. In AD, reversing potentially mal-adaptive compensations, such as a decline in glycolysis flux or in the drive to maintain mitochondrial mass (79), would seem to represent logical therapeutic goals.
Figure 10.
Suggested regulation of mitochondrial biogenesis and autophagy in AD and MCI cybrids. (A) Respiratory chain inefficiency induces oxidative stress, energy stress and a decreased NAD+/NADH ratio. The lowered NAD+/NADH ratio reduces glycolysis, which adds to the energy stress. AMPK and p38 are activated, while SIRT1 input falls. Depending on the balance of these signals, PGC1α activity can increase or decrease. (B) Energy stress activates AMPK, reduces mTOR activity and initiates autophagy. Enhanced fission of dysfunctional mitochondria also contributes to an increase in autophagy markers. ROS, reactive oxygen species.
MATERIALS AND METHODS
Human subjects and creation of cybrid cell lines
Subject participation was approved by the Kansas University Medical Center's Institutional Review Board. Subjects for this study were recruited from the University of Kansas Alzheimer's Disease Center (KU ADC). Each subject was determined, based on cognitive testing and by a memory disorders subspecialist clinician, to meet criteria for normal cognition (control status), MCI or sporadic AD. After providing informed consent, sporadic AD (n = 8), MCI (n = 7) and age-matched control (n = 7) subjects underwent a 10 ml phlebotomy using tubes containing acid–citrate–dextrose as an anticoagulant. The age of the AD subject platelet donors was 71.5 ± 9.7 years, the age of the MCI platelet donors was 72.3 ± 6.6 and the age of the control subject platelet donors was 73.9 ± 7.7.
Cybrid cell lines were created on an SH-SY5Y cell nuclear background (80) by the KU ADC Mitochondrial Genomics and Metabolism Core. To generate the cybrid lines used in these studies, platelets from human subjects were mixed with SH-SY5Y cells previously depleted of endogenous mtDNA (ρ0 cells) as previously described (81).
During the overall cybrid generation procedure, several different types of media were used. For each medium, Dulbecco's modified Eagle's medium (DMEM) was obtained from Gibco-Invitrogen, while non-dialyzed or dialyzed fetal bovine serum (FBS) was obtained from Sigma. SH-SY5Y ρ0 cell growth medium consisted of DMEM supplemented with 10% non-dialyzed FBS, 200 ug/ml sodium pyruvate, 150 ug/ml uridine and 1% penicillin–streptomycin solution. SH-SY5Y cybrid selection medium consisted of DMEM supplemented with 10% dialyzed FBS and 1% penicillin–streptomycin solution. The selection process lasted 6 weeks. After cell line selection was completed, each line was continuously maintained in a cybrid growth medium containing DMEM supplemented with 10% non-dialyzed FBS and 1% penicillin-streptomycin solution for over 2 months prior to biochemical and molecular assays.
Cytochrome oxidase Vmax assays
Whole cell cytochrome oxidase (COX) Vmax activities were determined as previously described (43). The total protein contained in each assayed cuvette was estimated using the DC Protein assay (BioRad, Hercules, California, USA). The relative amount of a COX protein subunit, COX4I1, was determined for each cell line by western blot (see in what follows). COX activities in sec−1/mg protein values were calculated. The sec−1/mg protein Vmax activity for each line was also normalized to the amount of COX4I1 protein in the line.
Respiration and glycolysis analyses
Approximately 80 000 cells from each cybrid line were used to seed the wells of Seahorse XF cell culture microplates (Seahorse Bioscience, Billerica, MA, USA). A standard manufacturer-recommended two-step seeding procedure was utilized. After achieving cell adherence, microplates were placed overnight in a 37o, 5% CO2 incubator.
For respiratory analyses, on the assay day shortly before placing culture microplates in the Seahorse Analyzer, the cells were washed in buffered DMEM and the medium was then changed to the actual assay medium, which consisted of buffered DMEM containing no pyruvate and no BSA. Oxygen consumption rate (OCR) respiration measurements were made using a 3 min mix, 2 min wait, and 3 min read cycling protocol. During the first four reading periods, total cell OCRs were determined, with the fourth reading providing the value used for the analysis. After the fourth reading, wells were injected with 500 nM oligomycin and the resulting OCR was measured over three reading cycles. The third post-oligomycin reading cycle provided the post-oligomycin OCR. After this, we injected a mixture of rotenone (1 uM) and antimycin A (200 nM) and measured the resultant OCR over three reading cycles. The third post-rotenone/antimycin reading cycle provided the non-mitochondrial OCR. The non-mitochondrial OCR was subtracted from the total OCR to yield the mitochondrial OCR, and from the post-oligomycin OCR to yield the mitochondrial proton leak OCR. After OCR measurements were completed, the cell mass in each well was estimated by measuring the total protein in each well (DC Protein Assay, BioRad). OCR values from each well were normalized to the amount of protein in that well to yield a final corrected OCR in pmol O2/minute/mg protein.
