Abstract
We assessed the efficacy of Seneca Valley virus (SVV-001), a neuroendocrine cancer-selective oncolytic picornavirus, in primary heterotransplant mouse models of small cell lung cancer (SCLC), including three lines each of classic and variant SCLC. Half-maximal effective concentrations for cell lines derived from three variant heterotransplants ranged from 1.6×10–3 (95% confidence interval [CI] = 1×10–3 to 2.5×10–3) to 3.9×10–3 (95% CI = 2.8×10–3 to 5.5×10–3). Sustained tumor growth inhibition in vivo was only observed in variant lines (two-sided Student t test, P < .005 for each). Doses of 1014 vp/kg were able to completely and durably eradicate tumors in a variant SCLC heterotransplant model in two of six mice. Gene expression profiling revealed that permissive lines are typified by lower expression of the early neurogenic transcription factor ASCL1 and, conversely, by higher expression of the late neurogenic transcription factor NEUROD1. This classifier demonstrates a sensitivity of .89, specificity of .92, and accuracy of .91. The NEUROD1 to ASCL1 ratio may serve as a predictive biomarker of SVV-001 efficacy.
Small cell lung cancer (SCLC) is an exceptionally aggressive cancer (1, 2). Extensive-stage SCLC, representing 60% of cases, is associated with median survival of nine months and five-year survival less than 1% (3). More effective treatments for SCLC are urgently needed. SCLC is notable for frequent expression of neuroendocrine markers. Within the diagnostic category of SCLC there is considerable heterogeneity, in terms of both the extent of neuroendocrine marker expression and the fraction of cells with small cell morphology. SCLC can be classified as either classic (approximately 70%) or variant (approximately 30%) subtypes based on histologic appearance and differential gene expression (4). Variant SCLCs are characterized by higher variation in cell size, more prominent nucleoli, and increased expression of genes associated with neuronal differentiation (4,5). Variant histology has been associated with a shorter doubling time, higher cloning efficiency, and primary and acquired therapeutic resistance (4,6–9).
Seneca Valley virus (SVV-001) is a naturally occurring oncolytic picornavirus that selectively replicates in SCLC lines in vitro and can durably eradicate permissive SCLC xenografts in mice (10). About half of SCLC lines tested are permissive for SVV-001. Rare non–small cell lung cancer (NSCLC) lines, of large cell neuroendocrine histology, also support SVV-001 replication (10). SVV-001 has been shown in mice to durably eradicate certain pediatric neuroendocrine tumors and prevent spread of invasive retinoblastoma (11–13). A phase I clinical trial demonstrated high levels of SVV-001 replication in a subset of SCLC patients, and safety at intravenous doses up to 1011 vp/kg (14). These data have prompted subsequent studies in patients with SCLC and pediatric neuroendocrine cancers. Entry criteria for these trials include tumor expression of neuroendocrine markers NCAM1, CHGA, and SYP.
Although SVV-001 shows promise in a variety of preclinical models, reliable biomarkers of permissivity have not been identified. Several markers have been proposed, including the general markers of neuroendocrine differentiation (11,13), as well as markers based on putative binding motifs derived from the crystal structure of SVV-001 (15). Preliminary data using Cy5-labeled SVV-001 indicate that some nonpermissive lines fail to bind SVV-001, suggesting a cell surface receptor requirement (Supplementary Figure 1, available online). As yet, there is no evidence that any of these markers have utility as predictors of SVV-001 efficacy.
We have previously described three primary heterotransplant models of variant SCLC (LX22, LX33, and LX36) derived by direct transfer of human tumors into immunosuppressed mice (16,17). We have subsequently generated additional human classic SCLC heterotransplant models (LX44, LX47, and LX48) using the same approach. In this study, we tested SVV-001 in vivo efficacy against multiple heterotransplant lines and used these results as a basis for identifying potential predictors of SVV-001 permissivity.
