Abstract
The mammalian gut microbiota influences both sides of the energy balance equation, salvaging energy from undigested nutrients and directing the host to accumulate adipose tissue. Semova et al. (2012) use zebrafish to demonstrate that the gut microbiota also promotes dietary lipid absorption, emphasizing the many host-microbial interactions contributing to adiposity.
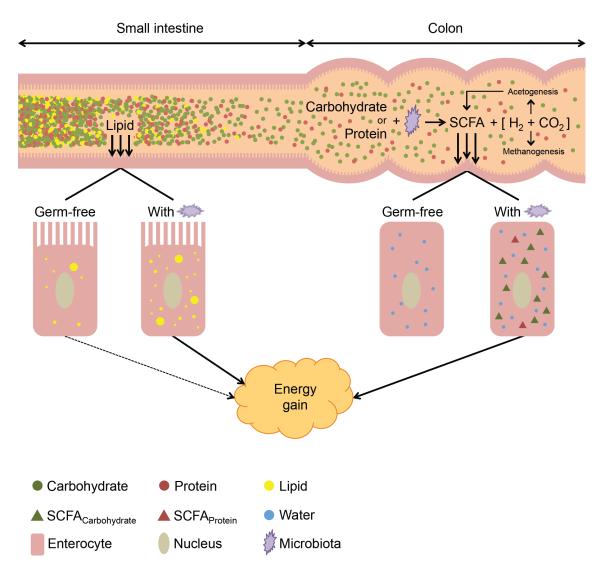
The rapid rise in the global incidence of obesity has prompted efforts to more carefully consider the many genetic and environmental factors that influence dietary energy harvest. Recent studies have emphasized that human nutrition may be better understood as a series of complex interactions between diet, host genotype, and the trillions of microorganisms that inhabit our gastrointestinal tracts, collectively referred to as the gut microbiota. Notably, germ-free mice that are inoculated with a gut microbiota harvested from conventionally raised mice show a marked increase in body fat within two weeks despite a reduction in chow consumption and higher overall levels of metabolic expenditure (Bäckhed et al., 2004), a response enhanced upon colonization with a microbial community harvested from obese donor animals (Turnbaugh et al., 2009). The various mechanisms contributing to the microbial stimulation of adiposity are thought to include the fermentation of complex plant polysaccharides to short-chain fatty acids that can be assimilated by the host (Martens et al., 2011), acetogenesis (the production of acetate from H2 and CO2) (Rey et al., 2010), methanogenesis (the production of methane from H2 and CO2) (Samuel and Gordon, 2006), and microbial factors that direct the host to increase fat deposition (Bäckhed et al., 2004) (Figure 1).
Figure 1. An expanded model for the contribution of the gut microbiota to energy harvest from dietary lipids, carbohydrates, and proteins.
In this issue, Semova et al. (2012) show that the gut microbiota stimulates lipid absorption in the zebrafish proximal intestine. Compared with germ-free animals fed the same diet, enterocytes of animals conventionalized with a gut microbiota accumulated larger and more numerous lipid droplets. Increased lipid accumulation was also observed in the liver of conventionalized animals, suggesting greater uptake of lipids into systemic circulation. Microbial processes in the distal gut (colon) are also known to contribute to host energy gain. Carbohydrates and protein that resist digestion in the small intestine are fermented by the colonic microbiota, producing short-chain fatty acids (SCFA) that can be assimilated and used as energy by the host (Martens et al., 2011). The H2 and CO2 released during fermentation are consumed as substrates by some microbes in acetogenesis (Rey et al., 2010), which produces a secondary source of SCFA, as well as methanogenesis (Samuel and Gordon, 2006). Consumption of H2 and CO2 reaction products helps to drive the continued production of SCFA.
Despite these compelling links between host body fat and the gut microbiota, it remains unclear to what degree gut microorganisms, or microbial consortia, can influence the absorption, metabolism, and/or systemic distribution of dietary lipids. One reason for this oversight is that the emulsification, digestion, and subsequent absorption of lipids in the small intestine is thought to be highly efficient, suggesting little room for a microbial contribution. Secondly, it can be difficult to distinguish dietary, host-derived, and microbe-derived lipids in vivo when studying human cohorts or rodent models. Thirdly, the study of model commensals, e.g. Bacteroides thetaiotaomicron, have provided a detailed view of carbohydrate metabolism (Martens et al., 2011), whereas relatively little is known about the genes and metabolic pathways required for microbial lipid metabolism in the human gut.
New research by Semova et al. (2012) indicates that the absorption of dietary lipids, and potentially their impact on energy balance, may have more to do with the gut microbiota than previously appreciated. The authors took advantage of a recently developed method that enables the tracking of fatty acids during absorption by the intestinal epithelium in live zebrafish, coupled to methods for rearing germ-free zebrafish. They fed fluorescent fatty acids to transparent zebrafish larvae and then visualized the extent of lipid assimilation and lipid droplet (LD) formation in the intestinal epithelium and liver under various dietary and microbial colonization scenarios. They found that ‘conventionalized’ zebrafish exposed to a gut microbiota displayed increased fatty acid uptake and LD formation in their enterocytes and liver compared with germ-free animals (Figure 1). The increased uptake in conventionalized zebrafish was enhanced in fed relative to starved animals, coincident with an expansion in the relative abundance of the Firmicutes phylum. Interestingly, Semova et al. (2012) found evidence of at least two distinct mechanisms for the microbial promotion of lipid assimilation, as colonization of animals with a representative Firmicutes strain (genus: Exiguobacterium) resulted in increased LD number whereas colonization with individual strains of either Bacteroidetes or Proteobacteria resulted in increased LD size.