For glycolysis analyses, on the assay day, the cells were washed in unbuffered DMEM and following this the cells were placed in unbuffered DMEM containing no pyruvate, no bovine serum albumin (BSA) and no glucose. The microculture plates were then de-gassed in a non-CO2 incubator at 37°C for 1 h before being placed into the Seahorse Analyzer. The wells were analyzed according to the procedure described in the Seahorse Glycolysis Stress Test kit. Briefly, initial measurements taken in the absence of glucose, and again after an injection of 2-deoxyglucose (to a final concentration of 100 mm), provide a non-glycolysis extracellular acidification rate (ECAR). In between these two baseline ECAR measurements, glucose was added to each well at a concentration of 10 mm. The resulting ECAR minus the non-glycolysis ECAR (in these experiments, we specifically subtracted the post-2-deoxyglucose non-glycolysis ECAR) provided the glycolysis ECAR. Next, oligomycin was added to each well (in this case, at a 1 µm concentration). The resulting ECAR minus the non-glycolysis ECAR provided the glycolysis capacity ECAR. Subtracting the glycolysis ECAR from the glycolysis capacity ECAR yielded the glycolysis spare reserve capacity.
ADP/ATP assay
ADP/ATP ratios were determined using the EnzyLight™ ADP/ATP Ratio Assay Kit (Bioassay Systems, CA, USA). Briefly, 1 × 105 cells per well were plated in 96-well plates. In the first step, cells were lysed to release ATP and ADP. ATP reacts with the kit substrate in an initial fluorescence reaction, which provided a direct indication of the ATP concentration. Next, we followed the change in fluorescence that occurred when ADP was converted to ATP. The difference between the initial and final fluorescences indicated the ADP concentration.
NAD+/NADH assay
Cells were harvested and assayed for NAD+/NADH levels using the Fluorescent NAD/NADH Detection Kit (Cell Technology, Inc., CA, USA). Briefly, 2 × 106 cells were harvested and divided into two different vials, one for NAD+ and another for NADH. Cells were then re-suspended in the appropriate extraction and lysis buffers. After vortexing, the lysates were incubated at 60°C for 15 min. Immediately following this, the lysates were cooled and reaction buffer was added. Samples were centrifuged at 8000g for 5 min to clarify the supernatants. After this, centrifugation samples were assayed for NAD+/NADH with fluorescence measurements taken at excitation 530 nm and emission 590 nm.
Immunoblotting
Cells were harvested with trypsin, centrifuged at 500g for 5 min, re-suspended in phosphate-buffered saline (PBS) and centrifuged again. To prepare whole cell lysates, the washed cell pellet was suspended in M-PER Mammalian Protein Extraction Reagent (Pierce, Rockford, IL, USA) supplemented with Halt Protease Inhibitor Cocktail (Pierce) and gently shaken for 5 min as directed. To prepare pure nuclear and cytosolic lysates, the washed cell pellet was processed using an N-PER Nuclear and Cytoplasmic Extraction Reagent Kit (Pierce) as directed. To prepare mitochondrial fractions, cells were harvested and the pellet processed using a Mitochondrial Isolation Kit for Cultured Cells (MitoSciences, Abcam, MA, USA). Protein concentrations for all lysates were determined using a DC Protein Assay Kit (BioRad).
