Abstract
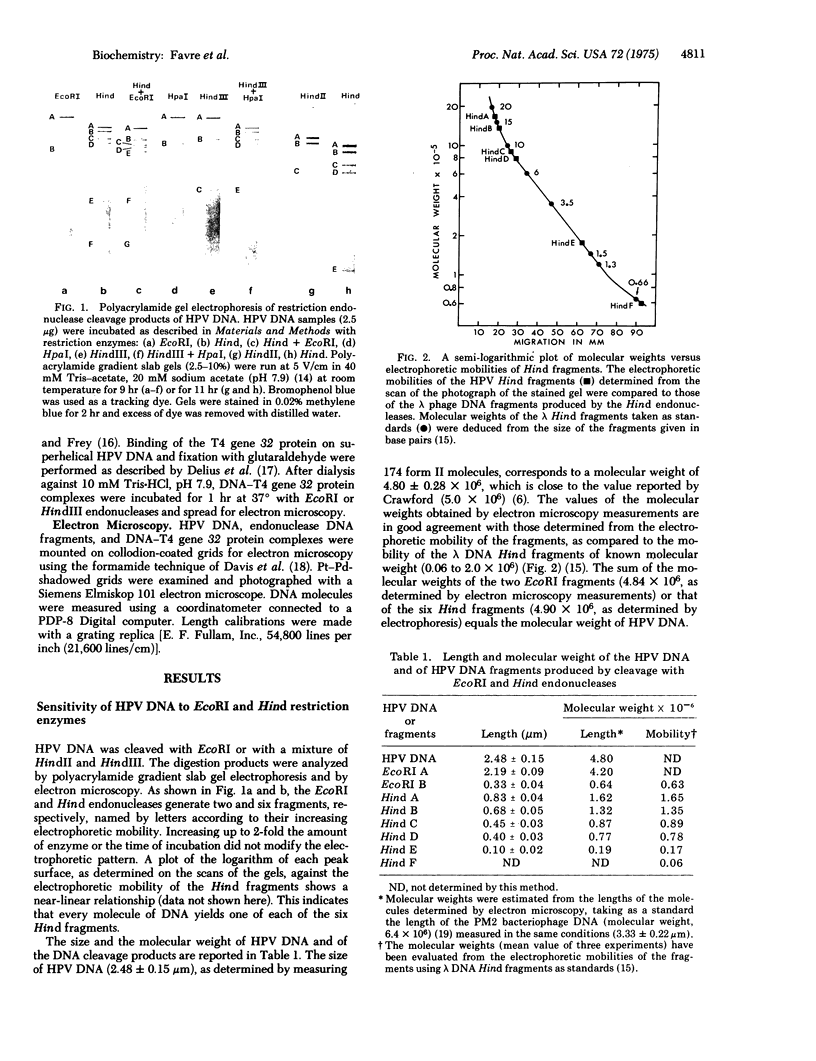
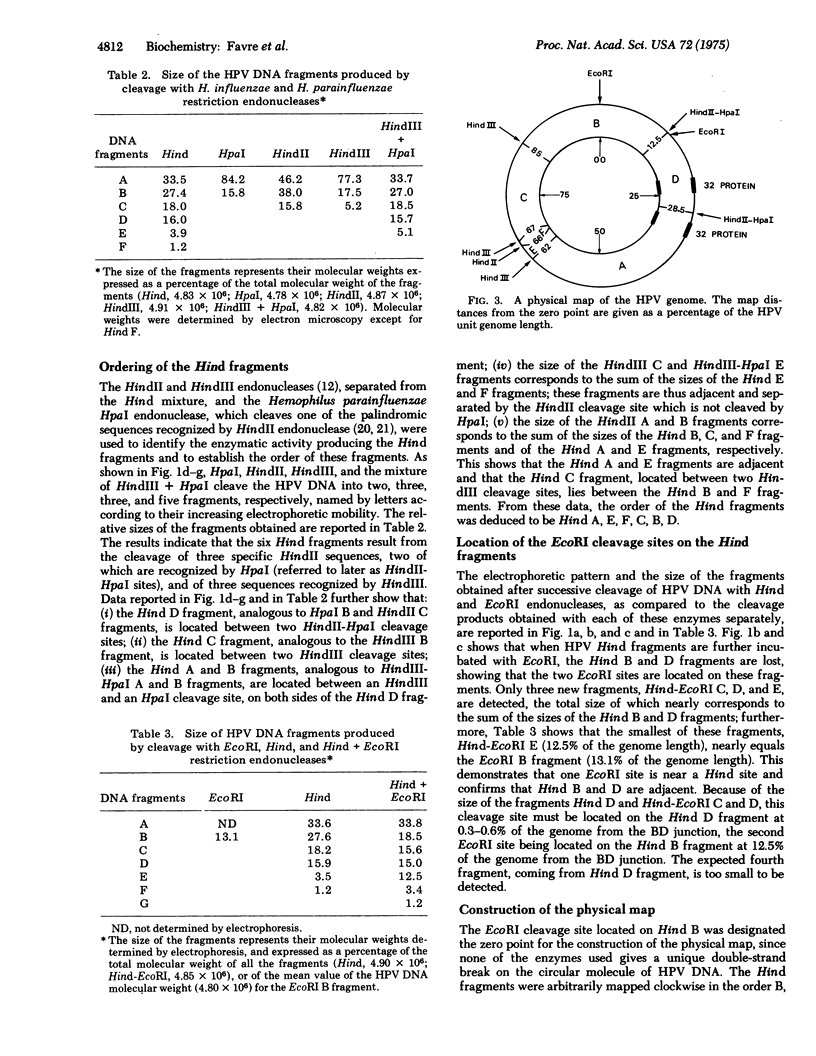
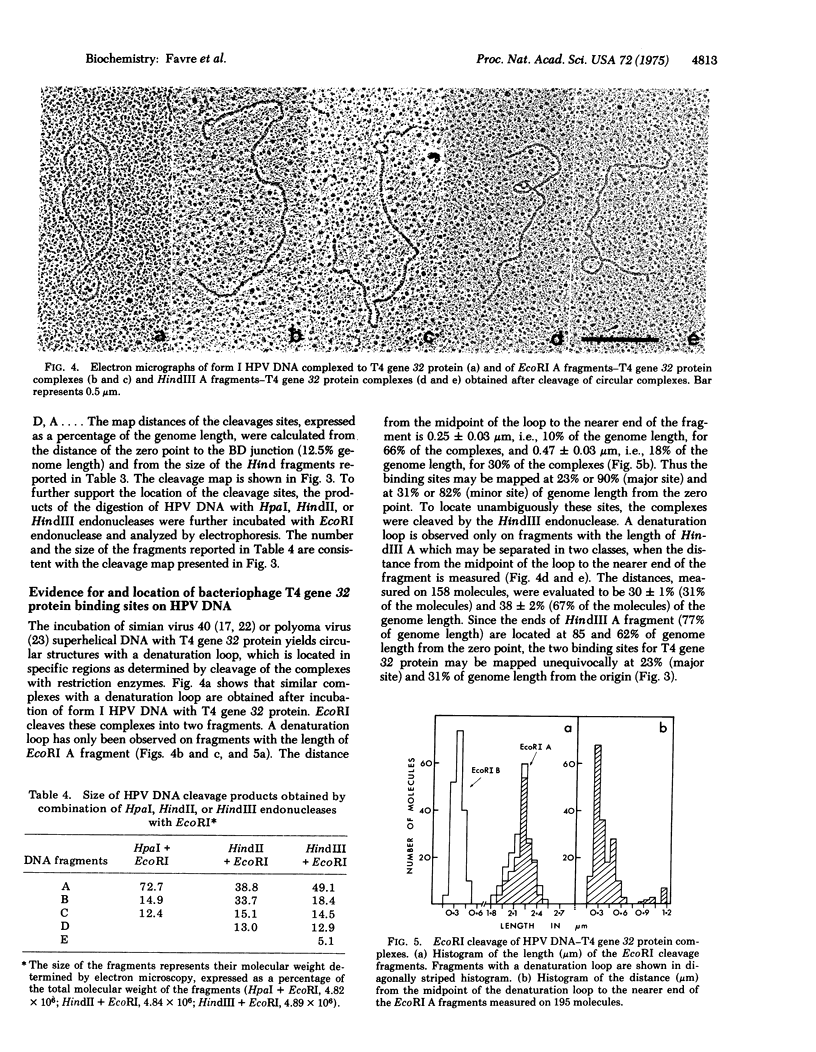
Human papillomavirus (HPV) DNA form I (supercoiled) was prepared from plantar warts. HPV DNA was cleaved with restriction enzymes obtained from the following sources: escherichia coli (EcoRI), Hemophilus influenzae strain Rd (both unfractionated Hind and aeparated HindII and HindIII enzymes) and Hemophilus parainfluenzae (HpaI). The cleavage products were analyzed by polyacrylamide gradient slab gel electrophoresis and electron microscopy. HPV DNA was cleaved into two fragments by EcoRI (87% and 13% of the genome) and into six fragments, ranging in size from 33.5 to 1.2% of the genome, by Hind endonucleases. The six Hind fragments result from the cleavage of three sequences recognized by HindII, two of which are also cleaved by HpaI, and of three sequence recognized by HindIII. The order of these fragments was determined by comparing their size with that of the fragments obtained with HindII, HindIII, HpaI, and the mixture of HindIII + Hpal. The two EcoRI cleavage sites were located on two adjacent Hind fragments and one of these sites has been taken for the zero point to construct a physical map. The treatment of superhelical HPV DNA with bacteriophage T4 gene 32 protein yields circular structures with a denaturation loop. The cleavage of these complexes with EcoRI and HindIII has shown two easily denatured regions which were located on the cleavage map.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M., Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970 Sep 26;227(5265):1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- Allet B., Bukhari A. I. Analysis of bacteriophage mu and lambda-mu hybrid DNAs by specific endonucleases. J Mol Biol. 1975 Mar 15;92(4):529–540. doi: 10.1016/0022-2836(75)90307-1. [DOI] [PubMed] [Google Scholar]
- Butel J. S. Studies with human papilloma virus modeled after known papovavirus systems. J Natl Cancer Inst. 1972 Feb;48(2):285–299. [PubMed] [Google Scholar]