Three variant SCLC cell lines derived from heterotransplants were infected with recombinant SVV–green fluorescent protein (GFP) reporter virus and monitored for expression of GFP by epifluorescence microscopy and flow cytometry (18). H446, a highly permissive line, and H460, a resistant NSCLC line, were included as positive and negative controls, respectively. In H446 and all three heterotransplant-derived cell lines, cytopathic effect concurrent with a high percentage of GFP-expressing cells was observed (Figure 1A).
Figure 1.
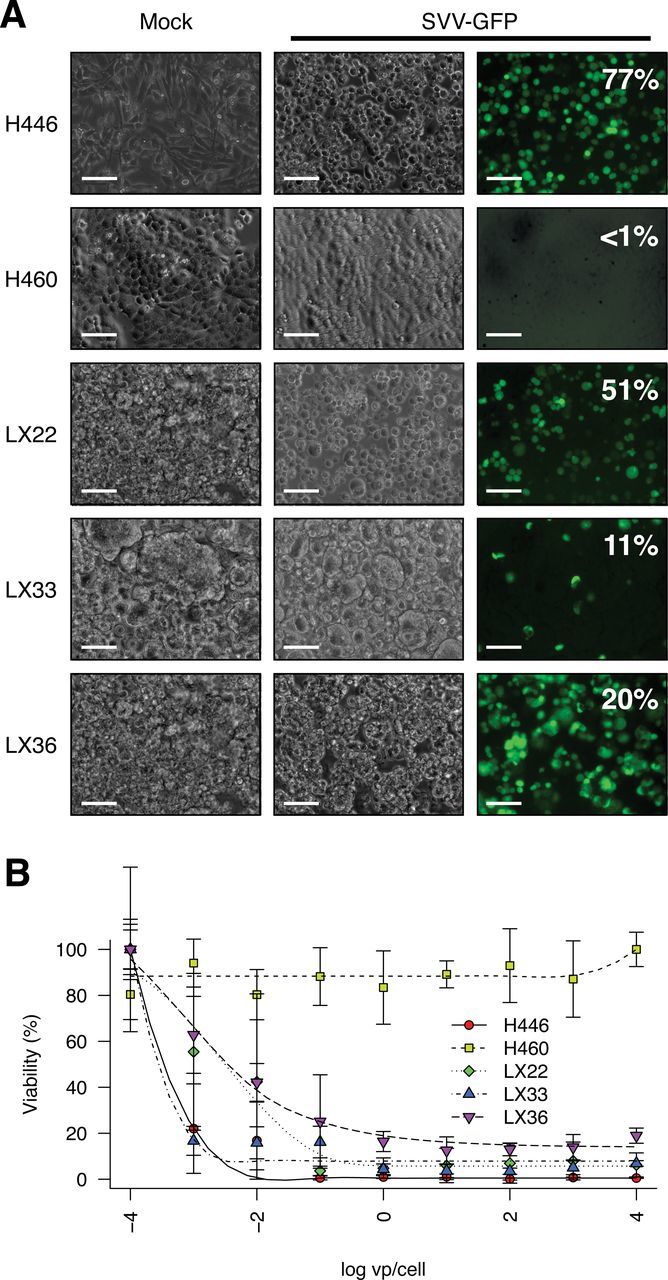
In vitro analysis of Seneca Valley virus (SVV-001) permissivity in cell lines derived from SCLC heterotransplant models. SVV-001 was serially diluted and administered from frozen stocks provided by Neotropix, Inc (Malvern, PA) as previously described (10). SVV-GFP is a derivative of SVV-001 expressing green fluorescent protein (18). Cell lines were plated in Opti-MEM containing 1 vp/cell SVV-GFP in 96-well plates. After 6 to 8 hours, cells were imaged by epifluorescence microscopy or manually triturated and analyzed by flow cytometry on a FACSCalibur (BD Biosciences). A) Fluorescence microscopy of H446 (positive control), H460 (negative control), and cell lines derived from LX22, LX33, and LX36 infected with SVV-GFP reporter virus demonstrates a high fraction of infected cells. Scale bars = 100 µm. B) The half-maximal effective concentration of wild-type SVV-001 was determined for each cell line by conversion of 3-(4,5-dimethylthiazol-2-yl)-5- (3-carboxymethoxyphenyl)-2-(4-sulfo-phenyl)-2H-tetrazolium) as previously described (10). Points represent mean values of five independent wells and uncertainty is summarized by standard deviation. Data shown are representative of four independent experiments.