For western blot analyses, samples were boiled, diluted 1:5 in sample buffer, resolved by electrophoresis in pre-cast 4–12% gels (BioRad) and transferred to polyvinylidene difluoride (PVDF) membranes. Non-specific binding was blocked by gently agitating the membranes in 5% non-fat milk or 5% BSA, as recommended, and 0.1% Tween in PBS for 1 h at room temperature. Blots were subsequently incubated in buffer containing a designated primary antibody. Primary antibodies purchased from Cell Signaling Technology (Beverly, MA, USA) included antibodies to phospho-sirtuin-1 (Ser47), sirtuin 1, tubulin, HIF1α, phospho-p38, total p38, mTOR, phospho-AMPK (Thr172), total AMPK, phospho-Drp1, total Drp1 and COX4; these antibodies were each used at a 1:1000 dilution. Primary antibodies purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA) included antibodies to Tom20 (used at a 1:1000 dilution) and PGC1α (used at a 1:500 dilution). An antibody to COX2 was purchased from Molecular Probes (Molecular Probes, Eugene, OR, USA) and used at a 1:500 dilution. An antibody to HDAC1 was purchased from Thermo Scientific (Pierce, Rockford, IL, USA) and used at a 1:1000 dilution. An antibody to Opa1 was purchased from BD Biosciences (San Jose, CA, USA) and used at a 1:1000 dilution. An antibody to TFAM was purchased from Aviva Systems Biology (San Diego, CA, USA) and used at a 1:1000 dilution. An antibody to PARIS (ZNF746) was obtained from the UC Davis/NIH NeuroMab Facility (Antibodies Inc., PO Box 1560, Davis, CA) and used at a 1:1000 dilution.
After addition of a primary antibody, membranes were incubated overnight at 4°C with gentle agitation. Blots were washed with PBS containing 0.1% Tween three times (each time for 10 min), and incubated with the appropriate horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature with gentle agitation. After three washes, the blots were incubated with SuperSignal West Femto Maximum Sensitivity Substrate (Pierce). Chemiluminescence signals were detected using a Bio-Rad ChemiDoc Imager and band densities determined using Quantity One Software.
Purification of genomic DNA
Genomic DNA was extracted using a QIAamp DNA Mini Kit (QIAGEN, Valencia, CA, USA). Briefly, 2 × 106 cells were harvested with trypsin. The cell pellets were resuspended in PBS and Proteinase K was added. Cells were lysed using AL Buffer and purification was performed as directed in the QIAamp DNA Handbook. At the end of the purification process DNA was eluted from the columns using nuclease-free water. The resulting DNA was stored at −80°C prior to use.
Purification of total RNA
Total RNA was extracted using a QIAamp RNA Blood Mini Kit (QIAGEN). Briefly, 2 × 106 cells were harvested with trypsin. RTL buffer was added to help lyse the cells and then cell lysates were homogenized in QIAshredder spin columns. The resultant flow-through was placed in QIAamp spin columns and the purification procedure was performed according to the QIAamp RNA Handbook. RNA was eluted from columns using nuclease-free water and the resulting RNA was stored at −80°C prior to use.
cDNA reaction
cDNA was obtained using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). cDNA reverse transcription reactions were prepared with 10 ul of 2× RT master mix to which we added 1 µg of total RNA and sufficient nuclease-free water to bring the total reaction volume to 20 µl.
Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was performed using the TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA) and ready-to-use TaqMan Gene Expression Assays (Applied Biosystems). We quantified mRNA levels deriving from the following genes: HIF1, PGC1α, PGC1β, PRC, TFAM, COX4, cytochrome c, sirtuin1 and Bax. GAPDH was used as an internal loading control. qPCR amplification was determined utilizing an Applied Biosystems StepOnePlus Real-Time PCR System (Applied Biosystems).
To quantify mtDNA, we used the TaqMan Gene Expression Assay Kit (Applied Biosystems) and primers towards two mtDNA-encoded genes, NADH dehydrogenase subunit 1 (ND1) and the 16S RNA gene, as well as to the nuclear 18s rRNA gene. The relative mtDNA to nuclear DNA copy number ratio was determined using the comparative ΔΔCT method, in which ND1/18S and 16S/18S ratios were calculated.
Data analysis
Data are expressed as means ± standard error of the means (SEM). To compare means between three groups, we used one-way analysis of variance (ANOVA) followed by Fisher's LSD post-hoc testing. To compare means between two groups, we used two-way, unpaired Student's t-tests, with P-values of < 0.05 considered significant.
FUNDING
This work was supported by the University of Kansas Alzheimer's Disease Center (P30AG035982), the Portuguese Foundation for Science and Technology (FCT), the Frank and Evangeline Thompson Alzheimer's Therapy Program and the Morgan Family Foundation.