- Crawford L. V. Nucleic acids of tumor viruses. Adv Virus Res. 1969;14:89–152. doi: 10.1016/s0065-3527(08)60558-8. [DOI] [PubMed] [Google Scholar]
- Danna K. J., Sack G. H., Jr, Nathans D. Studies of simian virus 40 DNA. VII. A cleavage map of the SV40 genome. J Mol Biol. 1973 Aug 5;78(2):363–376. doi: 10.1016/0022-2836(73)90122-8. [DOI] [PubMed] [Google Scholar]
- Delius H., Mantell N. J., Alberts B. Characterization by electron microscopy of the complex formed between T4 bacteriophage gene 32-protein and DNA. J Mol Biol. 1972 Jun 28;67(3):341–350. doi: 10.1016/0022-2836(72)90454-8. [DOI] [PubMed] [Google Scholar]
- Favre M. Structural polypeptides of rabbit, bovine, and human papillomaviruses. J Virol. 1975 May;15(5):1239–1247. doi: 10.1128/jvi.15.5.1239-1247.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follett E. A., Crawford L. V. Electron microscope study of the denaturation of human papilloma virus DNA. II. The specific location of denatured regions. J Mol Biol. 1967 Sep 28;28(3):461–467. doi: 10.1016/s0022-2836(67)80096-2. [DOI] [PubMed] [Google Scholar]
- Franklin R. M. Structure and synthesis of bacteriophage PM2, with particular emphasis on the viral lipid bilayer. Curr Top Microbiol Immunol. 1974;(68):107–159. doi: 10.1007/978-3-642-66044-3_5. [DOI] [PubMed] [Google Scholar]
- Garfin D. E., Goodman H. M. Nucleotide sequences at the cleavage sites of two restriction endonucleases from Hemophilus parainfluenzae. Biochem Biophys Res Commun. 1974 Jul 10;59(1):108–116. doi: 10.1016/s0006-291x(74)80181-6. [DOI] [PubMed] [Google Scholar]
- Hudson H. C., Holcomb F. L., Gates W. Giant condyloma acuminatum of the penis: case reports and review. J Urol. 1973 Sep;110(3):301–302. doi: 10.1016/s0022-5347(17)60195-2. [DOI] [PubMed] [Google Scholar]
- Jablonska S., Dabrowski J., Jakubowicz K. Epidermodysplasia verruciformis as a model in studies on the role of papovaviruses in oncogenesis. Cancer Res. 1972 Mar;32(3):583–589. [PubMed] [Google Scholar]