The half-maximal effective concentration (EC50) for each of these cell lines was determined as previously described (10). The estimated EC50 for H446 of 3.0×10–4 vp/cell (95% confidence interval [CI] = 2.6×10–4 to 3.5×10–4) was consistent with the previously described high susceptibility of this line to SVV-001 (10), whereas those determined for LX22, LX33, and LX36 cell lines were 1.6×10–3 (95% CI = 1×10–3 to 2.5×10–3) vp/cell, 3.1×10–4 (95% CI = 2.5×10–4 to 3.6×10–4) vp/cell, and 3.9×10–3 (95% CI = 2.8×10–4 to 5.5×10–3) vp/cell, respectively (Figure 1B). These values are among the lowest EC50 values reported for SVV-001.
SVV-001 was tested for efficacy against three classic and three variant SCLC heterotransplants at a single intraperitoneal dose of 1012 vp/kg. Marked inhibition of tumor growth was observed in all three variant lines tested (LX22, LX33, and LX36) with sustained tumor growth inhibition over the duration of the experiment (Figure 2). Growth inhibition in the variant xenografts was consistent with the in vitro sensitivity of their matching cell lines. In contrast, the three classic SCLC lines, LX44, LX47, and LX48, were evidently refractory to SVV-001, demonstrating no statistically significant difference in tumor growth between treated mice and controls (Figure 2). A P value of less than .05 was considered statistically significant.
Figure 2.
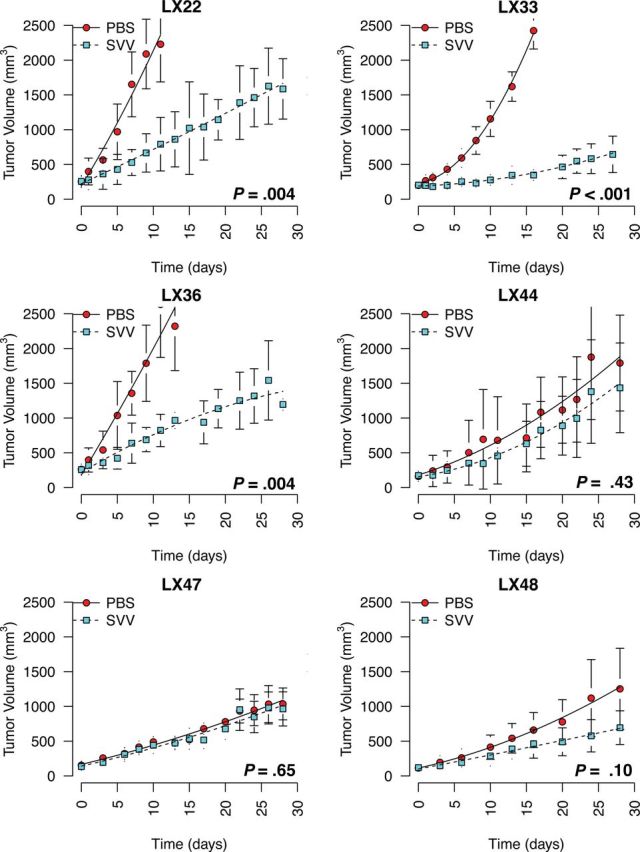
In vivo efficacy of Seneca Valley virus (SVV-001) in small cell lung cancer primary heterotransplant models. All primary heterotransplant models (LX22, LX33, LX36, LX44, LX47, and LX48) were generated as previously described (16). Clinical specimens were disrupted into single-cell suspensions in Matrigel and injected subcutaneously into NOG mice. Subsequent passages were carried in nonobese diabetic/severe combined immunodeficient (SCID) mice. Female CB-17 SCID mice less than 24 weeks of age (Charles River Laboratories) were injected subcutaneously with a suspension of 2.5×106 cells in phosphate-buffered saline (PBS) and Matrigel (BD Biosciences). Mice were distributed at random between control and treatment groups (n = 5 per group) when tumors reached a volume greater than 150mm3. Mice were treated with either PBS or SVV-001 as a single injection on day one of each study. Virus was serially diluted from frozen stocks and administered via intraperitoneal injection in 100 µL PBS. Tumor dimensions were measured by external calipers and volume was estimated by the formula V = (L x W2)/2, where L is the largest diameter and W is the diameter perpendicular to L. Tumor data for each group are presented as mean tumor volume with variance summarized by standard deviation. Statistical significance was determined by two-sided unpaired Student t test at the final time point where survival was 100% for both groups. Mouse experiments were performed according to protocols approved by the Johns Hopkins University Institutional Animal Care and Use Committee.