ACKNOWLEDGEMENTS
Human subjects were clinically characterized by the Clinical Core of the University of Kansas Alzheimer's Disease Center. Cybrid cell lines were generated by the Mitochondrial Genomics and Metabolism Core of the University of Kansas Alzheimer's Disease Center.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Swerdlow R.H. Mitochondria and cell bioenergetics: increasingly recognized components and a possible etiologic cause of Alzheimer's disease. Antioxid. Redox Signal. 2012;16:1434–1455. doi: 10.1089/ars.2011.4149. doi:10.1089/ars.2011.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valla J., Schneider L., Niedzielko T., Coon K.D., Caselli R., Sabbagh M.N., Ahern G.L., Baxter L., Alexander G., Walker D.G., et al. Impaired platelet mitochondrial activity in Alzheimer's disease and mild cognitive impairment. Mitochondrion. 2006;6:323–330. doi: 10.1016/j.mito.2006.10.004. doi:10.1016/j.mito.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen R.C. Clinical practice. Mild cognitive impairment. N. Engl. J. Med. 2011;364:2227–2234. doi: 10.1056/NEJMcp0910237. doi:10.1056/NEJMcp0910237. [DOI] [PubMed] [Google Scholar]
- 4.Pellerin L., Magistretti P.J. Food for thought: challenging the dogmas. J. Cereb. Blood Flow Metab. 2003;23:1282–1286. doi: 10.1097/01.WCB.0000096064.12129.3D. doi:10.1097/01.WCB.0000096064.12129.3D. [DOI] [PubMed] [Google Scholar]
- 5.Swerdlow R.H., Burns J.M., Khan S.M. The Alzheimer's disease mitochondrial cascade hypothesis. J. Alzheimers Dis. 2010;20:S265–S279. doi: 10.3233/JAD-2010-100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirai K., Aliev G., Nunomura A., Fujioka H., Russell R.L., Atwood C.S., Johnson A.B., Kress Y., Vinters H.V., Tabaton M., et al. Mitochondrial abnormalities in Alzheimer's disease. J. Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nixon R.A. Autophagy, amyloidogenesis and Alzheimer disease. J. Cell Sci. 2007;120:4081–4091. doi: 10.1242/jcs.019265. doi:10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- 8.Henriques A.G., Vieira S.I., da Cruz E.S.E.F., da Cruz E.S.O.A. Abeta promotes Alzheimer's disease-like cytoskeleton abnormalities with consequences to APP processing in neurons. J. Neurochem. 2010;113:761–771. doi: 10.1111/j.1471-4159.2010.06643.x. doi:10.1111/j.1471-4159.2010.06643.x. [DOI] [PubMed] [Google Scholar]
- 9.Silva D.F., Esteves A.R., Arduino D.M., Oliveira C.R., Cardoso S.M. Amyloid-beta-induced mitochondrial dysfunction impairs the autophagic lysosomal pathway in a tubulin dependent pathway. J. Alzheimers Dis. 2011;26:565–581. doi: 10.3233/JAD-2011-110423. [DOI] [PubMed] [Google Scholar]
- 10.de la Monte S.M., Luong T., Neely T.R., Robinson D., Wands J.R. Mitochondrial DNA damage as a mechanism of cell loss in Alzheimer's disease. Lab Invest. 2000;80:1323–1335. doi: 10.1038/labinvest.3780140. doi:10.1038/labinvest.3780140. [DOI] [PubMed] [Google Scholar]
- 11.Nagy Z., Esiri M.M., LeGris M., Matthews P.M. Mitochondrial enzyme expression in the hippocampus in relation to Alzheimer-type pathology. Acta Neuropathol. 1999;97:346–354. doi: 10.1007/s004010050997. doi:10.1007/s004010050997. [DOI] [PubMed] [Google Scholar]
- 12.Su B., Wang X., Bonda D., Perry G., Smith M., Zhu X. Abnormal mitochondrial dynamics–a novel therapeutic target for Alzheimer's disease? Mol. Neurobiol. 2010;41:87–96. doi: 10.1007/s12035-009-8095-7. doi:10.1007/s12035-009-8095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santos R.X., Correia S.C., Wang X., Perry G., Smith M.A., Moreira P.I., Zhu X. A synergistic dysfunction of mitochondrial fission/fusion dynamics and mitophagy in Alzheimer's disease. J. Alzheimers Dis. 2010;20:S401–S412. doi: 10.3233/JAD-2010-100666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoppins S., Lackner L., Nunnari J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. doi:10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 15.Wang X., Su B., Lee H.G., Li X., Perry G., Smith M.A., Zhu X. Impaired balance of mitochondrial fission and fusion in Alzheimer's disease. J. Neurosci. 2009;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. doi:10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manczak M., Calkins M.J., Reddy P.H. Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer's disease: implications for neuronal damage. Hum. Mol. Genet. 2011;20:2495–2509. doi: 10.1093/hmg/ddr139. doi:10.1093/hmg/ddr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Su B., Siedlak S.L., Moreira P.I., Fujioka H., Wang Y., Casadesus G., Zhu X. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc. Natl Acad. Sci. USA. 2008;105:19318–19323. doi: 10.1073/pnas.0804871105. doi:10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Twig G., Shirihai O.S. The interplay between mitochondrial dynamics and mitophagy. Antioxid. Redox Signal. 2011;14:1939–1951. doi: 10.1089/ars.2010.3779. doi:10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva D.F., Santana I., Esteves A.R., Baldeiras I., Arduino D.M., Oliveira C.R., Cardoso S.M. Prodromal metabolic phenotype in MCI cybrids: implications for Alzheimers disease. Curr. Alzheimer Res. 2012;10:180–190. doi: 10.2174/1567205011310020008. doi:10.2174/1567205011310020008. [DOI] [PubMed] [Google Scholar]
- 20.Trimmer P.A., Keeney P.M., Borland M.K., Simon F.A., Almeida J., Swerdlow R.H., Parks J.P., Parker W.D., Jr, Bennett J.P., Jr Mitochondrial abnormalities in cybrid cell models of sporadic Alzheimer's disease worsen with passage in culture. Neurobiol. Dis. 2004;15:29–39. doi: 10.1016/j.nbd.2003.09.011. doi:10.1016/j.nbd.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Cardoso S.M., Santana I., Swerdlow R.H., Oliveira C.R. Mitochondria dysfunction of Alzheimer's disease cybrids enhances Abeta toxicity. J. Neurochem. 2004;89:1417–1426. doi: 10.1111/j.1471-4159.2004.02438.x. doi:10.1111/j.1471-4159.2004.02438.x. [DOI] [PubMed] [Google Scholar]
- 22.Semenza G.L. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. doi:10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Handschin C., Spiegelman B.M. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 24.Shin J.H., Ko H.S., Kang H., Lee Y., Lee Y.I., Pletinkova O., Troconso J.C., Dawson V.L., Dawson T.M. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson's disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. doi:10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Canto C., Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr. Opin. Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. doi:10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemoto S., Fergusson M.M., Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J. Biol. Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. doi:10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 27.Jager S., Handschin C., St-Pierre J., Spiegelman B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl Acad. Sci. USA. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. doi:10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feige J.N., Auwerx J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007;17:292–301. doi: 10.1016/j.tcb.2007.04.001. doi:10.1016/j.tcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Onyango I.G., Ahn J.Y., Tuttle J.B., Bennett J.P., Jr, Swerdlow R.H. Nerve growth factor attenuates oxidant-induced beta-amyloid neurotoxicity in sporadic Alzheimer's disease cybrids. J. Neurochem. 2010;114:1605–1618. doi: 10.1111/j.1471-4159.2010.06871.x. doi:10.1111/j.1471-4159.2010.06871.x. [DOI] [PubMed] [Google Scholar]
- 30.Onyango I.G., Tuttle J.B., Bennett J.P., Jr Altered intracellular signaling and reduced viability of Alzheimer's disease neuronal cybrids is reproduced by beta-amyloid peptide acting through receptor for advanced glycation end products (RAGE) Mol. Cell Neurosci. 2005;29:333–343. doi: 10.1016/j.mcn.2005.02.012. doi:10.1016/j.mcn.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 31.Scarpulla R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. doi:10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 32.Chan D.C. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. doi:10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- 33.Chen H., Chan D.C. Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum. Mol. Genet. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. doi:10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cereghetti G.M., Stangherlin A., Martins de Brito O., Chang C.R., Blackstone C., Bernardi P., Scorrano L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc. Natl Acad. Sci. USA. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. doi:10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cereghetti G.M., Costa V., Scorrano L. Inhibition of Drp1-dependent mitochondrial fragmentation and apoptosis by a polypeptide antagonist of calcineurin. Cell Death Differ. 2010;17:1785–1794. doi: 10.1038/cdd.2010.61. doi:10.1038/cdd.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho S.G., Du Q., Huang S., Dong Z. Drp1 dephosphorylation in ATP depletion-induced mitochondrial injury and tubular cell apoptosis. Am. J. Physiol. Renal Physiol. 2010;299:F199–F206. doi: 10.1152/ajprenal.00716.2009. doi:10.1152/ajprenal.00716.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Autret A., Martin S.J. Bcl-2 family proteins and mitochondrial fission/fusion dynamics. Cell Mol. Life Sci. 2010;67:1599–1606. doi: 10.1007/s00018-010-0286-x. doi:10.1007/s00018-010-0286-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheridan C., Delivani P., Cullen S.P., Martin S.J. Bax- or Bak-induced mitochondrial fission can be uncoupled from cytochrome C release. Mol. Cell. 2008;31:570–585. doi: 10.1016/j.molcel.2008.08.002. doi:10.1016/j.molcel.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Cunningham J.T., Rodgers J.T., Arlow D.H., Vazquez F., Mootha V.K., Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. doi:10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 40.Schieke S.M., Phillips D., McCoy J.P., Jr, Aponte A.M., Shen R.F., Balaban R.S., Finkel T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. doi:10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- 41.Stephan B.C., Hunter S., Harris D., Llewellyn D.J., Siervo M., Matthews F.E., Brayne C. The neuropathological profile of mild cognitive impairment (MCI): a systematic review. Mol. Psychiatry. 2012;17:1056–1076. doi: 10.1038/mp.2011.147. doi:10.1038/mp.2011.147. [DOI] [PubMed] [Google Scholar]
- 42.Parker W.D., Jr, Filley C.M., Parks J.K. Cytochrome oxidase deficiency in Alzheimer's disease. Neurology. 1990;40:1302–1303. doi: 10.1212/wnl.40.8.1302. doi:10.1212/WNL.40.8.1302. [DOI] [PubMed] [Google Scholar]
- 43.Swerdlow R.H., Parks J.K., Cassarino D.S., Maguire D.J., Maguire R.S., Bennett J.P., Jr, Davis R.E., Parker W.D., Jr Cybrids in Alzheimer's disease: a cellular model of the disease? Neurology. 1997;49:918–925. doi: 10.1212/wnl.49.4.918. doi:10.1212/WNL.49.4.918. [DOI] [PubMed] [Google Scholar]
- 44.Soucek T., Cumming R., Dargusch R., Maher P., Schubert D. The regulation of glucose metabolism by HIF-1 mediates a neuroprotective response to amyloid beta peptide. Neuron. 2003;39:43–56. doi: 10.1016/s0896-6273(03)00367-2. doi:10.1016/S0896-6273(03)00367-2. [DOI] [PubMed] [Google Scholar]
- 45.Silverman D.H., Small G.W., Chang C.Y., Lu C.S., Kung De Aburto M.A., Chen W., Czernin J., Rapoport S.I., Pietrini P., Alexander G.E., et al. Positron emission tomography in evaluation of dementia: Regional brain metabolism and long-term outcome. JAMA. 2001;286:2120–2127. doi: 10.1001/jama.286.17.2120. doi:10.1001/jama.286.17.2120. [DOI] [PubMed] [Google Scholar]
- 46.Matsuda H. The role of neuroimaging in mild cognitive impairment. Neuropathology. 2007;27:570–577. doi: 10.1111/j.1440-1789.2007.00794.x. doi:10.1111/j.1440-1789.2007.00794.x. [DOI] [PubMed] [Google Scholar]
- 47.Semenza G.L. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell Dev. Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. doi:10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y., Liu F., Iqbal K., Grundke-Iqbal I., Gong C.X. Decreased glucose transporters correlate to abnormal hyperphosphorylation of tau in Alzheimer disease. FEBS Lett. 2008;582:359–364. doi: 10.1016/j.febslet.2007.12.035. doi:10.1016/j.febslet.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grammas P., Samany P.G., Thirumangalakudi L. Thrombin and inflammatory proteins are elevated in Alzheimer's disease microvessels: implications for disease pathogenesis. J. Alzheimers Dis. 2006;9:51–58. doi: 10.3233/jad-2006-9105. [DOI] [PubMed] [Google Scholar]
