Abstract
The introduction of clinical genome-wide sequencing raises complex issues regarding the management of incidental findings. However, there is a lack of empirical studies assessing views of providers involved in potential disclosure of such findings. In an anonymous survey of 279 clinical genetics professionals, we found that the vast majority agreed they were interested in knowing about clinically actionable incidental findings in themselves (96%) and their child (99%), and they reported that these types of findings should be disclosed in adult (96%) and minor (98%) patients. Approximately three-fourths agreed they were personally interested in knowing about an adult-onset clinically actionable disease (78%), and a childhood-onset non-clinically actionable disease (75%) in their child. A similar percentage of participants (70%) felt these two types of findings should be disclosed to patients. Forty-four percent wanted to know about an incidental finding that indicates an adult-onset non-clinically actionable condition in themselves and 31% wanted to know about this type of information in their child. Findings from this study revealed participant views highly dependent on clinical actionability. Further research is needed with a broader population of geneticists to increase generalizability, and with diverse patients to assess their perspectives about results disclosure from clinical sequencing.
Keywords: Clinical genetics, ethics, genetics professionals, genomic sequencing, incidental findings, policy, return of results, secondary findings, whole genome sequencing
Introduction
Human genome sequencing has the potential to predict disease risk, identify disease causation, and refine therapeutic interventions. With improving accuracy and decreasing cost, whole genome sequencing (WGS) and whole exome sequencing (WES) have entered clinical practice as diagnostic tests for patients with rare diseases (1-4) and cancer (5, 6). It is predicted that these tests will also become used in healthy individuals to help guide healthcare decisions throughout life (1, 7), although a number of service delivery challenges have been raised (8, 9).
The growth in clinical use of genome-wide sequencing also introduces a number of ethical and social questions. One issue under debate is whether, and to what degree, molecular laboratories and clinicians should seek out, interpret, and communicate incidental findings from genome sequencing. From a clinical genetics perspective, incidental findings refer to gene variants associated with phenotypes that are not believed to be related to the disease under investigation. Ethical questions raised by clinical WGS and WES results include such issues as “How should decisions be made about which incidental findings to return?” “Do these incidental findings need to be “clinically actionable?” “How should incidental findings be managed in children (less than 18 years of age)?” To begin to address some of these questions, It will be important to understand and consider multiple stakeholder views toward management and disclosure of various types of incidental findings discovered during the course of clinical genome sequencing. At present, there is little information regarding the views of clinical genetics professionals who are, or will be, involved in the process of generating and returning these findings.
While there is scant information about stakeholder opinions concerning the risks and benefits of disclosing incidental findings in the clinical genetics setting, views regarding returning incidental findings in the genetic research context have been examined-- revealing varying opinions for the management of these findings. For example, some studies assessing views of the public and research participants suggest strong interest in receiving a variety of incidental findings discovered during genetic research (10-13). However, in a qualitative research study of families sequenced for a rare genetic condition, Miller syndrome, research participants described more ambivalence toward receiving incidental findings--those results that were not related to Miller syndrome (14). With respect to other key stakeholders, in a U.S. study of human genetic researchers, and a parallel study of institutional review board professionals, the majority of participants in each group (82% and 78% respectively) reported that researchers have an ethical obligation to return individual research results that would affect a participant's health or health care (15, 16).
A number of guidelines and policy recommendations have been proposed for the management of incidental findings in genetic research (17-23) and more recently are being developed for clinical practice. To foster further discussion on this topic, the National Human Genome Research Institute has convened a group of experts to form a Return of Results Consortium to assess practical and ethical issues related to the return of incidental findings from genomic research (24). While guidelines and lessons learned about return of incidental findings from genomic research can inform clinical practice, they might not address issues unique to the clinical setting. Currently, there are no evidence-based guidelines for return of incidental findings from clinical sequencing, although there have been proposals for various categories of results disclosure based on clinical validity and actionability (1, 25-27). The American College of Medical Genetics has recently issued a policy statement, “Points to Consider in the Clinical Application of Genomic Sequencing” and recommends that laboratories and clinics utilizing WGS/WES have clear policies in place related to disclosure of incidental, or secondary, findings (28). However, specific examples of findings recommended for disclosure are not delineated in this policy.