Lipid droplets were once believed to be inert lipid depots, but research over the last decade has shown them to be dynamic organelles serving a range of cellular functions with energetic implications: regulation of intracellular level of free fatty acids and cholesterol, lipid synthesis and metabolism, protein storage and degradation, as well as viral replication (Walther and Farese, 2012). Whether microbial promotion of LD size versus number in enterocytes influences the energetic potential of the assimilated dietary lipids remains to be determined; however, many LD functions depend on interactions that happen at interface between the hydrophobic interior of the LD and the surrounding aqueous cytoplasm. Since small but numerically abundant LDs have a higher surface area to volume ratio, they may be more efficient at lipid mobilization, including delivery of lipids from the intestinal enterocytes into systemic circulation. Washout experiments performed by Semova et al. (2012) may provide some initial support for this hypothesis, as lipid export from the intestinal epithelium reduced the proportion of small LDs compared with medium and large LDs.
More extensive experiments with culturable microbial isolates from zebrafish may help reveal the extent to which the ability to stimulate LD size or number is conserved among members of the same bacterial phylum, in addition to elucidating the underlying mechanisms responsible. Of note, both gram-negative strains stimulated increased LD size, potentially suggesting a role for lipopolysaccharides or other components of the bacterial outer membrane. Interestingly, the response to the Firmicutes strain Exiguobacterium did not require live bacteria, suggesting that there may be a secreted factor that stimulates LD number.
The zebrafish larvae employed by Semova et al. (2012) are an elegant model system. Apart from their transparency, which allows real-time visualization of lipid assimilation, the aqueous environment in which the larvae develop can be tightly controlled, allowing microbial composition in the gut versus the environment to be evaluated separately across the range of feeding conditions. For instance, the authors found that the increased relative abundance of Firmicutes associated with feeding occurred solely within the gut and not the surrounding water, suggesting that factors secreted by the zebrafish gut, or its nutritional composition, promoted the expansion of the Firmicutes. This ability to control the microbial and nutritional properties of the environment precisely lends itself to future studies. For example, monitoring the disappearance of fluorescent fatty acids from the environment could help to discriminate whether observed increases in lipid uptake were due to more efficient assimilation or, alternatively, higher overall levels of lipid ingestion among conventionalized zebrafish. Such monitoring would also allow accountancy for very small LDs, beginning at ~12 nm diameter, which may be present in high quantities but are below the resolution limit of microscopy (Suzuki et al., 2011). It would thus permit the total amount of lipid uptake to be quantified, which is likely to be related to overall host energy harvest.
Extending such inquiry to other model organisms will also prove useful in interpreting these data in the context of human digestion. Recent work has demonstrated clear functional analogues between sections of the zebrafish gut and the small and large intestines of humans (Wang et al., 2010). Yet the zebrafish gut differs structurally from that of humans, existing as a simple tapered tube that bears no stomach. Moreover, microorganisms are thought to be distributed evenly throughout the zebrafish gut, whereas the vast majority of the human gut microbiota resides within the colon. The lower relative abundance of microorganisms in the human proximal small intestine could imply that the gut microbiota can less readily influence lipid absorption in humans than in zebrafish. However, even if their presence at the site of lipid absorption is limited, the human gut microbiota could conceivably influence total lipid assimilation through mechanisms initiated in the colon, e.g. effects on transit time.
Lipids represent a substantial caloric fraction of global human diets and differences in fatty acid intake have been implicated in the development of obesity, diabetes, and coronary heart disease (Micha and Mozaffarian, 2009). Illuminating the mechanisms of host-microbial interactions in lipid absorption could thus have profound economic and social benefits. We anticipate that the exciting results of Semova et al. (2012) will likely stimulate much additional research in this direction, with the end goal of identifying conserved microbial mechanisms that might be exploited to promote fattier fish and leaner Homo sapiens.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micha R, Mozaffarian D. Trans fatty acids: effects on metabolic syndrome, heart disease and diabetes. Nature Reviews Endocrinology. 2009;5:335–344. doi: 10.1038/nrendo.2009.79. [DOI] [PubMed] [Google Scholar]
- Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, Newgard CB, Gordon JI. Dissecting the in vivo metabolic potential of two human gut acetogens. J Biol Chem. 2010;285:22082–22090. doi: 10.1074/jbc.M110.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. P Natl Acad Sci USA. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semova I, Carten JD, Stombaugh J, Mackey LC, Knight R, Farber SA, Rawls JF. Cell host & microbe TBD. TBD; 2012. Microbiota and diet regulate fatty acid absorption in the zebrafish intestine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Shinohara Y, Ohsaki Y, Fujimoto T. Lipid droplets: size matters. Journal of Electron Microscopy. 2011;60:S101–A116. doi: 10.1093/jmicro/dfr016. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:1–10. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RVJ. Lipid droplets and cellular lipid metabolism. Annual Review of Biochemistry. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Du J, Lam SH, Mathavan S, Matsudaira P, Gong Z. Morphological and molecular evidence for functional organization along the rostrocaudal axis of the adult zebrafish intestine. BMC Genomics. 2010;11:392. doi: 10.1186/1471-2164-11-392. [DOI] [PMC free article] [PubMed] [Google Scholar]