To further characterize the effective dose range of SVV-001 in a permissive model of variant SCLC, SVV-001 was titrated down over a three-log dose range from 1011 vp/kg to 109 vp/kg in LX36, resulting in similar inhibition of tumor growth (Figure 3A). Conversely, to test whether complete responses could be achieved with high initial virus dose, SVV-001 was administered at 1014 vp/kg, resulting in durable growth inhibition, including complete response in two of six mice (Figure 3B).
Figure 3.
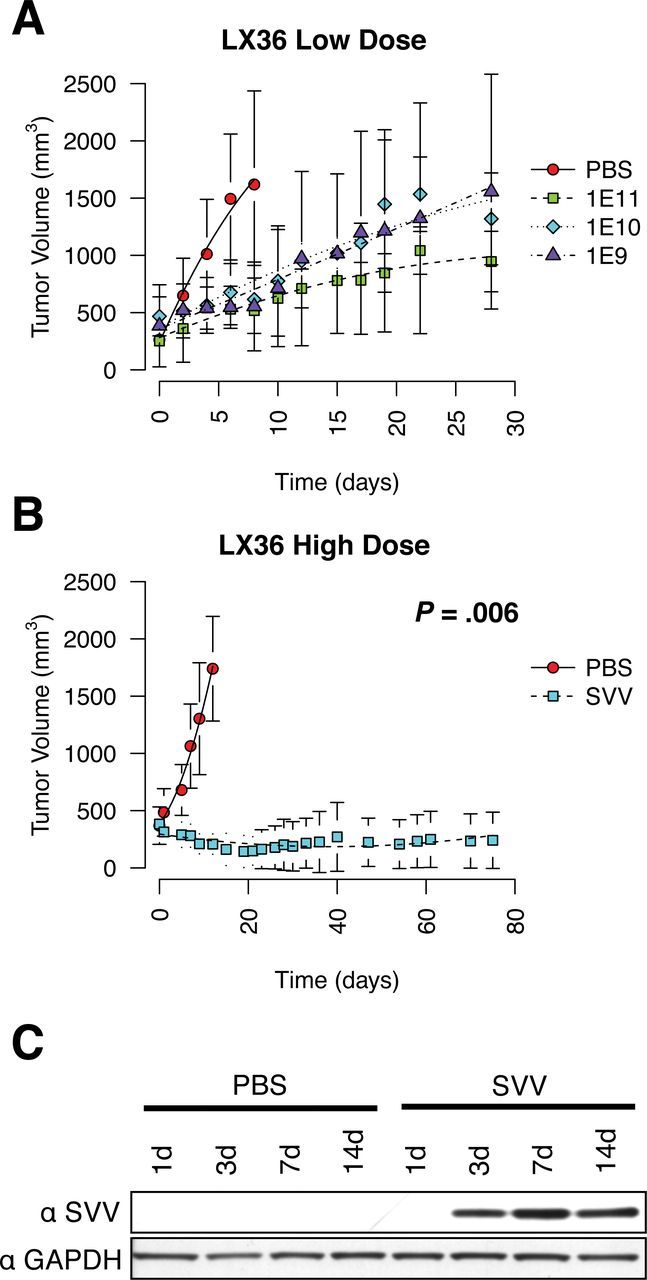
Dose modification of Seneca Valley virus (SVV-001) and time course of tumor growth in LX36 mice. LX36 xenografts were established in female CB-17 severe combined immunodeficient mice less than 24 weeks of age. Tumor data for each group are presented as mean tumor volume with variance summarized by standard deviation. Statistical significance was determined by two-sided unpaired Student t test at the final time point where survival was 100% for both groups. A) LX36 tumor-bearing mice were treated with either phosphate-buffered saline (PBS) or 1×1011, 1×1010, or 1×109 vp/kg of SVV-001 by single intraperitoneal injection on day 1 (n = 5 per group). B) LX36 mice were treated with either PBS or 1×1014 vp/kg (n = 6 per group). Two of six tumors were completely and durably eradicated whereas the remaining four tumors had a complete cytostatic effect. C) Western blot of pooled LX36 tumor lysates (n = 3 per time point) using rabbit hyperimmune serum generated against SVV-001 or glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Cells were collected and lysed in radioimmunoprecipitation assay (RIPA) buffer containing complete protease inhibitors (Sigma) for 30 minutes on ice, then clarified by low-speed centrifugation. Protein extracts were quantitated by bicinchoninic acid assay (Pierce) and normalized using RIPA buffer. Protein extracts were resolved on a 10% Bis-Tris acrylamide gel with MOPS running buffer (Invitrogen) and