Management of incidental findings in genome sequencing has been reported in two exploratory studies of genetics providers in the U.S. and Canada, and other countries are weighing in on the future delivery of this service. In a formative study of 16 U.S. genetic specialists’ views toward disclosure of incidental findings for both adults and minor children in the context of WGS/WES, the participants indicated varying views in their choices about return of genomic information and in the relative value of different criteria used for making disclosure decisions (29). Ten genetics health professionals in a Canadian tertiary hospital participated in a focus group to assess views toward disclosure of incidental findings and they favored patient-clinician discussions prior to testing that focused on medical relevance and targeted analysis to limit data management (30). To begin to standardize and improve patient care in this area, the PHG Foundation in the UK has drafted an extensive report on the implications of clinical whole genome sequencing within the National Health Service (31).
In order to develop effective guidelines and procedures for the management of findings from clinical genome sequencing, it will be important to evaluate and understand the perspectives of key stakeholders, including clinical geneticists. However, larger empirical studies assessing these stakeholder views are lacking and currently there is no accepted standard of care or clinical guidelines that specifically outline best practices in the return of results from clinical sequencing. Therefore, in an effort to involve one important group of stakeholders and inform an evolving practice of clinical care, we conducted an anonymous survey of clinical genetics professionals to assess their views toward genome sequencing and the management of incidental findings in adults and children. This report presents our survey research findings.
Materials and Methods
Sample and recruitment
Attendees of the American College of Medical Genetics and Genomics (ACMG) Next Generation Sequencing Workshop held in March, 2012, in Charlotte, North Carolina, were invited to participate in a voluntary, anonymous survey before and after the workshop (32). The ACMG is an organization composed of biochemical, clinical, cytogenetic, medical and molecular geneticists, genetic counselors and other health care professionals committed to the practice of medical genetics (33). This organization conducts an annual clinical genetics conference and specialized workshops focusing on key content areas of interest to their membership.
Survey development and data collection
A 23-item survey was developed and informed by relevant literature as well as extensive internal and external expert review. Eight cognitive interviews of individuals of similar backgrounds to the study participants were conducted in order to increase readability and validity of the survey tool (34). The Tailored Design Method was also used as a general guide in the survey development (35). The survey was pilot-tested using the Turning Technologies audience response system (36), with 39 attendees of a seminar at the Medical College of Wisconsin, to identify any potential technical difficulties using this type of response system. Turning Technologies software utilizes a PowerPoint imbedded survey and allows for anonymous data to be collected via participant hand-held response devices. The survey questions were read aloud from the PowerPoint slides, the participants answered them through their individual response system devices, and the anonymous data were recorded instantly.
The survey assessed four domains: 1) participant views about genome sequencing for themselves and types of incidental findings they would want to know; 2) participant views about genome sequencing for their children and types of incidental findings they would want to know; 3) participant views toward management of incidental findings in adults and children in the clinical care setting; and 4) participant characteristics. In the survey instrument, we defined an incidental finding as “information that WGS can detect that is not related to your/your child's health concern. It may or may not be of clinical or personal interest to the person who had testing.” An example of an incidental finding was provided for each question and it was explained that these findings may indicate varying degrees of disease association (risk) and a variety of conditions for which there may or may not be treatments or interventions. “Clinically actionable” was defined as established therapeutic or preventive interventions, or other available actions, that have the potential to change the clinical course of the disease. A four-point Likert scale, rating level of agreement, and a fifth “undecided” category was utilized in the majority of response categories for questions assessing opinions. Categorical response options were used in the background/demographic questions. The survey is available as an online supplement. Because of the anonymous nature of the survey, this study was determined to be “non-human subject” research by the Medical College of Wisconsin's Human Research Protections Program.