- Jeppesen P. G. A method for separating DNA fragments by electrophoresis in polyacrylamide concentration gradient slab gels. Anal Biochem. 1974 Mar;58(1):195–207. doi: 10.1016/0003-2697(74)90458-8. [DOI] [PubMed] [Google Scholar]
- KLUG A., FINCH J. T. STRUCTURE OF VIRUSES OF THE PAPILLOMA-POLYOMA TYPE. I. HUMAN WART VIRUS. J Mol Biol. 1965 Feb;11:403–423. doi: 10.1016/s0022-2836(65)80066-3. [DOI] [PubMed] [Google Scholar]
- Morrow J. F., Berg P. Cleavage of Simian virus 40 DNA at a unique site by a bacterial restriction enzyme. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3365–3369. doi: 10.1073/pnas.69.11.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oriel J. D., Almeida J. D. Demonstration of virus particles in human genital warts. Br J Vener Dis. 1970 Feb;46(1):37–42. doi: 10.1136/sti.46.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth G., Jeanteur P., Croissant O. Evidence for and localization of vegetative viral DNA replication by autoradiographic detection of RNA-DNA hybrids in sections of tumors induced by Shope papilloma virus. Proc Natl Acad Sci U S A. 1971 Aug;68(8):1876–1880. doi: 10.1073/pnas.68.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowson K. E., Mahy B. W. Human papova (wart) virus. Bacteriol Rev. 1967 Jun;31(2):110–131. doi: 10.1128/br.31.2.110-131.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Nathans D. Letter: A suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J Mol Biol. 1973 Dec 15;81(3):419–423. doi: 10.1016/0022-2836(73)90152-6. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Wilcox K. W. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J Mol Biol. 1970 Jul 28;51(2):379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- Yaniv M., Croissant O., Cuzin F. Location of the T4 gene 32 protein-binding site on polyoma virus DNA. Biochem Biophys Res Commun. 1974 Apr 23;57(4):1074–1079. doi: 10.1016/0006-291x(74)90806-7. [DOI] [PubMed] [Google Scholar]




