SUMMARY
Nonalcoholic fatty liver disease is the most common chronic liver disorder in developed countries. Its pathogenesis is poorly understood, and therapeutic options are limited. Here we show that SIRT7, an NAD+-dependent H3K18Ac deacetylase, functions at chromatin to suppress ER stress and prevents the development of fatty liver disease. SIRT7 is induced upon ER stress and is stabilized at the promoters of ribosomal proteins through its interaction with the transcription factor Myc to silence gene expression and to relieve ER stress. SIRT7 deficient mice develop chronic hepatosteatosis resembling human fatty liver disease. Myc inactivation or pharmacological suppression of ER stress alleviates fatty liver caused by SIRT7 deficiency. Importantly, SIRT7 suppresses ER stress and reverts the fatty liver disease in diet-induced obese mice. Our study identifies SIRT7 as a cofactor of Myc for transcriptional repression and delineates a druggable regulatory branch of the ER stress response that prevents and reverts fatty liver disease.
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) affects one-third of adults and an increasing number of children in developed countries and is strongly associated with obesity and insulin resistance (Browning and Horton, 2004; Browning et al., 2004; Cohen et al., 2012). NAFLD begins with aberrant accumulation of triglyceride in the liver (steatosis). Hepatic steatosis can proceed to nonalcoholic steatohepatitis (NASH), a condition associated with hepatocyte injury, inflammation, and fibrosis. Steatohepatitis can further progress to cirrhosis and liver cancer (Argo and Caldwell, 2009; Starley, 2010).
The ER stress response (also known as the unfolded protein response (UPRER)), a signal transduction pathway that is activated in response to the accumulation of unfolded proteins in the ER, has emerged as a critical regulator of lipid homeostasis in the liver (Basseri and Austin, 2012; Cnop et al., 2012; Fu et al., 2012; Hotamisligil, 2010; Ozcan and Tabas, 2012). The initial phase of the UPRER is suppression of protein translation and increased production of molecular chaperones to promote protein folding, allowing the cells to cope with an increased protein-folding demand and restore protein homeostasis (Hetz, 2012; Walter and Ron, 2011). Prolonged ER stress has been implicated in the development of numerous diseases, including fatty liver disease (Hotamisligil, 2010; Ozcan and Tabas, 2012). Identification of crucial UPR regulators with precise ER-stress-relieving properties that are amenable for therapeutic targeting represents attractive opportunities for pharmacological intervention of fatty liver and a wide spectrum of human diseases.
The sirtuin family of nicotinamide adenine dinucleotide (NAD+)-dependent deacetylases is profoundly implicated in metabolic regulation (Bellet et al., 2012; Finkel et al., 2009; Gillum et al.; Hirschey et al., 2011; Houtkooper et al.; Imai and Guarente, 2010). Sirtuins are well sought-after drug targets for metabolic disorders, as their enzymatic activities are amenable for regulation (Baur et al., 2012). SIRT7, a histone H3 lysine 18 (H3K18) deacetylase that binds to the promoters of a specific set of gene targets for transcriptional repression (Barber et al., 2012), is the only mammalian sirtuin whose function in metabolic regulation remains unknown.
We set out to fill this gap in knowledge by asking whether SIRT7 governs metabolic homeostasis under physiological conditions. Here we show that SIRT7 has a physiological function in metabolic regulation, which occurs through a chromatin-dependent signaling pathway that maintains metabolic homeostasis. Finally, we show that SIRT7 can be targeted to restore metabolic homeostasis in animals with metabolic disorders.
RESULTS
SIRT7 deficient mice develop steatosis resembling human fatty liver disease
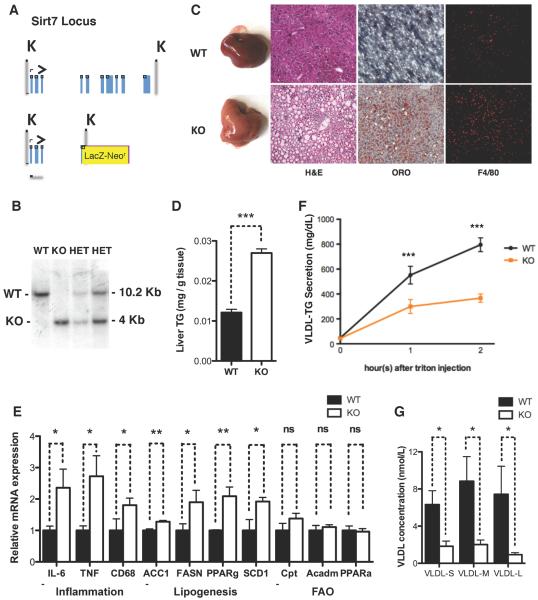
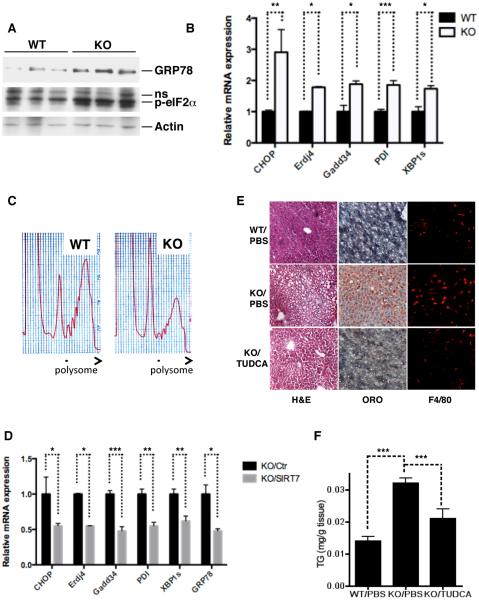
Among the metabolic tissues, SIRT7 is the most highly expressed in the liver (Ford et al., 2006). To gain insight into SIRT7 function, we generated SIRT7 knockout (KO) mice (Figure 1A, B, Figure S1A). Livers of SIRT7 KO mice fed a chow diet appeared paler and slightly larger than the WT controls with 100% penetrance (Figure 1C, Figure S1B). H&E staining showed that SIRT7 KO hepatocytes were markedly vacuolated, with the accumulated material staining positive for fat with the Oil Red O stain (Figure 1C). Quantification of the triglyceride extracted from the livers by a colorimetric assay showed that SIRT7 KO livers had a 2.5-fold increase in triglyceride content (Figure 1D). Compared to the WT controls, the livers of SIRT7 KO mice had increased expression of inflammatory markers (Figure 1E), and immunohistochemistry showed increased staining for F4/80 (Figure 1C), a marker for tissue macrophages, indicating the progression to steatohepatitis. Notably, although fatty liver disease is often associated with obesity (Cohen et al., 2012; Ozcan et al., 2006), SIRT7 KO mice were leaner than littermate controls (Figure S1C, D). Thus, SIRT7 deficiency results in fatty liver without obesity.