Statistical analysis
Data were downloaded from the Turning Technologies software and exported to SPSS (version 20) and StatXact (Cytel Studio version 8) for statistical analysis (37, 38). Descriptive statistics were used to summarize responses to all questions. For the Likert scale, four categories were collapsed to two, combining the “strongly” and “somewhat strongly” categories at either end of the scale to facilitate analysis and interpretation. Percentages reported reflect the valid percent (excludes missing answers). For cross-tabulations of independent samples, such as age and questions, a Chi-square and Fisher-Halton-Freeman Exact Test were used. The McNemar Test was used for correlated data. Exact P values were calculated and a value of ≤ 0.05 was considered statistically significant for all tests. Because participants were allowed to skip individual items, the sample size varied by question.
Results
Three hundred fifty-five individuals were registered to attend the ACMG Next Generation Sequencing Workshop. Three hundred Turning Technologies hand-held devices were available for use and were randomly distributed on the chairs of attendees prior to the beginning of the workshop. Two hundred eighty-eight of the 300 workshop attendees who were provided with the audience response devices (~90% uptake rate) elected to participate in the pre-workshop survey study. Because some attendees came late or left early during the survey administration, we excluded participants who answered <50% of the non-demographic questions in the analysis. Due to a significant drop-off of participants after the 6 hour workshop, only pre-workshop survey data is presented in this report. In addition, of the data we were able to collect from both pre- and post- workshop surveys, findings revealed no significant changes in views toward disclosure of incidental findings from clinical sequencing. Therefore, the final sample presented includes 279 pre-workshop survey participants. The majority (75%) of participants were geneticists, genetic counselors and clinical laboratory professionals. Thirty-one percent were involved in delivery of genome sequencing (whole or exome) and 51% were planning to offer this service in the future. Sixty-seven percent of the survey participants were female and 65% percent reported that they have children. Table 1 provides a more detailed overview of the participant characteristics.
Table 1.
Participant Characteristics
| Response Categories | Percent |
|---|---|
| Gender (n=252) | |
| Male | 33% |
| Female | 67% |
| Age in years (n=255) | |
| 18-35 | 28% |
| 36-45 | 29% |
| 46-55 | 26% |
| 55+ | 17% |
| Have children (n=254) | |
| No | 35% |
| Yes | 65% |
| Primary area of work (n=259) | |
| Clinical geneticist | 24% |
| Genetic counselor | 13% |
| Clinical laboratory | 36% |
| Basic science laboratory | 5% |
| Bioinformatics | 3% |
| Trainees/students | 7% |
| Other | 12% |
| Years in primary area of work (n=260) | |
| Still in training | 9% |
| 0-5 | 26% |
| 6-10 | 16% |
| 11-15 | 15% |
| 16-20 | 17% |
| 20+ | 18% |
| Involved in delivery of clinical genome sequencing (n=254) | |
| No | 18% |
| Yes | 31% |
| Planning to in future | 51% |
| Have had genome sequenced and analyzed (n=256) | |
| No | 90% |
| Yes | 2% |
| Planning to in future | 9% |
Genome sequencing for participant and participant's child
With respect to interest in genome sequencing (no particular presenting scenario, or diagnostic indication, was provided), approximately half (52%) of participants (somewhat and strongly) agreed that they would like to have their genome sequenced and analyzed. The younger the participant, the more likely they were to agree to this statement (49% in age group 18-35, decreasing to 18% in age group 55 and older, p< 0.028). In comparison, 29% of participants indicated that they would like to have their child's genome sequenced. No age association was detected with agreement to this latter statement.