transferred to a polyvinylidene difluoride membrane. Membranes were blotted with rabbit hyperimmune serum against SVV-001 or commercial primary antibodies against GAPDH (Santa Cruz Biosciences), and the V5 epitope tag (Invitrogen), and detected using horseradish peroxidase–conjugated secondary antibodies and chemiluminescence (GE Life Sciences).
The heterotransplant models also offer the opportunity to better characterize SVV-001 kinetics in a permissive tumor. LX36 xenograft tumors were harvested at 1, 3, 7, and 14 days after infection. SVV-001 was assessed by western blotting for the SVV-001 structural protein VP1. VP1 was first detected at day three, peaking at day seven and continuing in a similar range at day 14 (Figure 3C). Our previous work using recombinant SVV-GFP reporter virus has shown that SVV-001 will home to tumors and begin to replicate in focal regions within 24 hours, but appears not to reach detectable levels by western blot analysis until day three (19). This result is consistent with clinical experience with SVV-001 in which patients often experience flu-like symptoms on day three after treatment, and in which persistence of intratumoral viral protein has been documented several weeks after single-dose administration (14).
Because no molecular determinants of permissivity have yet been identified, we analyzed gene expression profiles of a panel of SCLC lines characterized for SVV-001 permissivity for which microarray data were publicly available. Principal component analysis suggested that permissive and nonpermissive lines clustered separately, suggesting broad differences in transcription (Figure 4A). We hypothesized that potential biomarkers regulating global patterns of gene expression or differentiation status could be predictive. Because SVV-001 has selective tropism for neuroendocrine cancer types, we focused our analysis on differentially expressed genes involved in neurogenesis (gene ontology term GO:0022008).
Figure 4.