Figure 1. SIRT7 prevents the development of fatty liver disease.
(A) Schematic representation of the SIRT7 locus and the SIRT7 KO targeting vector.
(B) Southern blot confirming the generation of SIRT7 KO mice.
(C) Morphology, H&E staining, Oil Red O staining, and F4/80 staining showing increased lipid accumulation and inflammation in the livers of SIRT7 KO mice compared to WT controls.
(D) Quantification of triglyceride extracted from livers in a colorimetric assay showing increased triglyceride content in SIRT7 KO livers. n=6.
(E) Gene expression analysis by quantitative PCR showing increased inflammation and lipogenesis but not fatty acid oxidation in SIRT7 KO livers. n=4.
(F) A VLDL-TG secretion assay showing defective VLDL-TG secretion for SIRT7 KO mice. n=6.
(G) Quantification of VLDL particle concentration in the blood using a gas-phase differential electrical mobility analyzer showing reduced VLDL particle concentration in the blood of SIRT7 KO mice. n=6.
Error bars represent S. E. M. *: p<0.05. **: p<0.01. ***:p<0.001. ns: p>0.05.
See also Figure S1.
Next, we investigated the lipid metabolic pathways in SIRT7 KO livers. The expression of lipogenic genes was increased in SIRT7 KO livers compared to the WT controls, while the expression of genes in the fatty acid oxidation pathway was unchanged (Figure 1E), suggesting that increased lipogenesis may be a contributing factor for hepatosteatosis. Despite increased lipid content in the livers of SIRT7 KO mice (Figure 1D), the levels of plasma triglyceride were 4-fold lower in SIRT7 KO mice compared to WT controls (Figure S1E). Reduced plasma triglycerides in SIRT7 KO mice is not due to reduced food intake or malabsorption of lipid (Figure S1F, G), suggesting that SIRT7 KO mice may have reduced very-low-density lipoprotein (VLDL) secretion, the lipoprotein responsible for hepatic lipid export. In a well-established VLDL-TG secretion assay, SIRT7 KO mice had a 50% reduction in VLDL-TG secretion (Figure 1F). Quantification of the VLDL particles (size ranging from 250–550Å) using a gas-phase differential electrical mobility analyzer showed greatly reduced VLDL concentration in the blood of SIRT7 KO mice compared to the WT controls (Figure 1G). Together, these data indicate that SIRT7 KO mice develop fatty liver due to increased lipogenesis and reduced VLDL secretion.
Hepatic SIRT7 autonomously prevents the development of fatty liver
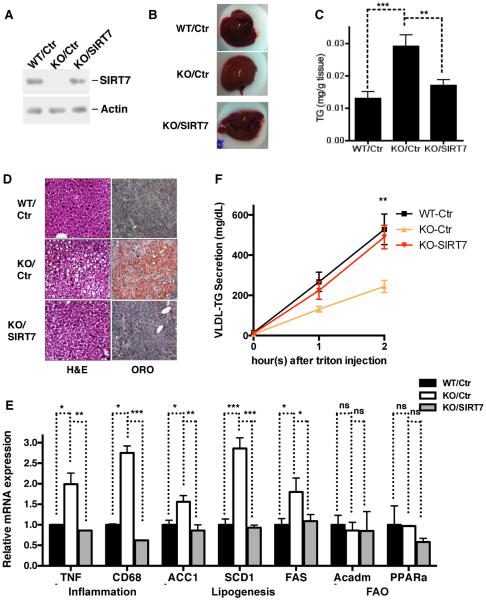
To investigate whether the fatty liver phenotype of SIRT7 KO mice is due to SIRT7 deficiency in the liver or systemic effects of SIRT7 deletion, we reintroduced SIRT7 specifically in the livers of SIRT7 KO mice via adeno-associated virus 8 (AAV8)-mediated gene transfer (Figure 2A). Strikingly, liver-specific reconstitution of SIRT7 in SIRT7 KO mice reversed the fatty liver phenotype (Figure 2B, C, D), suppressed hepatic inflammation and lipogenesis (Figure 2E), and rescued the VLDL-TG secretion defect (Fig. 2F). Importantly, this reversal of hepatosteatosis was not due to unphysiological levels of SIRT7 overexpression, since the reconstituted expression of SIRT7 was comparable to endogenous levels (Figure 2A). Together, these data indicate that hepatic SIRT7 autonomously prevents the development of fatty liver.
Figure 2. Hepatic SIRT7 prevents the development of fatty liver autonomously.
Data shown are the comparison for the livers of WT, SIRT7 KO, and SIRT7 KO mice expressing SIRT7 in livers via AAV8-mediated gene transfer
(A) Western analysis for SIRT7 expression.
(B) Liver morphology.
(C) Liver triglyceride quantification.
(D) H&E and Oil Red O staining.
(E) Gene expression by quantitative PCR showing SIRT7 expression in liver suppresses hepatic inflammation and lipogenesis of SIRT7 KO mice.
(F) In a VLDL-TG secretion assay, SIRT7 expression in liver rescued VLDL-TG secretion defects of SIRT7 KO mice.
n=4. Error bars represent S. E. M. *: p<0.05. **: p<0.01. ***:p<0.001. ns: p>0.05.