Participants were asked to assume that they, or their child, had whole genome sequencing for a particular diagnostic indication and an “incidental finding” was detected. They were then asked to think about what information they personally would want. The vast majority (96%) of participants wanted to know about incidental findings in themselves indicating an adult-onset disease that is “clinically actionable” and 78% wanted to know about this type of information in their child. The example provided was BRCA1 early-onset breast cancer. Slightly less than half (44%) of participants wanted to know about an incidental finding in themselves that indicates an adult-onset condition that is not clinically actionable (survey example: early-onset familial Alzheimer disease) and roughly one-third (31%) wanted to know about this type of information in their child. With respect to incidental findings in themselves with unknown or no clinical significance, 39% of participants were interested in this type of information for themselves and 32% interested in knowing this in their child.
Incidental findings in participants and adult patients
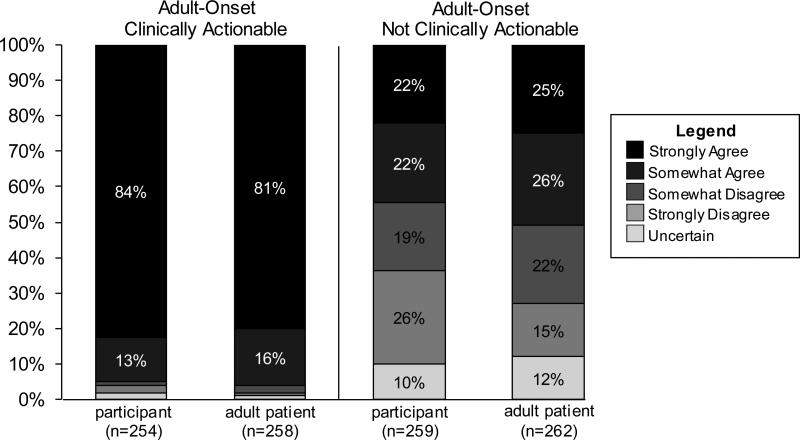
In addition to being asked about their own interest in receiving different types of incidental findings, participants were also asked to think about what types of incidental findings a clinical service provider should disclose to adult patients. The vast majority (96%) of participants wanted to know about incidental findings in themselves indicating an adult-onset disease that is “clinically actionable” and the same percentage felt this type of information should be disclosed to adult patients. Slightly less than half (44%) of participants wanted to know about an incidental finding in themselves that indicates an adult-onset condition that is not clinically actionable and roughly half (52%) felt this type of information should be disclosed to adult patients (See Fig. 1). Participants who reported interest in having their genome sequenced were more likely to agree that they wanted to know about an adult-onset, non-clinically actionable incidental finding; however, those who were undecided were more likely to disagree (p< 0.001). In contrast, a participant's interest in having their own genome sequenced did not correlate with agreement to disclose an adult-onset, non-clinically actionable incidental finding to an adult patient (p< 0.1).
Fig. 1.
Comparison of attitudes toward return of incidental findings in participants and in adult patients.
Incidental findings in participant's child and minor (under 18) patients
Participants were queried about their views toward their own child and what they felt a clinical service provider should do if a minor patient were found to have an incidental finding following genome sequencing. In comparing views about their child to a minor patient, the majority of participants wanted to know about an incidental finding in their child that indicates a childhood-onset disease that is clinically actionable (99%), a childhood-onset disease that is not clinically actionable (75%), and an adult-onset disease that is clinically actionable (78%). A similar percentage of participants felt this type of information should be disclosed in the clinical setting to parents of children who have sequencing performed (98%, 70%, and 70% respectively). See Fig. 2. Participants involved in the delivery of clinical genome sequencing, or planning to be involved in the future, were more likely to agree (p< .05) that incidental findings should be disclosed to parents that indicate a clinically adult-onset disease in a minor patient. Approximately one-third (32%) of participants were interested in an incidental finding about their child that indicates a genetic change for which there is unknown or no clinical significance and 31% were interested in an adult-onset, not clinically actionable incidental finding in their child.
Fig. 2.
Comparison of attitudes toward return of incidental findings in participant's child and in minor patients.