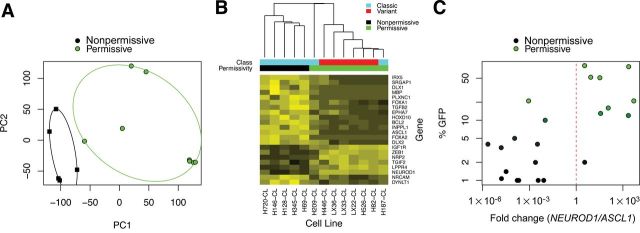
Prediction of Seneca Valley virus (SVV-001) permissivity by relative expression analysis of ASCL1 and NEUROD1. A) Microarray analysis was performed on publicly available data previously deposited in NCBI’s Gene Expression Omnibus (28) and accessible through GEO Series accession number GSE15240 (17). All analyses were performed using the Bioconductor suite for R (29). Raw intensities from.CEL files were normalized using the gcRMA algorithm and differential gene expression was assessed using the R/Bioconductor package (30). A linear model was fit for each probe to estimate expression differences between SVV-001 permissive and nonpermissive groups. The standard errors of log-2 fold change were moderated using an empirical Bayes approach. For each probe on the array, moderated t statistics, log odds ratios of differential expression (B statistics), and raw and adjusted P values (false discovery rate control by the Benjamini and Hochberg method) were calculated. Principal component analysis was done to distinguish SVV-001 permissive cell lines and nonpermissive lines. B) Hierarchical clustering of differentially expressed markers of neurogenesis (gene ontology term GO:0022008; adjusted P value cutoff <.025). C) Quantitative real-time polymerase chain reaction (PCR) of ASCL1 and NEUROD1 cDNA in a panel of small cell lung cancer cell lines and heterotransplants (n = 22). RNA was extracted from cells using TRIzol (Invitrogen) and cleaned up by RNeasy column purification (Qiagen) according to manufacturer protocols. Purified RNA was checked for degradation by ribosomal RNA integrity on a 1% agarose gel, and quantity and purity were determined by spectrophotometric measurements at A260 and A280. cDNA was synthesized using Superscript III (Invitrogen) and RNA was digested with RNase H. cDNA was used as a template for quantitative real-time PCR performed in quadruplicate using SYBR Green PCR Master Mix and a StepOnePlus thermal cycler with standard thermocycling conditions (Applied Biosystems). Primer sequences for ASCL1 (ID: 373) and NEUROD1 (ID: 116) have been previously described and are available in RTPrimerDB (31–33). Relative expression analysis was performed using the tspair package using 10% SVV–green fluorescent protein (GFP) positivity as a cutoff (34). The TSP score was defined as |Pr(X i > Y i|Class 1) − Pr(X i > Y i|Class 2)|, where X i is the gene expression measurement for the first gene of the pair for sample i and Y i is the gene expression measurement for the second gene of the pair for sample i. Leave-one-out cross-validation was performed using all samples. Log-transformed differential Ct is plotted on the horizontal axis and percentage of cells supporting infection by SVV-GFP on the vertical axis.
Among the highest-ranking genes differentially expressed between permissive and nonpermissive lines (Supplementary Table 1, available online) were genes for two transcription factors, Ascl1 and NeuroD1, that sequentially regulate neuroendocrine differentiation in multiple contexts, including hippocampus development (19), pulmonary neuroendocrine development (20–22), and gastrointestinal neuroendocrine tumors (23). ASCL1 in SCLC has low expression in variant relative to classic SCLC, and is among the best candidates for differentiating these SCLC subtypes (5,24). ASCL1 has been implicated in clonogenic and tumorigenic capacity as a primitive stem cell marker (25). We found ASCL1 to be highly expressed in nonpermissive lines, and conversely, NEUROD1 highly expressed in permissive lines (Figure 4B).
To validate these observations based on microarray analysis, ASCL1 and NEUROD1 expression was assessed by quantitative real-time polymerase chain reaction in a panel of 22 SCLC cell lines (Figure 4C). The ratio of NEUROD1 to ASCL1 was found to have a two-gene classification score of .81, sensitivity of .89, and specificity of .92, whereas the accuracy of classification by leave-one-out cross-validation was .91 (26).
Although NEUROD1 to ASCL1 ratio appears to be a strong predictive biomarker for SVV-001, this observation does not imply that expression of these factors is the sole determinant of permissivity. Exogenous expression of NEUROD1 in nonpermissive SCLC lines did not induce high-level permissivity, although some increase in SVV-001 replication was observed (Supplementary Figure 2, available online).
SVV-001 administration leads to prolonged tumor growth inhibition in variant SCLC heterotransplant models. Equivalent efficacy was observed over a dose range from 109 vp/kg to 1011. When the dose was increased to 1014 vp/kg, durable eradication of tumors was observed without apparent toxicity. This result is remarkable since this high level of prolonged efficacy has not been previously observed in SCLC heterotransplant models with cytotoxic or targeted therapeutics at maximally tolerated doses (27,28). The evident dose-response dependency for a replication-competent oncolytic virus is surprising. This effect may be due to inefficient SVV-001 extravasation, sequestration in nontumor tissues, or compartmentalization after the initial infection due to collapse of tumor microarchitecture and loss of vascularity.