SIRT7 activation is a critical event of the UPR to alleviate ER stress
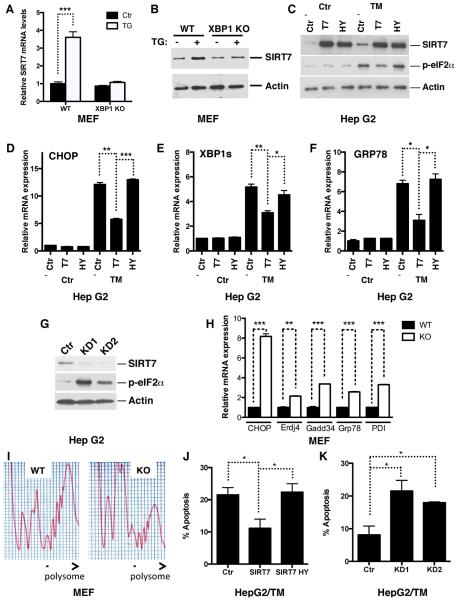
Activation of the UPR pathways induces inflammation (Hotamisligil, 2010), perturbs hepatic lipid metabolism by modulating lipogenesis and lipoprotein metabolism (Kammoun et al., 2009; Lee et al., 2008; Ota et al., 2008; Rutkowski et al., 2008; So et al., 2012; Wang et al., 2012; Zhang et al., 2011), and results in the development of fatty liver, reminiscent of essential aspects of SIRT7 KO phenotype in the liver (Figure 1). We therefore hypothesized that SIRT7 might be an essential regulator of the UPR. ER stress triggers finely regulated signaling events and transcriptional activation of ER stress response target genes with well-defined cis-elements (Hetz, 2012; Walter and Ron, 2011). Analysis of the SIRT7 promoter using MATInspector identified potential binding elements for XBP1, a key regulator of the UPR, which preferentially binds to sequences containing an ACGT core (Figure S2A) (Acosta-Alvear et al., 2007; Sha et al., 2009). Thus, SIRT7 may be transcriptionally upregulated upon ER stress. Indeed, treatment with chemical inducers of ER stress, tunicamycin and thapsigargin, increased the expression of SIRT7 at both the mRNA and protein levels in various cell types (Figure 3A, B, Figure S2B–D). However, ER stress did not induce SIRT7 expression in XBP1 KO mouse embryonic fibroblasts (MEFs) (Figure 3A, B). Furthermore, ectopic expression of the spliced XBP1 (XBP1s), the active form of XBP1, induced SIRT7 expression to the same degree as it induced Erdj4, a known XBP1s target (Figure S2E, F). Additionally, in a luciferase assay, XBP1s induced transcription driven by the SIRT7 promoter but not the SIRT3 promoter (Figure S2G). SIRT7 activity is dependent on NAD+, but tunicamycin treatment did not change the cellular NAD+ levels (Figure S2H). Thus, SIRT7 is induced transcriptionally by XBP1 upon ER stress.
Figure 3. SIRT7 suppresses ER stress.
(A, B) Gene expression analysis by quantitative PCR (A) and western blotting (B) showing increased SIRT7 expression upon treatment of ER stress inducer thapsigargin (TG) in WT but not XBP1 KO MEFs.
(C–F) Western blots (C) and quantitative PCR (D–F) showing reduced ER stress in tunicamycin-treated stable HepG2 cells overexpressing (OE) WT but not catalytically inactive (H187Y) SIRT7, as indicated by eIF2α phosphorylation levels and ER stress response gene expression.
(G) Western blots showing increased ER stress in SIRT7 knockdown (KD) stable HepG2 cells.
(H, I) Increased ER stress in SIRT7 KO MEFs. Quantitative PCR showing increased expression of ER stress-induced genes in SIRT7 KO MEFs (H). Polysome profiling showing reduced polysome-to-monosome ratio, an ER stress marker indicative of translational initiation blockade (I).
(J, K) SIRT7 prevents ER stress-induced cell death. Stable SIRT7 overexpression (OE) or knockdown (KD1 or KD2) cells used in Figure 3C, G were treated with tunicamycin (2ug/ml for J and 1ug/ml for K) for 24 hours. Apoptosis was scored with Annexin V staining.
Error bars represent S. E. M. *: p<0.05. **: p<0.01. ***:p<0.001.
See also Figure S2.
Next, we tested the effects of altered SIRT7 levels on ER stress. Overexpression of SIRT7 substantially reduced ER stress in response to tunicamycin, as evidenced by the reduced eIF2α phosphorylation and expression of ER stress response genes (Figure 3C–F, Figure S2I, J). A catalytically inactive SIRT7 mutant (H187Y) did not suppress ER stress (Figure 3C–F), linking the catalytic activity of SIRT7 to ER stress management. To determine whether endogenous SIRT7 prevents ER stress, we stably knocked down SIRT7 expression using two independent short hairpin RNAs that specifically target SIRT7. SIRT7 depletion led to constitutive induction of ER stress (Figure 3G and Figure S2K). Similarly, ER stress response genes were also upregulated in SIRT7 KO MEFs (Figure 3H). ER stress induces a reduction in the polysome-to-monosome ratio, indicative of translational initiation blockade (Kawai et al., 2004). Ribosomal profiling via a sucrose gradient showed that in contrast to WT MEFs, which had a high polysome-to-monosome ratio, SIRT7 KO MEFs had a reduction in polysomes and an increase in low-molecular-weight monosomes, consistent with increased ER stress in SIRT7 KO MEFs (Figure 3I). SIRT7 also suppressed ER stress-induced cell death (Figure 3J, K, Figure S2L, M) but not general apoptosis (Figure S2N, O). Together, these data suggest that SIRT7 activation is a critical event of the UPR to alleviate ER stress.
Myc recruits SIRT7 to repress the expression of ribosomal proteins and to suppress ER stress
How does SIRT7 regulate the UPR? The initial phase of the UPR is suppression of protein translation and increased production of molecular chaperones to reestablish homeostasis (Hetz, 2012; Walter and Ron, 2011). SIRT7 deacetylates H3K18Ac at specific gene promoters to repress transcription, and a major class of SIRT7 target genes are involved in protein translation and ribosome biogenesis (Barber et al., 2012). Thus, SIRT7 may alleviate ER stress by suppressing ribosome biogenesis and protein translation. Although SIRT7 lacks known DNA binding motifs, previous work has shown that it is recruited to a subset of its target promoters via interaction with the transcription factor ELK4 (Barber et al., 2012). However, ELK4 was dispensable for SIRT7 promoter occupancy at other target promoters, including the promoters of ribosomal protein genes, leaving open the question of how SIRT7 is recruited to such promoters.