Discussion
To our knowledge, this report represents the largest survey to date examining clinical genetic professionals’ opinions concerning return of incidental findings. This study explores the attitudes of genetics providers with respect to incidental findings identified through WGS/WES in themselves, their child, their adult patients and their minor patients. The participants were primarily female, were over 35 years old, and had children. The majority worked in a clinical environment for six or more years and was involved, or was planning to be involved, in delivery of clinical genome sequencing.
Our findings revealed that participants had certain views about incidental findings disclosure that were nearly uniform. For example, there was almost unanimous consensus to disclose incidental findings that are adult-onset and clinically actionable for themselves (96%) and their adult patients (96%). This held true for childhood-onset clinically actionable conditions in the participant's child (99%) and minor patients (98%). The consensus for return of results which are associated with a significant risk of disease --for which there is therapeutic or preventative measures-- is similar to results disclosure recommendations to study patients undergoing genetic research testing (17, 22). A similar high degree of interest (96%) in learning ancillary findings during pharmacogenetic testing for risk of serious but treatable disease was also reported in a telephone survey study of the U.S. public (39).
Overall, there was moderate consensus for disclosure of an incidental finding that was adult-onset and clinically actionable when found in the participant's child (78%) or in a minor patient (70%). There is a body of literature and opinion that states genetic testing for adult-onset conditions should not be sought in children because the child is denied the right to choose to find out this information at their majority (40-44). WGS/WES raises a subtly different issue. By the nature of the test, this type of information is obtained in the process of deriving the data. Diagnoses for adult disorders were not specifically requested but may be found as part of testing via WGS/WES. Therefore, further research into parent and child attitudes about risks and benefits of WGS/WES is essential. The issue of retaining results for minor patients for many years to allow them to decide whether they wish to find out about adult-onset risks includes a number of challenges, notably tracking the individual over time and retaining the large amount of data. Moderate consensus was also reported by participants for disclosure of an incidental finding that was not actionable. Seventy-five percent of participants reported interest in disclosure of a childhood-onset condition that was not actionable in their own children and 70% of participants favored disclosure of a childhood-onset condition that was not actionable to minor patients.
A number of our survey results revealed less participant consensus about return of certain types of incidental findings. For instance, 44% of participants indicated interest in disclosure of an adult-onset condition that was not actionable to themselves and 52% of participants favored disclosure of an adult-onset condition that was not actionable to their adult patients. Thirty-one percent of participants reported interest in disclosure of an adult-onset condition that was not actionable in their own children. Less consensus was also noted for incidental findings with unknown or no clinical significance-- roughly one-third of participants were interested in this type of information for themselves and for their child. Given the overall range of views of the genetic professionals surveyed in this study, it is foreseeable that the lack of consensus about determination about what incidental findings “can” or “should” be returned in various clinical WGS/WES scenarios could lead to inconsistent clinical practice.
This report provides a starting point for discussion about views of clinical genetics professionals toward incidental findings in genome sequencing. However, this study has several limitations. One limitation is that it is not known to what extent ACMG members are representative of the larger community of clinical genetics professionals. Therefore it is possible that our findings may be biased toward the opinions of those ACMG members and individuals who chose to attend the 2012 ACMG Next Generation Sequencing Workshop. In addition, sixty-seven percent of the study participants were women. Consequently, our findings may be more representative of women's views than men's on return of incidental findings. Our example of BRCA1 as a clinically actionable finding may also have resulted in some bias, given the larger number of women study participants. However, a strength of this study is the high participation rate, compared to mail and online surveys of health professionals. Another limitation is that the Turning Technologies survey method involves brief, closed-ended questions and does not allow for open-ended questions to further explore why participants answered the way they did. Also, due to the length of the workshop (6 hours) and people entering and leaving at different times, we had a number of surveys with incomplete data and did not have sufficient data for an unbiased pre- and post- analysis. Lastly, as with all empirical research utilizing hypothetical scenarios, our findings may not correlate precisely with actual future behavior.