- 50.Qin W., Haroutunian V., Katsel P., Cardozo C.P., Ho L., Buxbaum J.D., Pasinetti G.M. PGC-1alpha expression decreases in the Alzheimer disease brain as a function of dementia. Arch. Neurol. 2009;66:352–361. doi: 10.1001/archneurol.2008.588. doi:10.1001/archneurol.2008.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheng B., Wang X., Su B., Lee H.G., Casadesus G., Perry G., Zhu X. Impaired mitochondrial biogenesis contributes to mitochondrial dysfunction in Alzheimer's disease. J. Neurochem. 2012;120:419–429. doi: 10.1111/j.1471-4159.2011.07581.x. doi:10.1111/j.1471-4159.2011.07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vercauteren K., Pasko R.A., Gleyzer N., Marino V.M., Scarpulla R.C. PGC-1-related coactivator: immediate early expression and characterization of a CREB/NRF-1 binding domain associated with cytochrome c promoter occupancy and respiratory growth. Mol. Cell Biol. 2006;26:7409–7419. doi: 10.1128/MCB.00585-06. doi:10.1128/MCB.00585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swerdlow R.H. beta-Apptists and Tauists, it is time for a sermon from the book of biogenesis. J. Neurochem. 2012;120:347–349. doi: 10.1111/j.1471-4159.2011.07561.x. doi:10.1111/j.1471-4159.2011.07561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin Q., Yan T., Ge X., Sun C., Shi X., Zhai Q. Cytoplasm-localized SIRT1 enhances apoptosis. J. Cell Physiol. 2007;213:88–97. doi: 10.1002/jcp.21091. doi:10.1002/jcp.21091. [DOI] [PubMed] [Google Scholar]
- 55.Piantadosi C.A., Suliman H.B. Redox regulation of mitochondrial biogenesis. Free Radic. Biol. Med. 2012;53:2043–2053. doi: 10.1016/j.freeradbiomed.2012.09.014. doi:10.1016/j.freeradbiomed.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hensley K., Floyd R.A., Zheng N.Y., Nael R., Robinson K.A., Nguyen X., Pye Q.N., Stewart C.A., Geddes J., Markesbery W.R., et al. p38 kinase is activated in the Alzheimer's disease brain. J. Neurochem. 1999;72:2053–2058. doi: 10.1046/j.1471-4159.1999.0722053.x. doi:10.1046/j.1471-4159.1999.0722053.x. [DOI] [PubMed] [Google Scholar]
- 57.Sun A., Liu M., Nguyen X.V., Bing G. P38 MAP kinase is activated at early stages in Alzheimer's disease brain. Exp. Neurol. 2003;183:394–405. doi: 10.1016/s0014-4886(03)00180-8. doi:10.1016/S0014-4886(03)00180-8. [DOI] [PubMed] [Google Scholar]
- 58.Vingtdeux V., Davies P., Dickson D.W., Marambaud P. AMPK is abnormally activated in tangle- and pre-tangle-bearing neurons in Alzheimer's disease and other tauopathies. Acta Neuropath. 2011;121:337–349. doi: 10.1007/s00401-010-0759-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Julien C., Tremblay C., Emond V., Lebbadi M., Salem N., Jr, Bennett D.A., Calon F. Sirtuin 1 reduction parallels the accumulation of tau in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2009;68:48–58. doi: 10.1097/NEN.0b013e3181922348. doi:10.1097/NEN.0b013e3181922348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salminen A., Kaarniranta K., Haapasalo A., Soininen H., Hiltunen M. AMP-activated protein kinase: a potential player in Alzheimer's disease. J. Neurochem. 2011;118:460–474. doi: 10.1111/j.1471-4159.2011.07331.x. doi:10.1111/j.1471-4159.2011.07331.x. [DOI] [PubMed] [Google Scholar]
- 61.Hands S.L., Proud C.G., Wyttenbach A. mTOR's role in ageing: protein synthesis or autophagy? Aging. 2009;1:586–597. doi: 10.18632/aging.100070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O'Kane C.J., et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. doi:10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 63.Li X., Alafuzoff I., Soininen H., Winblad B., Pei J.J. Levels of mTOR and its downstream targets 4E-BP1, eEF2, and eEF2 kinase in relationships with tau in Alzheimer's disease brain. FEBS J. 2005;272:4211–4220. doi: 10.1111/j.1742-4658.2005.04833.x. doi:10.1111/j.1742-4658.2005.04833.x. [DOI] [PubMed] [Google Scholar]