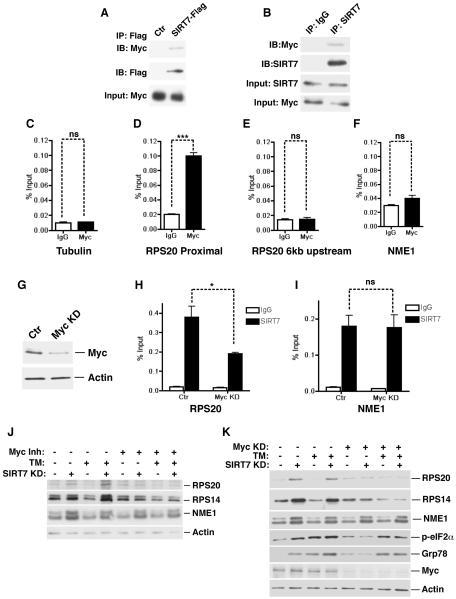
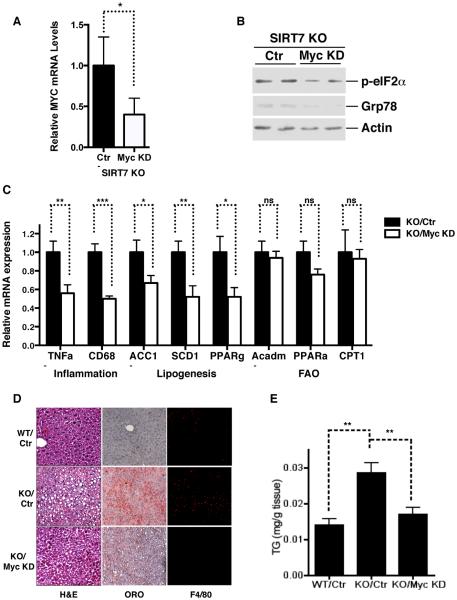
Recently, Myc has been shown to coordinate the transcriptional control of ribosomal components and serve as a master regulator of ribosome biogenesis (Kim et al., 2000; van Riggelen et al., 2010). Chromatin remodeling is believed to be central to Myc function in modulating the expression of its target genes (Knoepfler et al., 2006; van Riggelen et al., 2010). Myc binding on target chromatin is associated with the H3K18Ac mark, a substrate of SIRT7 (Martinato et al., 2008). We therefore hypothesized that Myc stabilizes SIRT7 at the promoters of ribosomal proteins to mediate chromatin remodeling and gene repression. To probe a potential connection between Myc and SIRT7, we tested whether SIRT7 physically interacts with Myc in vivo. Western blot analysis of the immunoprecipitates from Flag-tagged SIRT7 transfected cells revealed that Myc was associated with Flag-SIRT7 (Figure 4A). Western blot analysis of endogenous SIRT7 immunoprecipitates also revealed a specific interaction with Myc (Figure 4B). Thus, SIRT7 physically interacts with Myc in cells.
Figure 4. Myc recruits SIRT7 to repress the expression of ribosomal proteins and to suppress ER stress.
(A) Western analysis showing co-immunoprecipitation (co-IP) of Flag-tagged SIRT7 and endogenous Myc in 293T cells.
(B) Western blots showing co-IP of endogenous SIRT7 and Myc in Hep G2 cells.
(C–F) ChIP-qPCR (mean +/− S.E.M) showing Myc occupancy at the PRS20 proximal promoter but not 6kb upstream, compared to IgG negative control samples. Myc occupancy at the γ-tubulin and NME1 promoters was used as negative controls. All samples were normalized to input DNA.
(G) Western blots showing knockdown of Myc with siRNA in cells used in (H, I).
(H, I) Reduction of SIRT7 occupancy at the RPS20 but not the NME1 promoter in Myc knockdown cells determined by ChIP (mean +/− S.E.M).
(J) Western analysis showing Myc inactivation by a small molecule inhibitor abrogates increased expression of ribosomal proteins but not NME1 in stable SIRT7 KD cells used in Figure 3G.
(K) Western analysis showing Myc inactivation via siRNA abrogates ER stress and increased expression of ribosomal proteins but not NME1 in stable SIRT7 KD cells used in Figure 3G.
Error bars represent S. E. M. *: p<0.05. ***:p<0.001. ns: p>0.05.
See also Figure S3.
To investigate whether SIRT7 is stabilized at the promoters of ribosomal proteins through its interaction with Myc, we assessed SIRT7 and Myc co-occupancy at these promoter regions. Chromatin immunoprecipitation (ChIP) was performed with anti-SIRT7 and anti-Myc antibodies to assess the genomic occupancy of endogenous SIRT7 and Myc, compared to IgG negative background control. SIRT7 bound to the core promoters of ribosomal proteins (Barber et al., 2012). Myc was also detected at the same regions as SIRT7 on the core promoters of ribosomal proteins but not at the genomic region 6kb upstream of the transcription start site (Figure 4C–E, Figure S3A, B), consistent with the notion that Myc generally occupies the core promoter regions of actively transcribed genes (Lin et al., 2012). In contrast, Myc was not detected at the promoters of NME1 and COPS2, where SIRT7 binding is mediated through ELK4 (Figure 4F, S3C) (Barber et al., 2012). Myc depletion using siRNA led to a significant reduction in SIRT7 occupancy at the promoters of ribosomal proteins (Figure 4G, H, Figure S3D, E). However, Myc depletion had no effect on SIRT7 occupancy at the promoters of NME1 and COPS2 (Figure 4I, Figure S3F). These data suggest that Myc targets SIRT7 specifically to the promoters of ribosomal proteins.
The specific association of SIRT7 and Myc at the promoters of ribosomal proteins but not ELK4 target genes suggests that SIRT7 might specifically influence the expression of ribosomal proteins via Myc. Therefore, we examined the effects of Myc inhibition on SIRT7-mediated gene expression. As shown previously, SIRT7 KD led to increased expression of ribosomal proteins (Figure 4J, K)(Barber et al., 2012). Tunicamycin treatment induced SIRT7 expression (Figure 3A,B Figure S2B–D) and suppressed the expression of ribosomal proteins in control but not SIRT7 KD cells (Figure 4J, K), indicating that ER stress triggers the transcriptional silencing of ribosomal proteins and that this effect requires SIRT7. Strikingly, Myc inactivation via siRNA or by a specific inhibitor, 10058-F4, abrogated the elevated expression of ribosomal proteins but not NME1 in SIRT7 KD cells (Figure 4J, K, Figure S3G, H), demonstrating that the observations on ribosomal proteins are due to Myc-dependent effects of SIRT7 on gene expression. Together, these data suggest that Myc targets SIRT7 specifically to the promoters of ribosomal proteins for transcriptional silencing.