Genome sequencing, as a clinical service, will continue to grow as an important diagnostic tool for health care providers. The public and research participants have expressed interest in receiving a variety of incidental findings (12, 14, 45), however, little is known about how a wide variety of patients and service providers feel about the risks and benefits of disclosure of incidental findings from genome sequencing in the clinical setting. This cross-sectional study is one of the first to survey a large number of clinical geneticists to assess their views on the return of incidental findings from genome sequencing and illuminates a wide variety of views. This diversity of opinion, coupled with no formal guidance, illustrates the need for a more formal policy-making process to determine what types of incidental findings should or should not be returned in order to maximize benefit and reduce harms. The role of patients and consumers in healthcare decision-making is increasingly being recognized in the health policy community. Careful consideration of patient preferences and expectations about return of incidental findings will be essential to the development of effective and respectful practice guidelines. Future research will be needed to evaluate how the broader public and diverse groups of patients conceptualize the complexities of genome sequencing results disclosure in order to further inform evolving policies and best practices regarding incidental findings.
Acknowledgements
We gratefully acknowledge Richard Sharp and Gail Henderson for their survey construction advice and thank Holly Tabor for very helpful comments on an earlier draft of this article. We also thank the American College of Medical Genetics conference planning committee, as well as the Next Generation Sequencing Workshop facilitators Madhuri Hegde, Birgit Funke, Shashikant Kulkarni, and the survey study participants. This study was supported, in part, by the Advancing Healthier Wisconsin Initiative.
Footnotes
Conflict of Interest Statement: Amy Lemke, David Bick, David Dimmock, Pippa Simpson and Regan Veith have no conflicts of interest to disclose.
Post-acceptance of this manuscript, the American College of Medical Genetics published recommendations in the form of a minimal list of incidental findings to report from clinical sequencing. See https://www.acmg.net/docs/IF_Statement_Final_7.24.13.pdf.
References
- 1.Bick D, Dimmock D. Whole exome and whole genome sequencing. Curr Opin Pediatr. 2011;23:594–600. doi: 10.1097/MOP.0b013e32834b20ec. [DOI] [PubMed] [Google Scholar]
- 2.Ambry Genetics. Clinical Diagnostic Exome. from http://www.ambrygen.com/clinical-diagnostic-exome.
- 3.Baylor College of Medicine, Medical Genetics Laboratories Whole Exome Sequencing. from https://www.bcm.edu/geneticlabs/test_detail.cfm?testcode=1500.
- 4.GeneDx. XomeDx Whole Exome Sequencing. 2011 from http://www.genedx.com/test-catalog/xomedx/
- 5.Roychowdhury S, Iyer MK, Robinson DR, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3::111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welch JS, Westervelt P, Ding L, et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA. 2011;305:1577–1584. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drmanac R. The advent of personal genome sequencing. Genet Med. 2011;13:188–190. doi: 10.1097/GIM.0b013e31820f16e6. [DOI] [PubMed] [Google Scholar]
- 8.Brunham LR, Hayden MR. Medicine. Whole-genome sequencing: the new standard of care? Science. 2012;336:1112–1113. doi: 10.1126/science.1220967. [DOI] [PubMed] [Google Scholar]
- 9.Sharp RR. Downsizing genomic medicine: approaching the ethical complexity of whole-genome sequencing by starting small. Genet Med. 2011;13:191–194. doi: 10.1097/GIM.0b013e31820f603f. [DOI] [PubMed] [Google Scholar]