The findings of this study carry some limitations. The lack of immunocompetent mouse models of SCLC permissive to SVV-001 is problematic: sustained intratumoral viral replication in heterotransplant models could be attributed to a lack of viral clearance in immunodeficient mice. Although we have observed sustained SVV-001 titers in patient serum weeks after administration and persistent viral replication in liver metastases 28 days after SVV-001 administration (14), it remains to be seen whether the biomarkers of SVV-001 efficacy defined here will apply in a clinical context.
Efficacy of SVV-001 in SCLC primary heterotransplants appears to be limited to variant SCLC, which is characterized by frequent loss of classic neuroendocrine markers. This may have important implications in patient selection: ongoing trials are focused on patients whose tumors specifically express these markers. Here we identified two genes, ASCL1 and NEUROD1, the ratio of which may efficiently predict permissivity of SCLC to SVV-001. These data provide rationale for continued human trials of SVV-001 in SCLC and imply that permissivity to SVV-001 is dependent in part on the differentiation status of the tumor.
Funding
American Association of Clinical Research/Caring for Carcinoid Foundation (grant 109884); Flight Attendant Medical Research Institute Center of Excellence in Translational Research; Burroughs Wellcome Fund Clinical Scientist Award in Translational Research (all to CMR). The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Supplementary Material
We thank Dr Luis Diaz for deriving and providing the LX47 and LX48 heterotransplant lines.
References
- 1. Wistuba I, Gazdar AF, Minna JD. Molecular genetics of small cell lung carcinoma. Semin Oncol. 2001;28(2 suppl 4):3–13 [PubMed] [Google Scholar]
- 2. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29 [DOI] [PubMed] [Google Scholar]
- 3. Oze I, Hotta K, Kiura K, et al. Twenty-seven years of phase III trials for patients with extensive disease small-cell lung cancer: disappointing results. PLoS One. 2009;4(11):e7835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carney DN, Gazdar AF, Bepler G, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985;45(6):2913–2923 [PubMed] [Google Scholar]
- 5. Pedersen N, Mortensen S, Sorensen SB, et al. Transcriptional gene expression profiling of small cell lung cancer cells. Cancer Res. 2003;63(8):1943–1953 [PubMed] [Google Scholar]
- 6. Carney DN, Mitchell JB, Kinsella TJ. In vitro radiation and chemotherapy sensitivity of established cell lines of human small cell lung cancer and its large cell morphological variants. Cancer Res. 1983;43(6):2806–2811 [PubMed] [Google Scholar]
- 7. Kraus AC, Ferber I, Bachmann SO, et al. In vitro chemo- and radio-resistance in small cell lung cancer correlates with cell adhesion and constitutive activation of AKT and MAP kinase pathways. Oncogene. 2002;21(57):8683–8695 [DOI] [PubMed] [Google Scholar]
- 8. Radice PA, Matthews MJ, Ihde DC, et al. The clinical behavior of “mixed” small cell/large cell bronchogenic carcinoma compared to “pure” small cell subtypes. Cancer. 1982;50(12):2894–2902 [DOI] [PubMed] [Google Scholar]
- 9. Brambilla E, Moro D, Gazzeri S, et al. Cytotoxic chemotherapy induces cell differentiation in small-cell lung carcinoma. J Clin Oncol. 1991;9(1):50–61 [DOI] [PubMed] [Google Scholar]
- 10. Reddy PS, Burroughs KD, Hales LM, et al. Seneca Valley virus, a systemically deliverable oncolytic picornavirus, and the treatment of neuroendocrine cancers. J Natl Cancer Inst. 2007;99(21):1623–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morton CL, Houghton PJ, Kolb EA, et al. Initial testing of the replication competent Seneca Valley virus (NTX-010) by the pediatric preclinical testing program. Pediatr Blood Cancer. 2010;55(2):295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu L, Baxter PA, Zhao X, et al. A single intravenous injection of oncolytic picornavirus SVV-001 eliminates medulloblastomas in primary tumor-based orthotopic xenograft mouse models. Neuro Oncol. 2011;13(1):14–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wadhwa L, Hurwitz MY, Chevez-Barrios P, et al. Treatment of invasive retinoblastoma in a murine model using an oncolytic picornavirus. Cancer Res. 2007;67(22):10653–10656 [DOI] [PubMed] [Google Scholar]