The observation that ER stress represses ribosomal proteins in a SIRT7-dependent manner suggests that upon ER stress, the induction of SIRT7 might function to suppress ribosomal protein expression and protein translation, in order to relieve ER stress. To test this possibility, we assessed the effects of suppressing Myc-mediated expression of ribosomal proteins on SIRT7-associated ER stress management. Myc inactivation by siRNA or 10058-F4 abrogated the increased ER stress in SIRT7 KD cells (Figure 4K, Figure S3I). Myc inactivation also blunted the effects of SIRT7 on ER stress resistance (Figure S3J, K). Together, these results indicate that SIRT7 is targeted to the promoters of ribosomal proteins by interacting with Myc, and alleviates ER stress by countering Myc-dependent expression of these genes.
ER stress underlies the development of fatty liver caused by SIRT7 deficiency
We next determined whether ER stress underlies the development of fatty liver in SIRT7 deficient mice. SIRT7 KO livers showed increased ER stress, as evidenced by the induction of e-IF2α phosphorylation and ER stress response genes, as well as a reduction in the polysome-to-monosome ratio (Figure 5A–C). Reintroduction of SIRT7 in the livers of SIRT7 KO mice via AAV8-mediated gene transfer reduced ER stress (Figure 5D) and reverted the fatty liver phenotype (Figure 2A–E). Moreover, treatment of SIRT7 KO mice with TUDCA, a small molecule chaperone that has been shown to alleviate ER stress in vivo (Ozcan et al., 2006), partially rescued the fatty liver phenotype (Figure 5E, F). Finally, liver-specific Myc knockdown in SIRT7 KO mice via AAV8-mediated gene transfer reduced ER stress (Figure 6A, B), suppressed hepatic inflammation and lipogenesis (Figure 6C, D), and reversed the fatty liver phenotype (Figure 6D, E). Thus, SIRT7 prevents the development of fatty liver by suppressing ER stress.
Figure 5. SIRT7 prevents the development of fatty liver by suppressing ER stress.
(A, B, C) Western analysis (A), quantitative PCR (B), and polysome profiling (C) showing increased ER stress in SIRT7 KO livers.
(D) Quantitative PCR showing decreased expression of ER stress response genes in SIRT7 KO livers by reintroduction of SIRT7 via AAV8-mediated gene transfer.
(E, F) H&E, oil red O staining, and F4/80 staining of liver sections (E) and quantification of liver triglyceride (F) showing a small molecule chaperone TUDCA partially reverses fatty liver phenotype of SIRT7 KO mice. n=6.
Error bars represent S. E. M. *: p<0.05. **:p<0.01. ***:p<0.001.
Figure 6. SIRT7 suppresses ER stress and prevents the development of fatty liver by repressing Myc.
Data shown are the comparison of WT, SIRT7 KO, and SIRT7 KO mice with Myc knockdown in livers via AAV8-mediated gene transfer.
(A) Myc expression by quantitative PCR.
(B) ER stress in the livers by western analysis.
(C) Quantitative PCR analysis for the expression of inflammation, lipogenesis, and fatty acid oxidation genes.
(D) H&E, oil red O staining, and F4/80 staining of liver sections.
(E) Liver triglyceride quantification.
n=5. Error bars represent S. E. M. *: p<0.05. **:p<0.01. ***:p<0.001. ns: p>0.05.
See also Figure S4.
SIRT7 reverts fatty liver associated with diet-induced-obesity
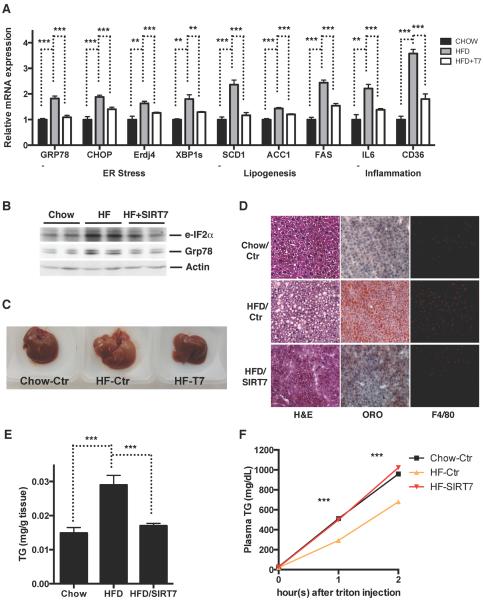
A high-fat high-calorie diet is associated with increased ER stress and the development of fatty liver disease (Oyadomari et al., 2008). We next asked whether SIRT7 could be targeted to alleviate high fat diet-induced ER stress and the development of fatty liver disease. We overexpressed SIRT7 specifically in the livers of mice fed a high fat diet via AAV8-mediated gene transfer. Consistent with previous reports (Oyadomari et al., 2008), high fat diet feeding led to an increase in ER stress markers (Figure 7A, B), accumulation of TG in the liver (Figure 7C–E) possibly due to decreased VLDL secretion and increased lipogenesis (Figure 7A, F), and increased inflammation (Figure 7A, D). Strikingly, SIRT7 overexpression in the livers of high fat diet fed mice suppressed ER stress (Figure 7A, B), VLDL secretion (Figure 7F), lipogenesis (Figure 7A), inflammation (Figure 7A, D), and rescued the fatty liver phenotype (Figure 7C–E). Thus, SIRT7 activation represents an attractive approach to revert ER stress-mediated fatty liver.
Figure 7. SIRT7 rescues high fat diet-induced fatty liver.
Data shown are comparison of livers of mice fed a chow diet, a high fat diet, and a high fat diet with SIRT7 reintroduced specifically in the liver via AAV8-mediated gene transfer.
(A) Quantitative PCR analyses for gene expression of ER stress, inflammation, and lipogenesis.