- 10.Beskow LM, Smolek SJ. Prospective biorepository participants’ perspectives on access to research results. J Empir Res Hum Res Ethics. 2009;4:99–111. doi: 10.1525/jer.2009.4.3.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaufman D, Geller G, Leroy L, Murphy J, Scott J, Hudson K. Ethical implications of including children in a large biobank for genetic-epidemiologic research: a qualitative study of public opinion. Am J Med Genet C Semin Med Genet. 2008;148C:31–39. doi: 10.1002/ajmg.c.30159. [DOI] [PubMed] [Google Scholar]
- 12.Lemke AA, Halverson C, Ross LF. Biobank participation and returning research results: perspectives from a deliberative engagement in South Side Chicago. Am J Med Genet A. 2012;158A:1029–1037. doi: 10.1002/ajmg.a.34414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy J, Scott J, Kaufman D, Geller G, LeRoy L, Hudson K. Public expectations for return of results from large-cohort genetic research. Am J Bioeth. 2008;8:36–43. doi: 10.1080/15265160802513093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabor HK, Stock J, Brazg T, et al. Informed consent for whole genome sequencing: A qualitative analysis of participant expectations and perceptions of risks, benefits, and harms. Am J Med Genet A. 2012;158A:1310–1319. doi: 10.1002/ajmg.a.35328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards KL, Lemke AA, Trinidad SB, et al. Attitudes toward genetic research review: results from a survey of human genetics researchers. Public Health Genomics. 2011;14:337–345. doi: 10.1159/000324931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemke AA, Trinidad SB, Edwards KL, Starks H, Wiesner GL, Consortium G. Attitudes toward genetic research review: results from a national survey of professionals involved in human subjects protection. J Empir Res Hum Res Ethics. 2010;5:83–91. doi: 10.1525/jer.2010.5.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bookman EB, Langehorne AA, Eckfeldt JH, et al. Reporting genetic results in research studies: summary and recommendations of an NHLBI working group. Am J Med Genet A. 2006;140:1033–1040. doi: 10.1002/ajmg.a.31195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caulfield T, McGuire AL, Cho M, et al. Research ethics recommendations for whole-genome research: consensus statement. PLoS Biol. 2008;6:e73. doi: 10.1371/journal.pbio.0060073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabsitz RR, McGuire A, Sharp RR, et al. Ethical and practical guidelines for reporting genetic research results to study participants: updated guidelines from a National Heart, Lung, and Blood Institute working group. Circ Cardiovasc Genet. 2010;3:574–580. doi: 10.1161/CIRCGENETICS.110.958827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knoppers BM, Deschenes M, Zawati MH, Tasse AM. Population studies: return of research results and incidental findings Policy Statement. European Journal of Human Genetics. 2012:1–3. doi: 10.1038/ejhg.2012.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf SM, Crock BN, Van Ness B, et al. Managing incidental findings and research results in genomic research involving biobanks and archived data sets. Genet Med. 2012;14:361–384. doi: 10.1038/gim.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolf SM, Lawrenz FP, Nelson CA, et al. Managing incidental findings in human subjects research: analysis and recommendations. J Law Med Ethics. 2008;36:219–248. 211. doi: 10.1111/j.1748-720X.2008.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zawati MH, Van Ness B, Knoppers BM. Incidental findings in genomic research: a review of international norms. GenEdit. 2011;9:1–8. [Google Scholar]
- 24.Mjoseth J. National Human Genome Research Institute (NHGRI). NHGRI funds return of results studies, forms expert consortium. 2011 from http://www.genome.gov/27545526.