- 64.Boland B., Kumar A., Lee S., Platt F.M., Wegiel J., Yu W.H., Nixon R.A. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J. Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. doi:10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nixon R.A., Wegiel J., Kumar A., Yu W.H., Peterhoff C., Cataldo A., Cuervo A.M. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J. Neuropathol. Exp. Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 66.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. doi:10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. doi:10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown A.M., Sheu R.K., Mohs R., Haroutunian V., Blass J.P. Correlation of the clinical severity of Alzheimer's disease with an aberration in mitochondrial DNA (mtDNA) J. Mol. Neurosci. 2001;16:41–48. doi: 10.1385/JMN:16:1:41. doi:10.1385/JMN:16:1:41. [DOI] [PubMed] [Google Scholar]
- 69.Coskun P.E., Beal M.F., Wallace D.C. Alzheimer's brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc. Natl Acad. Sci. USA. 2004;101:10726–10731. doi: 10.1073/pnas.0403649101. doi:10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rodriguez-Santiago B., Casademont J., Nunes V. Is mitochondrial DNA depletion involved in Alzheimer's disease? Eur. J. Hum. Genet. 2001;9:279–285. doi: 10.1038/sj.ejhg.5200629. doi:10.1038/sj.ejhg.5200629. [DOI] [PubMed] [Google Scholar]
- 71.Baloyannis S.J. Mitochondrial alterations in Alzheimer's disease. J. Alzheimers Dis. 2006;9:119–126. doi: 10.3233/jad-2006-9204. [DOI] [PubMed] [Google Scholar]
- 72.Baloyannis S.J. Mitochondria are related to synaptic pathology in Alzheimer's disease. Int. J. Alzheimers Dis. 2011;2011:305395. doi: 10.4061/2011/305395. doi:10.4061/2011/305395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manczak M., Park B.S., Jung Y., Reddy P.H. Differential expression of oxidative phosphorylation genes in patients with Alzheimer's disease: implications for early mitochondrial dysfunction and oxidative damage. Neuromolecular Med. 2004;5:147–162. doi: 10.1385/NMM:5:2:147. doi:10.1385/NMM:5:2:147. [DOI] [PubMed] [Google Scholar]
- 74.Morris J.C., Storandt M., Miller J.P., McKeel D.W., Price J.L., Rubin E.H., Berg L. Mild cognitive impairment represents early-stage Alzheimer disease. Arch. Neurol. 2001;58:397–405. doi: 10.1001/archneur.58.3.397. doi:10.1001/archneur.58.3.397. [DOI] [PubMed] [Google Scholar]
- 75.Swerdlow R.H., Khan S.M. The Alzheimer's disease mitochondrial cascade hypothesis: an update. Exp. Neurol. 2009;218:308–315. doi: 10.1016/j.expneurol.2009.01.011. doi:10.1016/j.expneurol.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Swerdlow R.H., Khan S.M. A ‘mitochondrial cascade hypothesis’ for sporadic Alzheimer's disease. Med. Hypotheses. 2004;63:8–20. doi: 10.1016/j.mehy.2003.12.045. doi:10.1016/j.mehy.2003.12.045. [DOI] [PubMed] [Google Scholar]
- 77.Swerdlow R.H. Mitochondria in cybrids containing mtDNA from persons with mitochondriopathies. J. Neurosci. Res. 2007;85:3416–3428. doi: 10.1002/jnr.21167. doi:10.1002/jnr.21167. [DOI] [PubMed] [Google Scholar]
- 78.Lu J., Wang K., Rodova M., Esteves R., Berry D., E L., Crafter A., Barrett M., Cardoso S.M., Onyango I., et al. Polymorphic variation in cytochrome oxidase subunit genes. J. Alzheimers Dis. 2010;21:141–154. doi: 10.3233/JAD-2010-100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Swerdlow R.H. Role and treatment of mitochondrial DNA-related mitochondrial dysfunction in sporadic neurodegenerative diseases. Curr. Pharm. Des. 2011;17:3356–3373. doi: 10.2174/138161211798072535. doi:10.2174/138161211798072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller S.W., Trimmer P.A., Parker W.D., Jr, Davis R.E. Creation and characterization of mitochondrial DNA-depleted cell lines with ʻneuronal-like’ properties. J. Neurochem. 1996;67:1897–1907. doi: 10.1046/j.1471-4159.1996.67051897.x. doi:10.1046/j.1471-4159.1996.67051897.x. [DOI] [PubMed] [Google Scholar]
- 81.Swerdlow R.H., Parks J.K., Miller S.W., Tuttle J.B., Trimmer P.A., Sheehan J.P., Bennett J.P., Jr, Davis R.E., Parker W.D., Jr Origin and functional consequences of the complex I defect in Parkinson's disease. Ann. Neurol. 1996;40:663–671. doi: 10.1002/ana.410400417. doi:10.1002/ana.410400417. [DOI] [PubMed] [Google Scholar]