(B) Western blotting for ER stress markers.
(C) Morphology.
(D) H&E staining, Oil Red O staining, and F4/80 staining.
(E) Quantification of liver triglyceride.
(F) A VLDL-TG secretion assay.
n=6. Error bars represent S. E. M. **:p<0.01. ***:p<0.001.
DISCUSSION
Our work uncovered a physiological role of SIRT7 in maintaining hepatic metabolic homeostasis, demonstrated the feasibility of targeting SIRT7 to restore metabolic homeostasis in animals with metabolic disorders, and revealed a regulatory branch of the UPR that is amenable for therapeutic targeting. The canonical UPR leads to the phosphorylation of eIF2α and the suppression of translation to alleviate ER stress(Ozcan and Tabas, 2012; Walter and Ron, 2011). We show that SIRT7 is induced by XBP1 upon ER stress, and is recruited to the promoters of ribosomal proteins via Myc to repress gene expression and to alleviate ER stress (Figure S4A). In SIRT7 deficient cells, failure to engage SIRT7-mediated ER stress management results in constitutive ER stress, which induces activation of the canonical UPR (phosphorylation of eIF2α, blockade of polysome assembly and translational initiation, and induction of ER stress response genes), apoptosis, inflammation, and specifically in the liver, increased lipogenesis and reduced VLDL secretion (Figure S4B). Importantly, SIRT7 upregulation alleviates ER stress and restores hepatic metabolic homeostasis in diet-induced obese animals, providing an avenue to treat fatty liver disease and likely other ER stress-associated pathologies.
Our observation that SIRT7 opposes Myc-dependent gene regulation is intriguing. In addition to its role in fatty liver pathology, the dynamic balance between SIRT7 and Myc activities could play a pivotal role in the context of tumorigenesis. SIRT7 may keep the Myc oncogene in check to prevent tumorigenesis. However, under certain conditions, Myc can also trigger apoptosis as an evolved tumor defense mechanism (Evan et al., 1992; Soucek and Evan, 2010). Our study suggests that unopposed Myc activation in the absence of SIRT7 results in ER stress. This may be an underlying mechanism for Myc-induced apoptosis in cancer cells, which are particularly sensitive to ER stress(Ozcan and Tabas, 2012). Indeed, we find that SIRT7 prevents ER stress-induced cell death in a Myc-dependent manner (Figure S3J, K). These observations are consistent with previous findings that SIRT7 can promote cancer cell survival and maintain oncogenic transformation (Barber et al., 2012).
Our findings regarding the role of SIRT7 in ER stress management might also be important for aging biology. Indeed, SIRT7 has several potential links to aging. SIRT7 KO mice have shortened lifespans and exhibit phenotypes linked to aging (Vakhrusheva et al., 2008). Furthermore, SIRT7 expression decreases in some aging tissues (Vakhrusheva et al., 2008). Suppression of ribosomal proteins and ER stress leads to lifespan extension in yeast (Steffen et al., 2012). Thus, the interplay between SIRT7, ER stress, and protein translation may represent an evolutionarily conserved phenomenon in aging.
EXPERIMENTAL PROCEDURES
Mice
Construction of the targeting vector and generation of SIRT7 deficient mice. Sirt7 −/−mice were produced by the VelociGene method (Valenzuela et al., 2003) (VelociGene Allele Identification Number: VG321). The SIRT7 KO targeting vector was constructed by replacing exons 4 to 11 with a LacZ gene inserted in frame after the first few bp of exon 4 (Figure 1A, B). Srf1 linearized targeting vector (30ug) was electroporated into CJ7,129SJ ES cells as described (Valenzuela et al., 2003). Positives clones were identified by Taqman analysis (Valenzuela et al., 2003) and confirmed by Southern blot analysis in which genomic DNA was digested with Kpn1 and the membranes were probed with a 500 bp fragment covering Exon 1–3. Chimeric mice were generated by injection of targeted ES clones into C57BL6/J blastocysts. Male chimeras were mated with 129Sv females to generate F1 heterozygous mice, which were interbred to generate homozygous KO mice, which were screened by Southern blotting as described above (Figure S1A).
All mice were housed on a 12:12-hr light:dark cycle at 25°C. Experiments were performed using 4–6 month old male littermates. Mice were randomized into each of the groups. Samples were processed blindly during the experiments and the outcome assessment. All animal procedures were in accordance with the animal care committee at the University of California, Berkeley. The High-Fat diet was provided by OpenSource Diets (D12079B). For TUDCA treatment, mice received intraperitoneal injections of 500mg/kg/day TUDCA or PBS twice daily for 20 days as described (Cnop et al., 2012). VLDL-TG secretion assay was performed as described (Haeusler et al., 2010). Blood was collected from the mouse tail vein and kept on ice until centrifugation (1500g, 15 min at 4°C). VLDL concentration was quantified as described using a gas-phase differential electrical mobility analyzer (Caulfield et al., 2008). Plasma triglyceride was measured according to manufacturer's instruction (Wako Diagnostics). Triglycerides were extracted from liver tissues as described and were extracted from feces as described (Zadravec et al., 2010). Extracted triglyceride was quantified according to manufacturer's instruction (Wako Diagnostics). Liver tissues were processed for H&E staining, Oil Red O staining, and F4/80 staining as described(Sun et al., 2012).
For AAV-mediated gene transfer to the mouse liver, SIRT7 or Myc knockdown target sequence was cloned into dsAAV-RSVeGFP-U6 vector. dsAAV8 was produced by triple-transfection and CsCl purification, and virus titer was determined as previously described (Gao et al., 2006). 3 × 1011 genome copies of virus were injected per mouse via tail vein. Mouse livers were harvested for analysis after 2 weeks.