- 25.Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet Med. 2011;13:499–504. doi: 10.1097/GIM.0b013e318220aaba. [DOI] [PubMed] [Google Scholar]
- 26.Mayer AN, Dimmock DP, Arca MJ, et al. A timely arrival for genomic medicine. Genet Med. 2011;13:195–196. doi: 10.1097/GIM.0b013e3182095089. [DOI] [PubMed] [Google Scholar]
- 27.Dimmock D. A personal perspective on returning secondary results of clinical genome sequencing. Genome Med. 2012;4:54. doi: 10.1186/gm353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Points to consider in the clinical application of genomic sequencing. Genet Med. 2012;14:759–761. doi: 10.1038/gim.2012.74. [DOI] [PubMed] [Google Scholar]
- 29.Green RC, Berg JS, Berry GT, et al. Exploring concordance and discordance for return of incidental findings from clinical sequencing. Genet Med. 2012;14:405–410. doi: 10.1038/gim.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Townsend A, Adam S, Birch PH, Lohn Z, Rousseau F, Friedman JM. “I want to know what's in Pandora's box”: Comparing stakeholder perspectives on incidental findings in clinical whole genomic sequencing. Am J Med Genet A. 2012 doi: 10.1002/ajmg.a.35554. [DOI] [PubMed] [Google Scholar]
- 31.Wright CD. Next steps in the sequence : the implications of whole genome sequencing for health in the UK. PHG Foundation; Cambridge: 2011. [Google Scholar]
- 32.American College of Medical Genetics and Genomics (ACMG) ACMG workshop. from http://www.acmgmeeting.net/acmg2012/public/Content.aspx?ID=531&sortMenu=103003.
- 33.American College of Medical Genetics and Genomics (ACMG) ACMG Terms and Conditions. from http://www.acmg.net/AM/Template.cfm?Section=Mission_Statement&Template=/CM/HTMLDisplay.cfm&ContentID=5676.
- 34.Willis GB. Cognitive interviewing : a tool for improving questionnaire design. Sage Publications; Thousand Oaks, CA: 2005. [Google Scholar]
- 35.Dillman DA. Mail and internet surveys : the tailored design method 2007 update with new internet, visual, and mixed-mode guide. Wiley; Hoboken, NJ: 2007. [Google Scholar]
- 36.Turning Technologies TurningPoint. from http://www.turningtechnologies.com.
- 37.IBM SPSS Statistics Professional. 2011 from http://www-142.ibm.com/software/products/us/en/spss-stats-pro.
- 38.Cytel StatXact statistical software. 2007 from http://www.cytel.com/software/statxact.
- 39.Haga SB, O'Daniel JM, Tindall GM, Lipkus IR, Agans R. Public attitudes toward ancillary information revealed by pharmacogenetic testing under limited information conditions. Genet Med. 2011;13:723–728. doi: 10.1097/GIM.0b013e31821afcc0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Points to consider: ethical, legal, and psychosocial implications of genetic testing in children and adolescents. American Society of Human Genetics Board of Directors, American College of Medical Genetics Board of Directors. Am J Hum Genet. 1995;57:1233–1241. [PMC free article] [PubMed] [Google Scholar]
- 41.American Academy of Pediatrics, Committee on Bioethics Ethical issues with genetic testing in pediatrics. Pediatrics. 2001;107:1451–1455. doi: 10.1542/peds.107.6.1451. [DOI] [PubMed] [Google Scholar]
- 42.Borry P, Fryns JP, Schotsmans P, Dierickx K. Carrier testing in minors: a systematic review of guidelines and position papers. European Journal of Human Genetics : EJHG. 2006;14:133–138. doi: 10.1038/sj.ejhg.5201509. [DOI] [PubMed] [Google Scholar]
- 43.Borry P, Stultiens L, Nys H, Cassiman JJ, Dierickx K. Presymptomatic and predictive genetic testing in minors: a systematic review of guidelines and position papers. Clin Genet. 2006;70:374–381. doi: 10.1111/j.1399-0004.2006.00692.x. [DOI] [PubMed] [Google Scholar]
- 44.Goldman JS, Hahn SE, Catania JW, et al. Genetic counseling and testing for Alzheimer disease: joint practice guidelines of the American College of Medical Genetics and the National Society of Genetic Counselors. Genet Med. 2011;13:597–605. doi: 10.1097/GIM.0b013e31821d69b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bollinger JM, Scott J, Dvoskin R, Kaufman D. Public preferences regarding the return of individual genetic research results: findings from a qualitative focus group study. Genet Med. 2012;14:451–457. doi: 10.1038/gim.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]