- 14. Rudin CM, Poirier JT, Senzer NN, et al. Phase I clinical study of Seneca Valley Virus (SVV-001), a replication-competent picornavirus, in advanced solid tumors with neuroendocrine features. Clin Cancer Res. 2011;17(4):888–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Venkataraman S, Reddy SP, Loo J, et al. Structure of Seneca Valley virus-001: an oncolytic picornavirus representing a new genus. Structure. 2008;16(10):1555–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hann CL, Daniel VC, Sugar EA, et al. Therapeutic efficacy of ABT-737, a selective inhibitor of BCL-2, in small cell lung cancer. Cancer Res. 2008;68(7):2321–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Daniel VC, Marchionni L, Hierman JS, et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69(8):3364–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poirier JT, Reddy PS, Idamakanti N, et al. Characterization of a full-length infectious cDNA clone and a GFP reporter derivative of the oncolytic picornavirus SVV-001. J Gen Virol. 2012;93(Pt 12):2606–2613 [DOI] [PubMed] [Google Scholar]
- 19. Kim EJ, Ables JL, Dickel LK, et al. Ascl1 (Mash1) defines cells with long-term neurogenic potential in subgranular and subventricular zones in adult mouse brain. PLoS One. 2011;6(3):e18472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Neptune ER, Podowski M, Calvi C, et al. Targeted disruption of NeuroD, a proneural basic helix-loop-helix factor, impairs distal lung formation and neuroendocrine morphology in the neonatal lung. J Biol Chem. 2008;283(30):21160–21169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Borges M, Linnoila RI, van de Velde HJ, et al. An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature. 1997;386(6627):852–855 [DOI] [PubMed] [Google Scholar]
- 22. Ito T, Udaka N, Yazawa T, et al. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development. 2000;127 (18):3913–3921 [DOI] [PubMed] [Google Scholar]
- 23. Shida T, Furuya M, Kishimoto T, et al. The expression of NeuroD and mASH1 in the gastroenteropancreatic neuroendocrine tumors. Mod Pathol. 2008;21(11):1363–1370 [DOI] [PubMed] [Google Scholar]
- 24. Westerman BA, Neijenhuis S, Poutsma A, et al. Quantitative reverse transcription-polymerase chain reaction measurement of HASH1 (ASCL1), a marker for small cell lung carcinomas with neuroendocrine features. Clin Cancer Res. 2002;8(4):1082–1086 [PubMed] [Google Scholar]
- 25. Jiang T, Collins BJ, Jin N, et al. Achaete-scute complex homologue 1 regulates tumor-initiating capacity in human small cell lung cancer. Cancer Res. 2009;69(3):845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Geman D, d’Avignon C, Naiman DQ, et al. Classifying gene expression profiles from pairwise mRNA comparisons. Stat Appl Genet Mol Biol. 2004;3:Article19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park KS, Martelotto LG, Peifer M, et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat Med. 2011;17(11):1504–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S. et al. , eds. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005:397–420 [Google Scholar]
- 31. Pattyn F, Robbrecht P, De Paepe A, et al. RTPrimerDB: the real-time PCR primer and probe database, major update 2006. Nucleic Acids Res. 2006;34(database issue):D684–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nissan X, Larribere L, Saidani M, et al. Functional melanocytes derived from human pluripotent stem cells engraft into pluristratified epidermis. Proc Natl Acad Sci U S A. 2011;108(36):14861–14866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Van Gele M, Boyle GM, Cook AL, et al. Gene-expression profiling reveals distinct expression patterns for classic versus variant Merkel cell phenotypes and new classifier genes to distinguish Merkel cell from small-cell lung carcinoma. Oncogene. 2004;23(15):2732–2742 [DOI] [PubMed] [Google Scholar]
- 34. Leek JT. The tspair package for finding top scoring pair classifiers in R. Bioinformatics. 2009;25(9):1203–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