Cell culture, RNAi and viral transduction
293T, Hepa 1–6, Hep G2 cells were acquired from the ATCC. WT and SIRT7 KO MEFs were generated according to standard procedures (Greber et al., 2007). XBP-1 KO MEFs were acquired from L. Glimcher (Lee et al., 2008). Cells were cultured in Advanced DMEM (Invitrogen) supplemented with 1% penicillin-streptomycin (Invitrogen) and 10% FBS (Invitrogen). For ER stress induction, cells were treated with tunicamycin (Sigma, 1ug/ml) or thapsigargin (Sigma, 0.1uM) for 24 hours for biochemical analysis or 48 hours for cell survival analysis. Cells were treated with 0.5 uM staurosporin for 48 hours for cell survival analysis. Cell survival was scored with trypan blue staining or Annexin V Staining (Biolegend). For Myc inactivation, cells were treated with 10058-F4 (Sigma, 2uM) for 24 hours before analysis. Cellular NAD+ concentration was quantified using EnzyChrom™ NAD/NADH Assay Kit (Bioassay System).
SIRT7 knockdown target sequences are as follows, as previously described (Barber et al., 2012):
S7KD1, 5'-CACCTTTCTGTGAGAACGGAA-3';
S7KD2, 5'-TAGCCATTTGTCCTTGAGGAA-3',
Myc knockdown target sequences are as follows, as previously described (Caulfield et al., 2008):
Myc KD (human), 5'- GGACTATCCTGCTGCCAAG -3'
Myc KD (mouse), 5'- CCCAAGGTAGTGATCCTCAAA – 3'
Double-stranded siRNAs were purchased from Thermo Scientific and were transfected into cells via RNAiMax (Invitrogen) according to manufacture's instruction.
For lentiviral packaging, 293T cells were co-transfected with packaging vectors (pCMV-dR8.2 dvpr and pCMV-VSV-G) and the pSiCoR-SIRT7 knockdown or empty construct. For retroviral packaging, 293T cells were co-transfected with packaging vectors (pVPack-VSV-G, pVPack-GP) and pBABE-SIRT7, pBABE-SIRT7 H187Y, or empty construct. Viral supernatant was harvested after 48 hours. For transduction, cells were incubated with virus-containing supernatant in the presence of 8 μg/mL polybrene. After 48 hours, infected cells were selected with puromycin (1μg/mL).
Co-immunoprecipitations
Co-immunoprecipitations (Co-IPs) were performed as previously described (Qiu et al., 2010) with Flag-resin (Sigma) or Protein A/G beads (Santa Cruz) for SIRT7 IP. Elution was performed with either Flag peptide (Sigma) or 100mM Glycine solution (pH 3) for SIRT7 IP. Antibodies are provided in Table S1.
ChIP and mRNA analysis
Cells were prepared for ChIP as previously described (Dahl and Collas, 2007), with the exception that DNA was washed and eluted using a PCR purification kit (Qiagen) rather than by phenol-chloroform extraction. RNA was isolated from cells or tissue using Trizol reagent (Invitrogen) and purified using the RNeasy Mini Kit (Qiagen). cDNA was generated using the qScript™ cDNA SuperMix (Quanta Biosciences). Gene expression was determined by real time PCR using Eva qPCR SuperMix kit (BioChain Institute) on an ABI StepOnePlus system. All data were normalized to ActB or GAPDH expression. Antibodies and PCR primer details are provided in Table S1–3.
Polysomal profiling
MEFs or liver tissues were collected for polysomal profiling as described (Zid et al., 2009). Briefly, 10 million MEF cells or 0.1g of liver were harvested and homogenized on ice in 400 μL of solublization buffer [300 mM NaCl, 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM EGTA, 200 μg/mL heparin, 1 mM DTT, 400 U/mL RNAsin plus (Promega), 1X complete, Mini Protease Inhibitor Cocktail (Roche), 0.2 mg/mL cycloheximide, 1% Triton X-100, 0.1% Sodium deoxycholate]. 600 μL of additional solubilization buffer was added to a total of 1 mL. Cell lysate was placed back on ice for 10 min before centrifuging at 16,000 g for 15 min at 4 °C. 1 mL of the supernatant was applied to the top of a 10–50% continuous sucrose gradient in high salt resolving buffer [140 mM NaCl, 25 mM Tris-HCl (pH 8.0), 10 mM MgCl2] and centrifuged in a Beckman SW41Ti rotor (Beckman Coulter, Fullerton, CA, USA) at 180,000 g for 90 min at 4 °C. Gradients were fractionated with continuous monitoring of absorbance at 254 nm.
Statistical Analysis
The number of mice chosen for each experiment is based on the principle that the minimal number of mice is used to have sufficient statistical power and is comparable to published literature for the same assays performed. No animals were excluded from the analyses. Student t test was used for data analysis. Error Bars represent standard errors. In all corresponding figures, * represents p<0.05. ** represents p<0.01. *** represents p<0.001. ns represents p>0.05.
Supplementary Material
Highlights
SIRT7 deficient mice develop hepatosteatosis resembling human fatty liver disease
SIRT7 activation is a critical event in UPRER to alleviate ER stress
Myc recruits SIRT7 to repress ribosomal protein genes and to suppress ER stress
SIRT7 reverts fatty liver associated with diet-induced-obesity
ACKNOWLEDGMENTS
We thank the Kay lab for reagents. This work was support by grants from the NIH (D.C. R01AG040990, K.F.C. R01AG028867; S.P. training grant 3T32DK007217-36S1; M.M. training grant T32 AG000266), Searle Scholars Program (D.C.), Ellison Medical Foundation (D.C., K.F.C.), The Glenn Foundation (D.C.), American Heart Association (D.C.), UCOP TRDRP (D.C.), Hellman Family Faculty Fund (D.C.), the NSF (J.S.), Siebel Stem Cell Institute (D.C., M.M., N.N. X.Q.), Department of Veterans Affairs (K.F.C., Merit Award), Italian Foundation for Cancer Research (L.V.). F.A. is an investigator of the Howard Hughes Medical Institute. AJM, DMV, and GDY are employees and shareholders of Regeneron Pharmaceuticals, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Argo CK, Caldwell SH. Epidemiology and natural history of non-alcoholic steatohepatitis. Clin Liver Dis. 2009;13:511–531. doi: 10.1016/j.cld.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basseri S, Austin RC. Endoplasmic reticulum stress and lipid metabolism: mechanisms and therapeutic potential. Biochem Res Int. 2012;2012:841362. doi: 10.1155/2012/841362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Ungvari Z, Minor RK, Le Couteur DG, de Cabo R. Are sirtuins viable targets for improving healthspan and lifespan? Nat Rev Drug Discov. 2012;11:443–461. doi: 10.1038/nrd3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellet MM, Orozco-Solis R, Sahar S, Eckel-Mahan K, Sassone-Corsi P. The time of metabolism: NAD+, SIRT1, and the circadian clock. Cold Spring Harb Symp Quant Biol. 2012;76:31–38. doi: 10.1101/sqb.2011.76.010520. [DOI] [PubMed] [Google Scholar]
- Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest. 2004;114:147–152. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, Grundy SM, Hobbs HH. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- Caulfield MP, Li S, Lee G, Blanche PJ, Salameh WA, Benner WH, Reitz RE, Krauss RM. Direct determination of lipoprotein particle sizes and concentrations by ion mobility analysis. Clin Chem. 2008;54:1307–1316. doi: 10.1373/clinchem.2007.100586. [DOI] [PubMed] [Google Scholar]
- Cnop M, Foufelle F, Velloso LA. Endoplasmic reticulum stress, obesity and diabetes. Trends Mol Med. 2012;18:59–68. doi: 10.1016/j.molmed.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2012;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Collas P. A quick and quantitative chromatin immunoprecipitation assay for small cell samples. Front Biosci. 2007;12:4925–4931. doi: 10.2741/2438. [DOI] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S, Watkins SM, Hotamisligil GS. The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab. 2012;15:623–634. doi: 10.1016/j.cmet.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Gao GP, Lu Y, Sun X, Johnston J, Calcedo R, Grant R, Wilson JM. High-level transgene expression in nonhuman primate liver with novel adeno-associated virus serotypes containing self-complementary genomes. J Virol. 2006;80:6192–6194. doi: 10.1128/JVI.00526-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum MP, Erion DM, Shulman GI. Sirtuin-1 regulation of mammalian metabolism. Trends Mol Med. 2010 doi: 10.1016/j.molmed.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber B, Lehrach H, Adjaye J. Fibroblast growth factor 2 modulates transforming growth factor beta signaling in mouse embryonic fibroblasts and human ESCs (hESCs) to support hESC self-renewal. Stem Cells. 2007;25:455–464. doi: 10.1634/stemcells.2006-0476. [DOI] [PubMed] [Google Scholar]
- Haeusler RA, Han S, Accili D. Hepatic FoxO1 ablation exacerbates lipid abnormalities during hyperglycemia. J Biol Chem. 2010;285:26861–26868. doi: 10.1074/jbc.M110.134023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Huang JY, Schwer B, Verdin E. SIRT3 regulates mitochondrial protein acetylation and intermediary metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:267–277. doi: 10.1101/sqb.2011.76.010850. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammoun HL, Chabanon H, Hainault I, Luquet S, Magnan C, Koike T, Ferre P, Foufelle F. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest. 2009;119:1201–1215. doi: 10.1172/JCI37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Fan J, Mazan-Mamczarz K, Gorospe M. Global mRNA stabilization preferentially linked to translational repression during the endoplasmic reticulum stress response. Mol Cell Biol. 2004;24:6773–6787. doi: 10.1128/MCB.24.15.6773-6787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Li Q, Dang CV, Lee LA. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc Natl Acad Sci U S A. 2000;97:11198–11202. doi: 10.1073/pnas.200372597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. Embo J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional Amplification in Tumor Cells with Elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS One. 2008;3:e3650. doi: 10.1371/journal.pone.0003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Gayet C, Ginsberg HN. Inhibition of apolipoprotein B100 secretion by lipid-induced hepatic endoplasmic reticulum stress in rodents. J Clin Invest. 2008;118:316–332. doi: 10.1172/JCI32752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan L, Tabas I. Role of endoplasmic reticulum stress in metabolic disease and other disorders. Annu Rev Med. 2012;63:317–328. doi: 10.1146/annurev-med-043010-144749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Gorgun CZ, Hotamisligil GS. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie Restriction Reduces Oxidative Stress by SIRT3-Mediated SOD2 Activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha H, He Y, Chen H, Wang C, Zenno A, Shi H, Yang X, Zhang X, Qi L. The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 2009;9:556–564. doi: 10.1016/j.cmet.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So JS, Hur KY, Tarrio M, Ruda V, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V, Lichtman AH, Iwawaki T, Glimcher LH, et al. Silencing of Lipid Metabolism Genes through IRE1alpha-Mediated mRNA Decay Lowers Plasma Lipids in Mice. Cell Metab. 2012;16:487–499. doi: 10.1016/j.cmet.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Evan GI. The ups and downs of Myc biology. Curr Opin Genet Dev. 2010;20:91–95. doi: 10.1016/j.gde.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starley BQ, Calcagno CJ, Harrison SA. Nonalcoholic fatty liver disease and hepatocellular carcinoma: a weighty connection. Hepatology. 2010;51:1820–1832. doi: 10.1002/hep.23594. [DOI] [PubMed] [Google Scholar]
- Steffen KK, McCormick MA, Pham KM, MacKay VL, Delaney JR, Murakami CJ, Kaeberlein M, Kennedy BK. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics. 2012;191:107–118. doi: 10.1534/genetics.111.136549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Miller RA, Patel RT, Chen J, Dhir R, Wang H, Zhang D, Graham MJ, Unterman TG, Shulman GI, et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med. 2012;18:934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang S, Chen Z, Lam V, Han J, Hassler J, Finck BN, Davidson NO, Kaufman RJ. IRE1alpha-XBP1s Induces PDI Expression to Increase MTP Activity for Hepatic VLDL Assembly and Lipid Homeostasis. Cell Metab. 2012;16:473–486. doi: 10.1016/j.cmet.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadravec D, Brolinson A, Fisher RM, Carneheim C, Csikasz RI, Bertrand-Michel J, Boren J, Guillou H, Rudling M, Jacobsson A. Ablation of the very-long-chain fatty acid elongase ELOVL3 in mice leads to constrained lipid storage and resistance to diet-induced obesity. FASEB J. 2010;24:4366–4377. doi: 10.1096/fj.09-152298. [DOI] [PubMed] [Google Scholar]
- Zhang K, Wang S, Malhotra J, Hassler JR, Back SH, Wang G, Chang L, Xu W, Miao H, Leonardi R, et al. The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. Embo J. 2011;30:1357–1375. doi: 10.1038/emboj.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.